Abstract
RhoG is a member of the Rho family of GTPases that activates Rac1 and Cdc42 through a microtubule-dependent pathway. To gain understanding of RhoG downstream signaling, we performed a yeast two-hybrid screen from which we identified kinectin, a 156-kDa protein that binds in vitro to conventional kinesin and enhances microtubule-dependent kinesin ATPase activity. We show that RhoGGTP specifically interacts with the central domain of kinectin, which also contains a RhoA binding domain in its C terminus. Interaction was confirmed by coprecipitation of kinectin with active RhoGG12V in COS-7 cells. RhoG, kinectin, and kinesin colocalize in REF-52 and COS-7 cells, mainly in the endoplasmic reticulum but also in lysosomes. Kinectin distribution in REF-52 cells is modulated according to endogenous RhoG activity. In addition, by using injection of anti-kinectin antibodies that challenge RhoG-kinectin interaction or by blocking anti-kinesin antibodies, we show that RhoG morphogenic activity relies on kinectin interaction and kinesin activity. Finally, kinectin overexpression elicits Rac1- and Cdc42-dependent cytoskeletal effects and switches cells to a RhoA phenotype when RhoG activity is inhibited or microtubules are disrupted. The functional links among RhoG, kinectin, and kinesin are further supported by time-lapse videomicroscopy of COS-7 cells, which showed that the microtubule-dependent lysosomal transport is facilitated by RhoG activation or kinectin overexpression and is severely stemmed upon RhoG inhibition. These data establish that kinectin is a key mediator of microtubule-dependent RhoG activity and suggest that kinectin also mediates RhoG- and RhoA-dependent antagonistic pathways.
Rho GTPases represent a distinct group of the Ras superfamily consisting of 21 members (41). Like other Ras-related proteins, Rho proteins can bind GDP and GTP, and their activities are up-regulated by guanine nucleotide exchange factors (GEFs), which promote GTP loading, and down-regulated by GTPase-activating proteins, which stimulate GTP hydrolysis (9). Once loaded with GTP, Rho GTPases are able to interact with and activate downstream effector proteins, which in turn directly or indirectly trigger the initiation of cellular effects (2).
Among Rho family members, Rac1, Cdc42, and RhoA have been extensively studied in many cell types, supporting the notion that Rac1 and Cdc42 facilitate the emergence of protrusive cell structures associated with focal complexes while RhoA has an opposed effect, leading to cell retraction and adhesion (3, 15). The situation is well documented in fibroblasts, in which Rac1 regulates ruffle and lamellipodium formation and is required for cell migration and Cdc42 regulates filopodium and microvillus formation and controls cell polarity, while RhoA regulates cell adhesion and contractility through stress fiber assembly (31). In neuronal cell lines, Rac1 and Cdc42 are required for growth cone dynamics and neurite outgrowth, whereas RhoA promotes growth cone collapse and neurite retraction (13).
We reported earlier that RhoG, a Rho family member related to the Rac/Cdc42 subgroup (42), triggers in fibroblasts the formation of both lamellipodia and filopodia through distinct pathways controlled by Rac1 and Cdc42 (14). A similar hierarchical situation has recently been described in neuronal PC12 cells, in which RhoG mediates NGF-dependent neurite outgrowth through pathways controlled by Rac1 and Cdc42 (18). The implication of RhoG activity in neuronal cells is further supported by the fact that RhoG is a specific target of Trio (8), a mammalian exchange factor whose homologues in Drosophila and Caenorhabditis elegans are involved in axon pathfinding (4, 5, 35).
RhoG displays several distinctive features in comparison with Rac1 and Cdc42. First, cells expressing an active RhoG mutant exhibit polarized lamellipodia and filopodia (14), while Rac1 and Cdc42 trigger the formation of these structures around most of the cell periphery (32). Second, RhoG morphogenic activity requires the microtubule network, whereas Rac1 and Cdc42 activities do not (14). Finally, RhoG is the only member of the Rac1/Cdc42 subgroup that does not bind Cdc42-Rac1 interactive binding domains (14). This supports the notion that RhoG might locally activate Rac1 and Cdc42 through specific effectors connected with microtubules. To address the nature of such effectors, we performed a yeast two-hybrid screen and identified kinectin as a major RhoG target. Kinectin, a 156-kDa protein inserted in endoplasmic reticulum (ER) membranes (37), has recently been shown to interact with the cargo binding site of conventional kinesin and activate its microtubule-stimulated ATPase activity (33). We demonstrate here that the binding of RhoG to kinectin is essential for RhoG activity.
MATERIALS AND METHODS
Plasmid constructs. (i) GTPases.
Yeast pLex and mammalian constructs encoding active Rho GTPases have been described elsewhere (8, 14, 34). pLex-Rac1G12VC186S and pVJL10-RhoBG14V were gifts from G. Zalcman and J. Camonis (Institut Curie, Paris, France). pBTM116 RhoGQ61LΔCAAX was produced by directed mutagenesis from pBTM116 RhoGwtΔCAAX with the GeneEditor kit (Promega).
(ii) Kinectin.
The construct expressing kinectin amino acids (aa) 1117 to 1362 (K10Δ) was obtained by deleting a 1.58-kb EcoRI/XbaI fragment from clone K10. Kinectin fragments were swapped from pGAD1318 yeast plasmids to pEGFP-C2 (Clontech, Palo Alto, Calif.). The kinectin coding sequence was reconstructed from KIAA0004 and A65N17 clones (T. Nagase, Kazusa DNA Research Institute). The stop codon was deleted by PCR, and the resulting insert was fused in frame with GFP as a 3.3-kb NcoI/AgeI fragment in pEGFP-NI (Clontech).
(iii) Exchange factors and Rho targets.
Vectors expressing the exchange factors TrioD1 and Tiam1, specifically activating RhoG and Rac1, respectively, as well as effector fragments interacting with Rac1, Cdc42, and RhoA, have been described previously (8, 12, 14).
Interaction screening of a human Jurkat T-cell cDNA library with RhoG.
Yeast L40 was cotransformed with pBTM116RhoGG12VΔ, expressing LexA fused to RhoGG12V with its CAAX box deleted, and an oligo(dT)-primed Jurkat cDNA library (7). Double transformants (107) were plated out on selective drop-out medium and allowed to grow for 3 days. His+ colonies (236) were patched on selective medium, replica plated on Whatman 40 filters, and assayed for β-galactosidase activity (11).
Antibodies.
Rat R2 anti-RhoG antibodies have been described elsewhere (8, 14). Monoclonal anti-Rac1 antibodies were from Transduction Laboratories. The anti-kinectin antibodies α-KiD1 and α-KiD2, directed against kinectin domains, were raised by immunizing rabbits with purified glutathione S-transferase (GST) fused to aa 673 to 939 (KiD1) and 1117 to 1362 (KiD2) of human kinectin. Monoclonal 1D3 anti-PDI antibody was from StressGen (Victoria, Canada). Rabbit anti-lysosome-associated protein 2 (Lamp-2) was from Zymed (San Francisco, Calif.). Monoclonal anti-kinesin H2 antibody directed against kinesin heavy chain (10) was a gift from G. Bloom. This antibody is thought to cross-link adjacent kinesins, thereby impairing their movement along microtubules. Monoclonal anti-β-tubulin antibody was a gift from P. Mangeat.
Cell culture, transfection, microinjection, and immunohistochemistry.
The conditions for cell culture and transfection have been described elsewhere (8, 14). For microtubule disruption, cells were incubated for 30 min in 2 μM nocodazole. For microinjection, anti-kinectin, anti-kinesin, and rabbit control immunoglobulin G (IgG) antibodies (1 mg · ml−1 in 10 mM HEPES [pH 7.2]) were introduced by cytoplasmic injection. Twenty hours after transfection and 6 h after microinjection, the cells were fixed and processed for immunohistochemistry as described previously (8, 14). F-actin was detected by rhodamine-phalloidin labeling. For simultaneous detection of RhoG, kinectin, and kinesin, rat R2 antibody was incubated with biotinylated anti-rat antibody followed by Cya5-streptavidin labeling, α-KiD1 was incubated with anti-rabbit fluorescein isothiocyanate antibody, and mouse H2 was incubated with anti-mouse tetramethyl rhodamine isothiocyanate. For simultaneous detection of GFP-tagged proteins, MYC-tagged proteins, and F-actin, anti-MYC 9E10 staining was detected at a 445-nm wavelength using 7-amino-4-methylcoumarin-3-acetic acid-conjugated donkey anti-mouse IgG (Jackson Immunoresearch Laboratories). Fixed cells were observed under a laser scanning confocal microscope (MRC-1024; Bio-Rad Laboratories) or DMR B microscope (Leica, Wetzlar, Germany) (tube factor, 1×) equipped with a PL APO 40× objective (NA, 1.00). For all experiments, at least 100 transfected cells were examined. Images (16 bit) were captured with a MicroMax 1300 Y/HS cooled (−10°C) charge-coupled device camera driven by the MetaMorph (version 4.11) controller program (RS-Princeton Instruments, Trenton, N.J.). For colocalization analysis, confocal images were processed using the colocalization software package Imarys, version 2.7 (BitPlane, Zürich, Switzerland).
Image deconvolution.
To examine the inner cell three dimensionally, stacks of optical sections (z step = 0.1 μm) were captured (exposure time, 1 s, with halogen lamp illumination) with a Leica DMR B microscope using a 63× (NA, 1.32) objective mounted on a piezoelectric stepping motor. The stacks were restored with Huygens software (Scientific Volume Imaging b.v, Hilversum, The Netherlands). Briefly, Huygens is an iterative program that encodes light as 32-bit gray levels and reassigns it at high probability to specific voxels in the stack using a point spread function. This results in removing blur contained in the stack. In the present study, the maximum-likelihood estimation algorithm was used throughout. The restored stacks were then further processed with Imarys for visualization. Huygens and Imarys were run on a four-processor Origin 2000 and a two-processor Octane (Silicon Graphics, Mountain View, Calif.), respectively.
Time-lapse video microscopy.
For live cell studies, a laboratory-made device maintained cells in a 37°C, 5% CO2, 80% relative humidity atmosphere. Epifluorescence illumination was ensured by a halogen light bulb (100 W). COS-7 cells were grown on 0.17-mm-thick glass coverslips and transfected with various GFP constructs. Eighteen hours after transfection, the lysosomes were stained by incubation in normal Ringer's solution (115 mM NaCl, 2.6 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 10 mM glucose, 10 mM HEPES [pH 7.2], and 0.5 mg of bovine serum albumin [BSA]/ml) supplemented with 50 nM LysoTracker DND99 (Molecular Probes). After 30 min, the cells were rinsed twice with normal Ringer's solution and observed with an inverted Leica DMIRBE microscope using a 63× (NA, 1.32) objective. Cell images were captured (exposure time, 500 ms) every 10 s for 10 min as time series of 16-bit files. Successive frames of the movie were processed with MetaMorph to produce merged stacks in which moving lysosomes appear as linear series of dots. The average velocity approximated 0.1 μm/s, which is in the range of published data (38).
Protein interaction.
Procedures for yeast two-hybrid interaction have been described in detail (8). For biochemical interaction, 105 COS-7 cells were transfected by a construct encoding hemagglutinin (HA)-tagged wild-type RhoG (14). Forty-eight hours after transfection, the cells were lysed in IP buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM MgCl2) supplemented with 1% Triton X-100. HA-RhoG was immunoprecipitated by the addition of monoclonal 12CA5 antibody and protein G-Sepharose (Amersham-Pharmacia). Beads were rinsed twice in IP buffer, and GTPγS or GDP loading was performed as described previously (43). The Sepharose beads were then incubated for 30 min at room temperature with [35S]Met-labeled in vitro-translated kinectin fragment (aa 584 to 1356). After three washes, the bound proteins were eluted, fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and revealed by autoradiography. For in vitro interaction studies with KiD1, GST-RhoG (0.2 nmol; 30% contaminated by free GST) bound to glutathione-Sepharose was loaded for 2 min at 37°C in 100 μl of loading buffer (50 mM HEPES/NaOH [pH 7.4], 100 mM KCl, 1 mM dithiothreitol) supplemented with 1 mM GTPγS or GDP and then blocked with 20 mM MgCl2. GST-RhoG was then incubated at 4°C for 2 h with maltose binding protein (MBP; 0.2 nmol) or MBP-KiD1 (0.12 nmol; purity, greater than 90%), 1 mg of BSA ml−1, and 0.1% Tween 20. For interaction interference with α-KiD1 antibody, MBP-KiD1 was preincubated on ice for 30 min with α-KiD1 antibody in 30 μl of loading buffer and then incubated for 2 h at 4°C with GTPγS- or GDP-loaded GST-RhoG in the presence of 1 mg of BSA ml−1 and 0.1% Tween 20. Sepharose beads were rinsed three times in ice-cold loading buffer supplemented with 2 mM MgCl2, 1 mg of BSA ml−1, and 0.1% Tween 20. Total and Sepharose-bound proteins were analyzed by Western blotting using anti-MBP and anti-GST antibodies.
Immunoprecipitation and immunoblotting.
COS-7 cells (2 × 107) were transfected with constructs encoding various GFP-RhoG mutant proteins. Forty-eight hours after transfection, the cells were lysed in RIPA buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 1% deoxycholate, 0.1% SDS) supplemented with 1 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin ml−1, and 1 μg of leupeptin ml−1. Cell lysate (800 μl) was incubated with 1 μl of anti-GFP antibody (Clontech) and 20 μl of protein A Sepharose beads for 2 h at 4°C. The beads were washed extensively in RIPA buffer, subjected to SDS-PAGE, and transferred to nitrocellulose membranes (Schleicher & Schuell). Kinectin and GFP-RhoG proteins were detected by incubating filters with anti-KiD1 and anti-GFP antibodies, respectively. The filters were incubated with horseradish peroxidase-conjugated anti-rabbit antibody and revealed using enhanced chemiluminescence reagent (NEN, Boston, Mass.).
RESULTS
Kinectin selectively binds the GTP-bound form of RhoG.
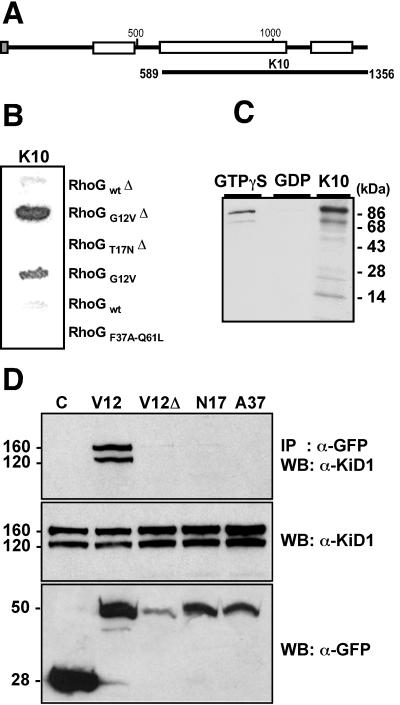
A yeast two-hybrid screen of a Jurkat T-cell cDNA library for RhoG-interacting proteins identified 172 positives; 45% corresponded to LyGDI/D4, a negative regulator acting on several members of the Rho family (22), 27% derived from three unknown independent mRNA sequences, and 28% were from kinectin. Most clones recovered from the screen proved to encode two-thirds of the protein, as exemplified by clone K10 (Fig. 1A). In yeast, all kinectin clones interacted only with the GTP-bound active RhoGG12V mutant; no signal was detected with the GDP-bound T17N mutant, the effector loop mutant RhoGF37AQ61L, or the wild-type proteins, as exemplified by clone K10 (Fig. 1B). Interaction between kinectin and GTP-bound RhoG was confirmed by in vitro binding analysis (Fig. 1C). Immunopurified HA-epitope-tagged wild-type RhoG protein was loaded with GTPγS or GDP and incubated with an 86-kDa [35S]Met-labeled kinectin fragment in vitro translated from clone K10. Kinectin associated with the GTPγS-bound RhoG protein, while only trace amounts were detected with GDP-loaded RhoG protein. RhoG interaction with endogenous kinectin was further established in living cells by immunoprecipitation experiments (Fig. 1D). Extracts from COS-7 cells expressing GFP alone or fused to RhoGG12V, RhoGG12V with its CAAX box deleted, RhoGT17N, or RhoGF37A (Fig. 1D, bottom) were immunoprecipitated with anti-GFP antibodies. Kinectin from total extracts appeared as a 160-kDa full-length species and a 120-kDa caspase-cleaved species (26) (Fig 1D, middle). As seen in Fig. 1D, top, kinectin was selectively coprecipitated with RhoGG12V. This establishes that only the GTP-bound form of RhoG binds to kinectin. At variance with two-hybrid data, RhoG CAAX box deletion totally abolished binding to kinectin, which suggests that RhoG subcellular distribution in COS-7 cells is crucial for the interaction.
FIG. 1.
Kinectin specifically binds the GTP-bound form of RhoG. (A) Full-length kinectin, with membrane-targeting signal sequence (shaded box) and regions with high probability of forming coiled-coil structures (open boxes). Most fragments recovered from the two-hybrid screen had their N termini between aa 589 and 673 and extended up to aa 1356. K10 represents the largest fragment. (B) Yeasts coexpressing K10 fragment fused to GAL4 activating domain and wild-type, G12V, and T17N RhoG mutants (with or without [Δ] the CAAX box) fused to LexA DNA binding domain were processed for β-galactosidase activity. (C) In vitro [35S]Met-labeled 86-kDa kinectin fragment from clone K10 was incubated with immunoprecipitated HA-tagged wild-type RhoG loaded with GTPγS or GDP. Shown is SDS-PAGE analysis of bound kinectin. (D) COS-7 cells were transfected to express GFP alone (lane C) or fused to RhoGG12V (lane V12), RhoGG12V with its CAAX box deleted (lane V12Δ), RhoGT17N (lane N17), or RhoGF37A (lane A37). Cell extracts were immunoprecipitated (IP) with anti-GFP antibodies and then analyzed by Western blotting (WB) with anti-kinectin α-KiD1 antibody (top). Total extracts were controlled by Western blotting for kinectin (middle) and for GFP fusion (bottom). Masses (in kilodaltons) are indicated on the left.
Two distinct kinectin domains specifically bind RhoG and RhoA in vivo.
Kinectin had been previously shown in the yeast two-hybrid system to weakly interact with other Rho GTPases (1, 17), delineating two binding domains located within aa 630 to 935 and 1153 to 1327. For clarity, we refer to these domains as KiD1 and KiD2 (Fig. 2A). To examine the binding specificity of KiD1 and KiD2, we coexpressed in yeast various dominant-active GTPase mutants with kinectin K10 (containing both KiD1 and KiD2), K66 (containing KiD1 only), or K10Δ (a version of K10 with deletions that only contains KiD2). Growth on histidine-free plates and quantitative β-galactosidase assays showed that RhoGG12V, RhoGQ61L, and, to a lesser extent, Rac1G12V interacted only with KiD1 (Fig. 2B, lanes K66), while RhoAG14V weakly but specifically bound KiD2 (lanes K10Δ). Cdc42G12V and RhoBG14V did not interact with any KiD domain. All GTPase constructs were expressed at comparable levels (not shown).
