Processing of DNA for nonhomologous end-joining by cell-free extract
- PMID: 15692565
- PMCID: PMC549622
- DOI: 10.1038/sj.emboj.7600563
Processing of DNA for nonhomologous end-joining by cell-free extract
Abstract
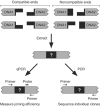
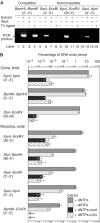
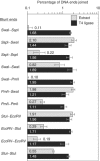
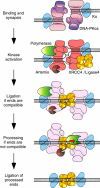
In mammalian cells, nonhomologous end-joining (NHEJ) repairs DNA double-strand breaks created by ionizing radiation and V(D)J recombination. We have developed a cell-free system capable of processing and joining noncompatible DNA ends. The system had key features of NHEJ in vivo, including dependence on Ku, DNA-PKcs, and XRCC4/Ligase4. The NHEJ reaction had striking properties. Processing of noncompatible ends involved polymerase and nuclease activities that often stabilized the alignment of opposing ends by base pairing. To achieve this, polymerase activity efficiently synthesized DNA across discontinuities in the template strand, and nuclease activity removed a limited number of nucleotides back to regions of microhomology. Processing was suppressed for DNA ends that could be ligated directly, biasing the reaction to preserve DNA sequence and maintain genomic integrity. DNA sequence internal to the ends influenced the spectrum of processing events for noncompatible ends. Furthermore, internal DNA sequence strongly influenced joining efficiency, even in the absence of processing. These results support a model in which DNA-PKcs plays a central role in regulating the processing of ends for NHEJ.
Figures




Similar articles
-
Assays for nonhomologous end joining in extracts.Methods Enzymol. 2006;408:430-44. doi: 10.1016/S0076-6879(06)08027-X. Methods Enzymol. 2006. PMID: 16793385
-
Processing of DNA for nonhomologous end-joining is controlled by kinase activity and XRCC4/ligase IV.J Biol Chem. 2007 Apr 20;282(16):11950-9. doi: 10.1074/jbc.M610058200. Epub 2007 Jan 31. J Biol Chem. 2007. PMID: 17272270
-
Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends.Proc Natl Acad Sci U S A. 2007 May 8;104(19):7851-6. doi: 10.1073/pnas.0702620104. Epub 2007 Apr 30. Proc Natl Acad Sci U S A. 2007. PMID: 17470781 Free PMC article.
-
Role and regulation of human XRCC4-like factor/cernunnos.J Cell Biochem. 2008 Aug 1;104(5):1534-40. doi: 10.1002/jcb.21726. J Cell Biochem. 2008. PMID: 18335491 Review.
-
Biochemical mechanisms of chromosomal translocations resulting from DNA double-strand breaks.DNA Repair (Amst). 2006 Sep 8;5(9-10):1199-212. doi: 10.1016/j.dnarep.2006.05.016. Epub 2006 Jul 5. DNA Repair (Amst). 2006. PMID: 16822725 Review.
Cited by
-
Strategies for improving the genome-editing efficiency of class 2 CRISPR/Cas system.Heliyon. 2024 Sep 27;10(19):e38588. doi: 10.1016/j.heliyon.2024.e38588. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39397905 Free PMC article. Review.
-
Pathogenic variants in human DNA damage repair genes mostly arose after the latest human out-of-Africa migration.Front Genet. 2024 Jun 14;15:1408952. doi: 10.3389/fgene.2024.1408952. eCollection 2024. Front Genet. 2024. PMID: 38948361 Free PMC article.
-
Characterizing the Repair of DNA Double-Strand Breaks: A Review of Surrogate Plasmid-Based Reporter Methods.Methods Mol Biol. 2023;2701:173-182. doi: 10.1007/978-1-0716-3373-1_11. Methods Mol Biol. 2023. PMID: 37574482 Review.
-
Holding it together: DNA end synapsis during non-homologous end joining.DNA Repair (Amst). 2023 Oct;130:103553. doi: 10.1016/j.dnarep.2023.103553. Epub 2023 Aug 8. DNA Repair (Amst). 2023. PMID: 37572577 Free PMC article. Review.
-
The flexible and iterative steps within the NHEJ pathway.Prog Biophys Mol Biol. 2023 Jul-Aug;180-181:105-119. doi: 10.1016/j.pbiomolbio.2023.05.001. Epub 2023 May 5. Prog Biophys Mol Biol. 2023. PMID: 37150451 Free PMC article.
References
-
- Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA (2003) The frameshift infidelity of human DNA polymerase lambda. Implications for function. J Biol Chem 278: 34685–34690 - PubMed
-
- Boe SO, Sodroski J, Helland DE, Farnet CM (1995) DNA end-joining in extracts from human cells. Biochem Biophys Res Commun 215: 987–993 - PubMed
-
- Chan DW, Lees-Miller SP (1996) The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem 271: 8936–8941 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials


