Novel Influenza D virus: Epidemiology, pathology, evolution and biological characteristics
- PMID: 28812422
- PMCID: PMC5810478
- DOI: 10.1080/21505594.2017.1365216
Novel Influenza D virus: Epidemiology, pathology, evolution and biological characteristics
Abstract
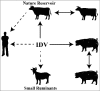
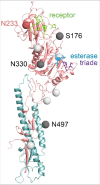
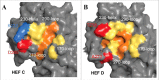
In 2011, a new virus was isolated from pigs with influenza-like symptoms and subsequently also from cattle, which are the main reservoir of the virus. It is similar to Influenza C virus (ICV), a (predominantly) human pathogen, causing respiratory disease in children. Since the virus is unable to reassort with ICV (and based on several other criteria as discussed in the text) it is now officially named as Influenzavirus D (IDV), a new genus of the Orthomyxoviridae. We summarize the epidemiology, pathology and evolution of IDV and its biological characteristics with emphasis on the only glycoprotein HEF. Based on the limited data available we finally consider whether IDV represent a public health threat.
Keywords: HEF; Influenzavirus D; bovine; cattle; epidemiology; evolution; pathology; zoonosis.
Figures


Similar articles
-
Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family.mBio. 2014 Mar 4;5(2):e00031-14. doi: 10.1128/mBio.00031-14. mBio. 2014. PMID: 24595369 Free PMC article.
-
Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle.J Virol. 2015 Jan 15;89(2):1036-42. doi: 10.1128/JVI.02718-14. Epub 2014 Oct 29. J Virol. 2015. PMID: 25355894 Free PMC article.
-
Detection of a New Genetic Cluster of Influenza D Virus in Italian Cattle.Viruses. 2019 Nov 30;11(12):1110. doi: 10.3390/v11121110. Viruses. 2019. PMID: 31801277 Free PMC article.
-
Influenza D in Domestic and Wild Animals.Viruses. 2023 Dec 15;15(12):2433. doi: 10.3390/v15122433. Viruses. 2023. PMID: 38140674 Free PMC article. Review.
-
Emerging Influenza D virus infection in European livestock as determined in serology studies: Are we underestimating its spread over the continent?Transbound Emerg Dis. 2021 May;68(3):1125-1135. doi: 10.1111/tbed.13812. Epub 2020 Sep 18. Transbound Emerg Dis. 2021. PMID: 32871031 Review.
Cited by
-
Rapid evolution leads to extensive genetic diversification of cattle flu Influenza D virus.Commun Biol. 2024 Oct 7;7(1):1276. doi: 10.1038/s42003-024-06954-4. Commun Biol. 2024. PMID: 39375524 Free PMC article.
-
Host Innate Antiviral Response to Influenza A Virus Infection: From Viral Sensing to Antagonism and Escape.Pathogens. 2024 Jul 3;13(7):561. doi: 10.3390/pathogens13070561. Pathogens. 2024. PMID: 39057788 Free PMC article. Review.
-
Receptor binding and immunogenic properties of the receptor binding domain of influenza D virus hemagglutinin-esterase-fusion protein expressed from Escherichia coli.Virology. 2024 Sep;597:110138. doi: 10.1016/j.virol.2024.110138. Epub 2024 Jun 12. Virology. 2024. PMID: 38880069
-
The forecasting power of the mucin-microbiome interplay in livestock respiratory diseases.Vet Q. 2024 Dec;44(1):1-18. doi: 10.1080/01652176.2024.2340003. Epub 2024 Apr 12. Vet Q. 2024. PMID: 38606662 Free PMC article.
-
Serological Evidence for Circulation of Influenza D Virus in the Ovine Population in Italy.Pathogens. 2024 Feb 11;13(2):162. doi: 10.3390/pathogens13020162. Pathogens. 2024. PMID: 38392900 Free PMC article.
References
-
- Sandbulte MR, Westgeest KB, Gao J, Xu X, Klimov AI, Russell CA, Burke DF, Smith DJ, Fouchier RA, Eichelberger MC. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20748-53. doi:10.1073/pnas.1113801108. PMID:22143798 - DOI - PMC - PubMed
-
- Yoon SW, Webby RJ, Webster RG. Evolution and ecology of influenza A viruses. Current topics in microbiology and Immunology. 2014;385:359-75. PMID:24990620 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