FIG. 2.
Two kinectin domains, KiD1 and KiD2, specifically bind RhoG and RhoA. (A) Structure of the fragments used. K10 (the largest fragment) and K66 (the shortest) were isolated from the two-hybrid screen. K10Δ was derived from K10. Shaded box, membrane-targeting signal sequence; open boxes, regions with high probability of forming coiled-coil structures. (B) Kinectin fragments (K10, K66, and K10Δ) and GTPases (RhoGG12V, RhoGQ61L, Rac1G12V, Cdc42G12V, RhoAG14V, and RhoBG14V) were introduced into the yeast two-hybrid system. Interactions were visualized by growth on His− plates (left) or by β-galactosidase activity (in arbitrary units) using an o-nitrophenyl-β-d-galactopyranoside liquid assay (right). Shown is the mean average of five independent experiments on distinct colonies. The standard error of the mean was less than 8%. Control interactions (lane C) were done with Por1 for Rac1 (39), WASP for Cdc42 (36), and p160ROCK for RhoA and RhoB (24). (C and D) REF-52 cells expressing MYC-RhoGG12V (C) or MYC-RhoAG14V (D) alone (a and d) or in combination with GFP-KiD1 (b and e) or GFP-KiD2 (c and f). Eighteen hours after transfection, the cells were fixed and observed for MYC epitope staining (a and insets of b and c), GFP fluorescence (b and c), and F-actin staining (d, e, and f). (E) REF-52 cells expressing MYC-Rac1G12V alone (a and c) or in combination with GFP-KiD1 (b and d). The cells were examined for MYC staining (a and b), GFP fluorescence (inset of b), and F-actin distribution (c and d). (F) REF-52 cells expressing GFP-KiD1 (a) or GFP-KiD2 (b) were examined for GFP fluorescence (insets) and F-actin distribution (a and b). For all experiments, the cells shown are representative of more than 100 observed cells. Bars, 10 μm.
We next addressed the specificity of KiD domains in vivo. GFP-fused KiD fragments were examined for the ability to inhibit the morphological effects of MYC-tagged active GTPases in REF-52 cells. RhoGG12V expression produced lamellipodia and a reduced content of stress fibers (Fig. 2C, a and d) which were both inhibited in 75% of cells coexpressing GFP-KiD1 (Fig. 2C, b and e), indicat that KiD1 competes the binding of RhoGG12V to endogenous targets. GFP-KiD2 expression had no inhibitory effect on RhoGG12V activity (Fig. 2C, c and f). A symmetrical situation occurred for RhoAG14V, whose effects on actin stress fiber assembly (Fig. 2D, a and d) were blocked upon KiD2 expression in 90% of expressing cells (Fig. 2D, c and f) but not upon KiD1 expression (Fig. 2D, b and e). In contrast with RhoGG12V, lamellipodia produced by Rac1G12V (Fig. 2E, a and c) remained unaffected by GFP-KiD1 (Fig. 2E, b and d) or GFP-KiD2 (not shown) coexpression. In agreement with two-hybrid data, KiD1 or KiD2 expression had no effect on Cdc42 activity (not shown). Interestingly, in more than 90% of transfected cells, GFP-KiD1 alone led to a twofold increase in actin stress fibers (Fig. 2F, a) while GFP-KiD2 alone triggered stress fiber disassembly and elongated cell morphology (Fig. 2F, b), which suggests that KiD1 and KiD2 also inhibit endogenous RhoG and RhoA proteins. Compared with the two-hybrid results, these in vivo data establish that KiD1 specifically interacts with RhoG and confirm the binding of KiD2 to RhoA.
Kinectin colocalizes with RhoG and kinesin in vivo.
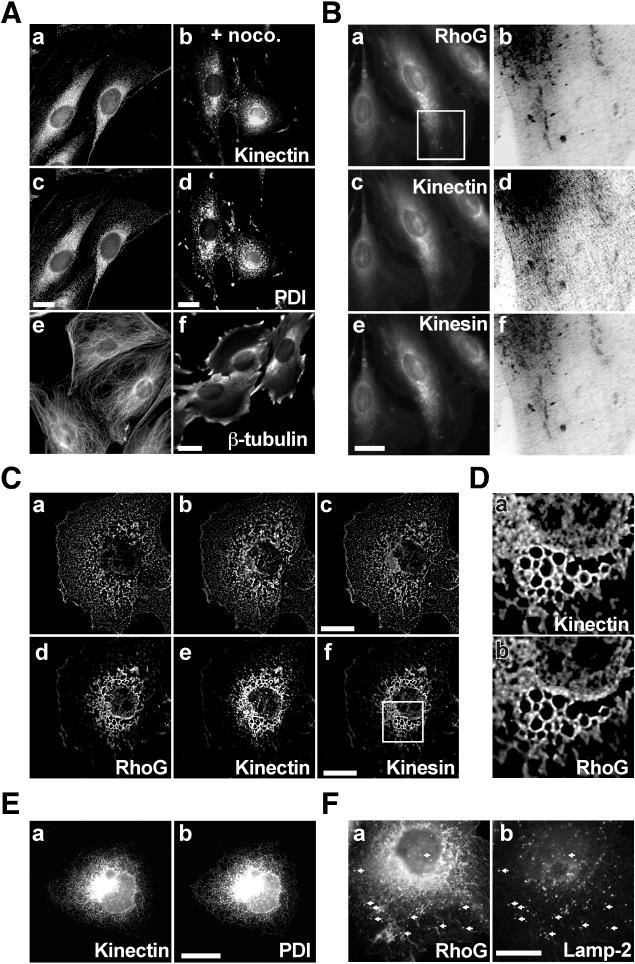
To establish a functional link between RhoG and kinectin, we examined their subcellular distributions. We first confirmed the localization of kinectin in the ER of REF-52 fibroblastic cells by comparing deconvolved staining of kinectin (Fig. 3A, a) and protein disulfide isomerase (PDI) (Fig. 3A, c), shown to be mainly located in the ER (40). Kinectin and PDI showed overall similar staining, in agreement with published data (37). A similar colocalization was also observed in nocodazole-treated cells, in which kinectin (Fig. 3A, b) and PDI (Fig. 3A, d) appeared as a more granular perinuclear staining. The extent of microtubule depolymerization upon nocodazole treatment is shown Fig. 3A, e and f. We next examined RhoG and kinectin distribution in conjunction with conventional kinesin, since the latter has been reported to bind kinectin directly (33). In all cells examined, endogenous RhoG, kinectin, and kinesin (Fig. 3B, a, c, and e) produced very similar staining, which labeled structures across the entire cytoplasm with perinuclear accumulation (Fig. 3A, a and c, show a single 0.2-μm-thick cell plane processed by image deconvolution, while Fig. 3B, a, c, and e, show unprocessed whole cells, which explains the difference in staining). In addition, anti-RhoG and anti-kinesin antibodies stained short tubular perinuclear structures, while anti-kinectin produced a more diffuse staining. Enlarged and deconvolved views (Fig. 3B, b, d, and f) of the region boxed in Fig. 3B, a, show that although more diffuse, kinectin accumulates in the same structures as those stained by RhoG and kinesin. We next examined the distribution of these proteins in COS-7 cells, in which we validated interaction