Selection on Optimal Haploid Value Increases Genetic Gain and Preserves More Genetic Diversity Relative to Genomic Selection
- PMID: 26092719
- PMCID: PMC4574260
- DOI: 10.1534/genetics.115.178038
Selection on Optimal Haploid Value Increases Genetic Gain and Preserves More Genetic Diversity Relative to Genomic Selection
Abstract
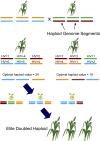
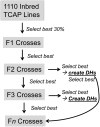
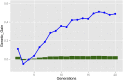
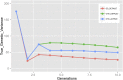
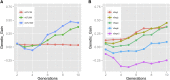
Doubled haploids are routinely created and phenotypically selected in plant breeding programs to accelerate the breeding cycle. Genomic selection, which makes use of both phenotypes and genotypes, has been shown to further improve genetic gain through prediction of performance before or without phenotypic characterization of novel germplasm. Additional opportunities exist to combine genomic prediction methods with the creation of doubled haploids. Here we propose an extension to genomic selection, optimal haploid value (OHV) selection, which predicts the best doubled haploid that can be produced from a segregating plant. This method focuses selection on the haplotype and optimizes the breeding program toward its end goal of generating an elite fixed line. We rigorously tested OHV selection breeding programs, using computer simulation, and show that it results in up to 0.6 standard deviations more genetic gain than genomic selection. At the same time, OHV selection preserved a substantially greater amount of genetic diversity in the population than genomic selection, which is important to achieve long-term genetic gain in breeding populations.
Keywords: GenPred; doubled haploid; genetic diversity; genetic gain; genomic selection; haplotype; shared data resource.
Copyright © 2015 by the Genetics Society of America.
Figures
Similar articles
-
Selection on Expected Maximum Haploid Breeding Values Can Increase Genetic Gain in Recurrent Genomic Selection.G3 (Bethesda). 2018 Mar 28;8(4):1173-1181. doi: 10.1534/g3.118.200091. G3 (Bethesda). 2018. PMID: 29434032 Free PMC article.
-
Improving Response in Genomic Selection with a Population-Based Selection Strategy: Optimal Population Value Selection.Genetics. 2017 Jul;206(3):1675-1682. doi: 10.1534/genetics.116.197103. Epub 2017 May 19. Genetics. 2017. PMID: 28526698 Free PMC article.
-
Haploids: Constraints and opportunities in plant breeding.Biotechnol Adv. 2015 Nov 1;33(6 Pt 1):812-29. doi: 10.1016/j.biotechadv.2015.07.001. Epub 2015 Jul 9. Biotechnol Adv. 2015. PMID: 26165969 Review.
-
Resource allocation for maximizing prediction accuracy and genetic gain of genomic selection in plant breeding: a simulation experiment.G3 (Bethesda). 2013 Mar;3(3):481-91. doi: 10.1534/g3.112.004911. Epub 2013 Mar 1. G3 (Bethesda). 2013. PMID: 23450123 Free PMC article.
-
Enhancing genetic gain in the era of molecular breeding.J Exp Bot. 2017 May 17;68(11):2641-2666. doi: 10.1093/jxb/erx135. J Exp Bot. 2017. PMID: 28830098 Review.
Cited by
-
Genomic-inferred cross-selection methods for multi-trait improvement in a recurrent selection breeding program.Plant Methods. 2024 Sep 2;20(1):133. doi: 10.1186/s13007-024-01258-4. Plant Methods. 2024. PMID: 39218896 Free PMC article.
-
PyBrOpS: a Python package for breeding program simulation and optimization for multi-objective breeding.G3 (Bethesda). 2024 Oct 7;14(10):jkae199. doi: 10.1093/g3journal/jkae199. G3 (Bethesda). 2024. PMID: 39158127 Free PMC article.
-
Genetic Gain and Inbreeding in Different Simulated Genomic Selection Schemes for Grain Yield and Oil Content in Safflower.Plants (Basel). 2024 Jun 6;13(11):1577. doi: 10.3390/plants13111577. Plants (Basel). 2024. PMID: 38891385 Free PMC article.
-
AI-assisted selection of mating pairs through simulation-based optimized progeny allocation strategies in plant breeding.Front Plant Sci. 2024 Mar 28;15:1361894. doi: 10.3389/fpls.2024.1361894. eCollection 2024. Front Plant Sci. 2024. PMID: 38817943 Free PMC article.
-
ChromaX: a fast and scalable breeding program simulator.Bioinformatics. 2023 Dec 1;39(12):btad691. doi: 10.1093/bioinformatics/btad691. Bioinformatics. 2023. PMID: 37991849 Free PMC article.
References
-
- Cole J. B., VanRaden P. M., 2010. Visualization of results from genomic evaluations. J. Dairy Sci. 93: 2727–2740. - PubMed
-
- Cole J. B., VanRaden P. M., 2011. Use of haplotypes to estimate Mendelian sampling effects and selection limits. J. Anim. Breed. Genet. 128: 446–455. - PubMed
-
- Cooper M., Messina C. D., Podlich D., Totir L. R., Baumgarten A., et al. , 2014. Predicting the future of plant breeding: complementing empirical evaluation with genetic prediction. Crop Pasture Sci. 65: 311–336.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources


