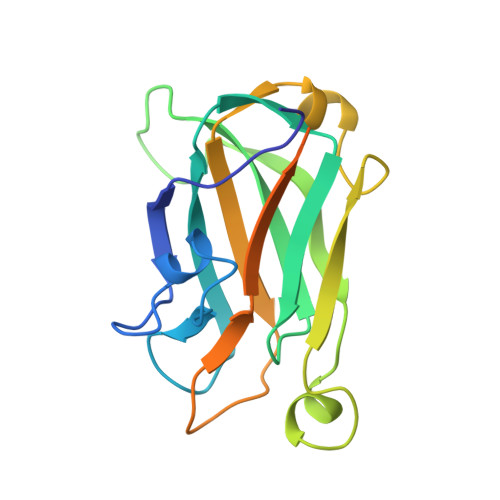
Solution structure of the single-strand break repair protein XRCC1 N-terminal domain.
Marintchev, A., Mullen, M.A., Maciejewski, M.W., Pan, B., Gryk, M.R., Mullen, G.P.(1999) Nat Struct Biol 6: 884-893
- PubMed: 10467102
- DOI: https://doi.org/10.1038/12347
- Primary Citation of Related Structures:
1XNA, 1XNT - PubMed Abstract:
XRCC1 functions in the repair of single-strand DNA breaks in mammalian cells and forms a repair complex with beta-Pol, ligase III and PARP. Here we describe the NMR solution structure of the XRCC1 N-terminal domain (XRCC1 NTD). The structural core is a beta-sandwich with beta-strands connected by loops, three helices and two short two-stranded beta-sheets at each connection side. We show, for the first time, that the XRCC1 NTD specifically binds single-strand break DNA (gapped and nicked). We also show that the XRCC1 NTD binds a gapped DNA-beta-Pol complex. The DNA binding and beta-Pol binding surfaces were mapped by NMR and found to be well suited for interaction with single-strand gap DNA containing a 90 degrees bend, and for simultaneously making contacts with the palm-thumb of beta-Pol in a ternary complex. The findings suggest a mechanism for preferential binding of the XRCC1 NTD to flexible single-strand break DNA.
Organizational Affiliation:
Department of Biochemistry, University of Connecticut Health Center, 263 Farmington Avenue, Farmington, Connecticut 06032, USA.