between RhoG and kinectin (Fig. 1D). Due to the thickness of COS-7 cells, we performed deconvolution of single cell planes. At the coverslip level (Fig. 3C, a to c), RhoG, kinectin, and kinesin showed nearly identical distributions, showing a tubulovesicular pattern that extended across the entire cell plane. At 0.7 μm above (Fig. 3C, d to f), all three proteins remained codistributed according to a reticular pattern (an enlarged view of the region boxed in Fig. 3C, f, shown in Fig. 3D, illustrates the codistribution of RhoG and kinectin). Analysis of additional cell planes showed that this pattern corresponds to the peripheries of large vesicles (not shown). As in REF-52 cells, the overall distribution of all three proteins remained very similar to PDI distribution, as illustrated by kinectin staining (Fig. 3E, a and b). Since kinectin and kinesin had been implicated in lysosome distribution (16, 33), we also examined whether RhoG, kinectin, and kinesin codistribute with these organelles. As illustrated in Fig. 3F, part of RhoG staining (a) colocalized with Lamp-2 (b). Similar colocalization was observed for kinesin and, to a lesser extent, for kinectin (not shown). These data indicate that in REF-52 and COS-7 cells, RhoG, kinectin, and kinesin show a high level of colocalization. This mainly concerns ER membranes but also lysosomes, in particular in COS-7 cells.
FIG. 3.
Colocalization of endogenous kinectin, RhoG, and kinesin. (A) REF-52 cells (a, c, and e) were treated for 30 min with 2 μM nocodazole (b, d, and f). The cells were fixed and stained for kinectin using rabbit α-KiD1 (a and b), for PDI using mouse 1D3 (c and d), or for β-tubulin (e and f). Images a to d represent single 0.2-μm-thick cell planes processed using image deconvolution (Huygens; see Materials and Methods). (B) REF-52 cells were fixed and stained for RhoG with rat R2 (a and b), for kinectin with rabbit α-KiD1 (c and d), and for kinesin with mouse H2 antibody (e and f). The cell area boxed in image a was processed using image deconvolution. Negative and enlarged views of the same cell area stained for RhoG, kinectin, and kinesin are shown in images b, d, and f, respectively. (C) COS-7 cells were fixed and stained for RhoG (a and d), kinectin (b and e), and kinesin (c and f). Single 0.2-μm-thick cell planes were processed using image deconvolution. Shown are a cell plane located at the coverslip level (a, b, and c) and a cell plane located 0.7 μm above it (d, e, and f). (D) Enlarged views of the cell area boxed in panel C, image f, stained for kinectin (a) and RhoG (b). (E) COS-7 cells were fixed and stained for kinectin (a) and PDI (b). The images shown represent staining of the whole cell volume. (F) COS-7 cells were fixed and stained for RhoG (a) and Lamp-2 (b). The images shown correspond to merged stacks of seven cell planes located up to 0.8 μm above the coverslip level. For all panels, the cells shown are representative of more than 100 observed cells. Bars, 10 μm.
Endogenous kinectin distribution depends on endogenous RhoG activity.
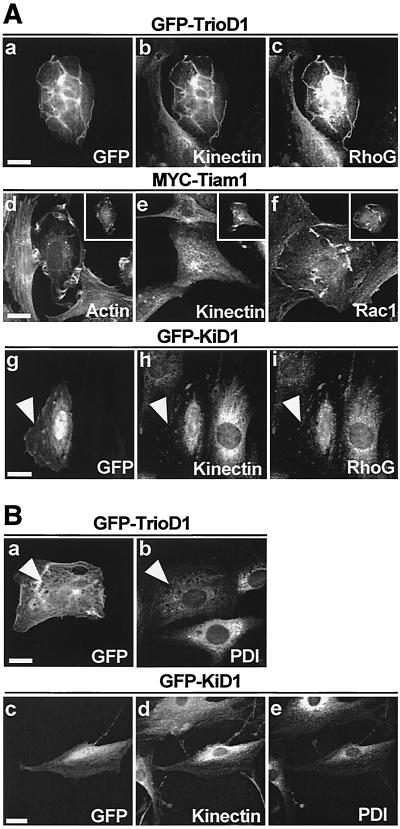
We next examined whether RhoG activation can affect kinectin distribution. With this aim, we first activated RhoG by expressing GFP fused to TrioD1, a GEF specific for RhoG (8). In more than 80% of transfected cells, GFP-TrioD1 triggered the formation of dorsal and peripheral ribbon-like ruffles, in which GFP-TrioD1, kinectin, and RhoG colocalized (Fig. 4A, a to c). To rule out the possibility that kinectin accumulates nonspecifically in ruffles, we examined its distribution in cells expressing Tiam1, a GEF specific for Rac1 (27). As expected, Tiam-1-expressing cells produced F-actin-stained lamellipodia and ruffles (Fig. 4A, d) in which Tiam1 and Rac1 (Fig. 4A, d and f) but not kinectin (Fig. 4A, e) were found to accumulate. We next examined whether RhoG inhibition had an effect on kinectin distribution by expressing GFP-KiD1, shown above to specifically inhibit RhoG activity (Fig. 2). In 70% of positive cells, KiD1 expression (Fig. 4A, g) led to a retraction of endogenous kinectin (Fig. 4A, h) and RhoG (Fig. 4A, i) proteins from the cell periphery, leading to a perinuclear codistribution. Similar results were obtained when a dominant-negative RhoGT17N mutant was expressed instead of KiD1 (not shown). These data indicate that kinectin intracellular distribution is sensitive to RhoG activity. They also indicate that active and inactive RhoG show different cellular distributions, but in each case, RhoG still colocalizes with kinectin, suggesting that both proteins are targeted to the same endomembranes independent of the GDP or GTP status of RhoG. We also examined whether changes in RhoG activity might have a general effect on ER membranes (Fig. 4B). TrioD1 expression elicited a dispersion of PDI staining throughout the cytoplasm, but no accumulation in dorsal ruffles was observed (Fig. 4B, a and b). Conversely, GFP-KiD1 expression led to a retraction of PDI staining (Fig. 4B, e), similar to that observed for kinectin (Fig. 4B, d).
FIG. 4.
Endogenous kinectin redistributes according to RhoG activity. (A) REF-52 cells expressing TrioD1 (a to c), MYC-Tiam1 (d to f), and GFP-KiD1 (g to i) were visualized for GFP (a and g) and revealed for kinectin (b, e, and h), RhoG (c and i), F-actin (d), Rac1 (f), and MYC (insets of d to f). The arrowheads in images g to i represent cell boundaries. (B) REF-52 cells expressing GFP-TrioD1 (a and b) or GFP-KiD1 (c to e) were visualized for GFP (a and c), PDI (b and e), and kinectin (revealed with α-KiD2 antibody; d). The arrowheads in images a and b signal the presence of GFP and the absence of PDI accumulation in dorsal lamellipodia. For all experiments, the cells shown are representative of more than 100 observed cells. Bars, 10 μm.
RhoG morphogenic activity is kinectin and kinesin dependent.
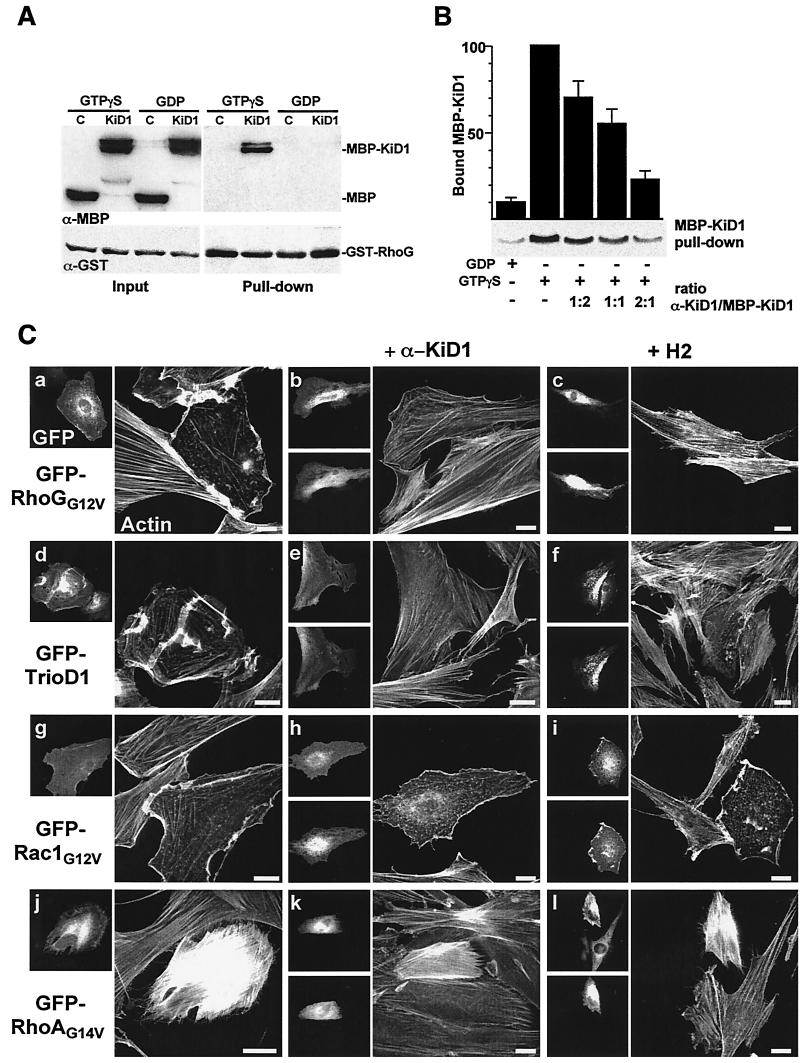
To address a direct implication of kinectin in RhoG downstream signaling, we next wished to analyze the consequences of inhibiting RhoG-kinectin interaction specifically. With this aim, we raised antibodies directed against KiD1 (α-KiD1) and examined their ability to hamper RhoG-KiD1 interaction. A Sepharose-bound GST-RhoG fusion protein was loaded with GTPγS or GDP and then incubated with a soluble affinity-purified MBP-KiD1 fusion protein (Fig. 5A). MBP-KiD1 specifically bound to GTPγS-loaded RhoG, demonstrating that active RhoG binds KiD1 directly. Preincubating MBP-KiD1 with increasing amounts of α-KiD1 inhibited the formation of GST-RhoG/MBP-KiD1 complex (Fig. 5B): up to 80% inhibition was observed with a twofold molar excess of α-KiD1 over MBP-KiD1. We thus anticipated that α-KiD1 microinjection in living cells might also prevent RhoG-kinectin interaction. In GFP-RhoGG12V (Fig. 5C, b)- and GFP-TrioD1 (Fig. 5C, e)-expressing cells, α-KiD1 microinjection led to a strong inhibition of the cytoskeletal changes (compare Fig. 5C, a and d). About 70% of injected cells maintained a high level of actin stress fibers and displayed no membrane ruffling; the remainder exhibited a clearly attenuated RhoGG12V or TrioD1 phenotype. In contrast, α-KiD1 antibody had no effect when microinjected in cells expressing GFP-Rac1G12V (compare Fig. 5C, g and h), GFP-RhoAG14V (compare Fig. 5C, j and k), or GFP-Cdc42G12V (not shown). This indicates that α-KiD1 microinjection specifically and efficiently inhibited the downstream signaling of RhoGG12V as well as that of endogenous RhoG. Similar inhibitory effects were obtained after injection of H2 anti-kinesin monoclonal antibody, previously shown to inhibit both anterograde and retrograde fast axonal transport (10). H2 microinjection inhibited the cellular effects of RhoGG12V (Fig. 5C, c) and TrioD1 (Fig. 5C, f) but was ineffective on Rac1G12V (Fig. 5C, i) or RhoAG14V (Fig. 5C, l). α-KiD1 and H2 antibodies had no major effect when microinjected in control REF-52 cells, except for a moderate increase in actin stress fibers in 30% of the cells (not shown). Microinjection of control rabbit Ig had no effect on REF-52 cells and did not inhibit the GFP-RhoGG12V phenotype (not shown). From these data, we conclude that the cellular effects of RhoG depend on RhoG-kinectin interaction and also require kinesin activity.
FIG. 5.
RhoG morphogenic activity is kinectin and kinesin dependent. (A) Glutathione-Sepharose-bound GST-RhoG was loaded with GTPγS or GDP and then incubated with MBP (lanes C) or MBP fused to K66 fragment (lanes KiD1). Total proteins (Input) and Sepharose-bound proteins (Pull-down) were analyzed by Western blotting using anti-MBP (α-MBP; top) and anti-GST (α-GST; bottom) antibodies. (B) MBP-KiD1 was first incubated with increasing relative amounts of α-KiD1 antibody as indicated and then incubated with Sepharose-bound GST-RhoG loaded with GTPγS. The amount of Sepharose-bound MBP-KiD1 was measured by Western blotting using anti-MBP antibodies. The background signal was estimated by incubating MBP-KiD1 with GST-RhoG loaded with GDP. Quantification of three independent experiments shows that a twofold α-KiD1 excess leads to an 80% reduction of RhoG-KiD1 interaction. The error bars indicate standard error of the mean. +, present; −, absent. (C) Cells expressing GFP-RhoGG12V (a to c), GFP-TrioD1 (d to f), GFP-Rac1G12V (g to i), and GFP-RhoAG14V (j to l) were microinjected with α-KiD1 (b, e, h, and k) or H2 anti-kinesin (c, f, i, and l). The cells were fixed 6 h after microinjection and observed for GFP fluorescence (upper left images), injected antibody (lower left images), and F-actin distribution (images on right). For each experiment, the cells shown are representative of more than 100 injected cells. Bars, 10 μm.
Kinectin overexpression elicits Rho-dependent phenotypes.
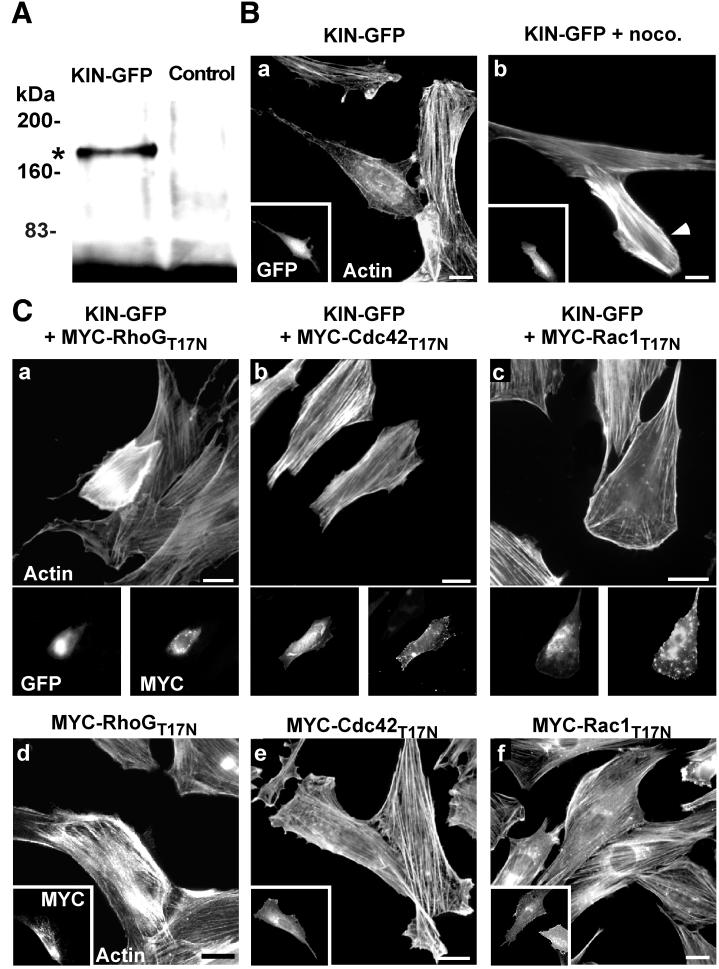
We next examined whether kinectin overexpression might elicit Rho-dependent cellular effects by itself. For this purpose, a full-length kinectin fused in its carboxy terminus to GFP (Kin-GFP) was constructed whose expression in COS-7 cells yielded a product of the expected size (180 kDa) (Fig. 6A). Expression of Kin-GFP modified the morphology of more than 90% of REF-52 cells, which exhibited an elongated shape associated with cortical F-actin accumulation, a marked reduction in the level of stress fibers, and occasional peripheral protrusions (Fig. 6B, a). Kin-GFP expression did not fully mimic RhoGG12V (in particular, no membrane ruffles were detected), indicating that additional RhoG effectors are probably involved in the establishment of RhoG morphogenic activity. Interestingly, nocodazole treatment (known to moderately activate the RhoA-dependent pathway [25]) not only led to cell retraction and loss of protrusions but also elicited a dramatic increase in actin stress fibers, far above the level of neighboring untransfected cells (Fig. 6A, b), reminiscent of the RhoAG14V phenotype (compare Fig. 2D, d). Thus, kinectin overexpression enhances the effect of microtubule disruption. A similar enhancing effect was observed when Kin-GFP was coexpressed with the dominant-negative RhoGT17N (Fig. 6C, a) or KiD1 (not shown), further supporting a functional link between RhoG and kinectin. Conversely, coexpression of the dominant-negative Cdc42T17N led to a simple reversion in 60% of cotransfected cells (Fig. 6C, b), while Rac1T17N had a weak inhibitory effect, mainly preventing the formation of peripheral protrusions (Fig. 6C, c). When expressed on their own, RhoGT17N (Fig. 6C, d), Cdc42T17N (Fig. 6C, e), and Rac1T17N (Fig. 6C, f) had little effect on cell morphology, except for a slight increase in actin stress fibers for RhoGT17N, as observed in KiD1-expressing cells (Fig. 2F, a). Taken together, these data indicate that kinectin can activate two opposed pathways, one leading to peripheral morphogenic activity and the other one leading to an increase in actin stress fibers. They also indicate that RhoG activity and the microtubule network are critical for redirecting kinectin activity to either pathway. Finally, they show that Cdc42 and, to a much lesser extent, Rac1 act in the pathway leading to peripheral morphogenic activity.
FIG. 6.
Overexpression of kinectin induces Rho-dependent changes in cell morphology. (A) COS-7 cells were transfected with a construct expressing full-length kinectin fused to the GFP. Anti-GFP antibody immunoprecipitates from transfected cells (KIN-GFP) or control cells (Control) were analyzed by Western blotting using the same antibody. The asterisk corresponds to the expected product size (180 kDa). (B) REF-52 fibroblasts expressing KIN-GFP, untreated (a) or treated for 60 min with 2 μM nocodazole (noco.) (b) were observed for F-actin (a and b) and GFP fluorescence (insets) 18 h after transfection. The arrowhead shows the transfected cell. (C) REF-52 cells were transfected with KIN-GFP in combination with MYC-RhoGT17N (a), MYC-Cdc42T17N (b), and MYC-Rac1T17N (c). As a control, the cells were transfected only with constructs expressing dominant-negative Rho mutants (d to f). The cells were fixed 18 h after transfection and observed for F-actin distribution (a to c, upper images, and d to f), GFP fluorescence (a to c, lower left), and MYC epitope expression (a to c, lower right, and d to f, insets). The cells shown are representative of more than 100 observed cells. Bars, 10 μm.
RhoG activity and kinectin overexpression influence microtubule-dependent transport.
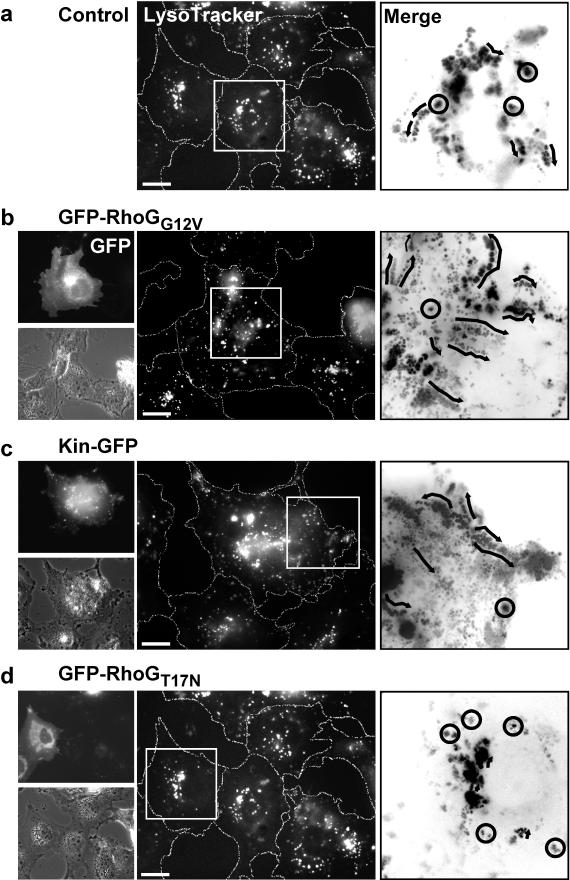
The functional links among RhoG, kinectin, kinesin, and microtubules called for a critical role of RhoG in kinesin-dependent vesicle transport. We showed that in COS-7 cells, RhoG coprecipitated with kinectin (Fig. 1D) and colocalized with lysosomal anti-Lamp-2 staining (Fig. 3F). In addition, it has been reported that lysosomes move towards the cell periphery in a kinesin-dependent manner (16, 29) and that expression of kinectin and kinesin inhibitory fragments blocks the peripheral distribution of lysosomes (33). We thus evaluated by time-lapse videomicroscopy the movement of lysosomes in control or transfected COS-7 cells stained with LysoTracker dye (Fig. 7). Stacks of time-lapse images were processed to track individual lysosomes and measure their velocities through images. Two classes were considered: SM, slow random movements (below 0.1 μm/s; appearing as dot clusters), and FM, fast movements directed towards the cell periphery (appearing as linear series of dots). In untransfected cells (Fig. 7a), lysosomes showed a broad perinuclear distribution and exhibited mainly slow movements (75% SM at a mean velocity of 0.01 μm/s; 25% FM at 0.25 μm/s) (Fig. 7a, merge panel, and Table 1). Cells expressing GFP-RhoGG12V (Fig. 7b) showed more diffuse and dispersed lysosome staining, consistent with an overall increase in vesicle velocity (79% SM at 0.02 μm/s; 21% FM at 0.42 μm/s) (Fig. 7b, merge panel, and Table 1). A similar positive effect was observed in kinectin-GFP cells (Fig. 7c), in which lysosome motions distributed as 78% SM at 0.03 μm/s and 22% FM at 0.43 μm/s. A clear contrasted situation occurred in GFP-RhoGT17N-expressing cells (Fig. 7d and Table 1), in which all observed lysosomes displayed random slow motions (0.01 μm/s). A similar inhibition was also observed in cells injected with α-KiD1, in which 95% of lysosomes were found to move slowly (Table 1). The same range of inhibitory effects was measured when lysosome redistribution was stimulated upon acidification (not shown). Control experiments evidenced no effect of GFP-RacT17N (Table 1), GFP-RacG12V, Cdc42G12V, and RhoAG14V (not shown) expression on lysosome velocity. These data indicate that changes in RhoG activity and kinectin expression have a direct and specific impact on lysosome movements towards the cell periphery, which further supports a role for both proteins in kinesin-dependent transport of vesicles.
FIG. 7.
Lysosome movements towards cell periphery are influenced by RhoG activity and kinectin expression. COS-7 cells (a) expressing GFP-RhoGG12V (b), Kinectin-GFP (c), and GFP-RhoGT17N (d) were incubated for 30 min with LysoTracker dye to stain lysosomes (middle images). Images were then acquired every 10 s for 10 min. Sections (boxed in middle panels) were processed using MetaMorph to produce merged stacks (images on right) in which randomly moving lysosomes appear as spot clusters (circles) whereas directed moving lysosomes appear as linear series of dots (arrows). Expressing cells were observed for GFP (b, c, and d, upper left images) and by phase-contrast (b, c, and d, lower left images). The cells shown are representative of more than 50 observed cells. Bars, 10 μm.
TABLE 1.
Expression of RhoG mutants and kinectin affects lysosome velocity
| Protein expressed | SM events (<0.1 μm · s−1)
|
FM events (>0.1 μm · s−1)
|
||
|---|---|---|---|---|
| Frequency (%) | Mean velocity (SEM) (μm · s−1) | Frequency (%) | Mean velocity (SEM) (μm · s−1) | |
| Control | 75 | 0.009 (0.001) | 25 | 0.256 (0.019) |
| GFP-RhoGG12V | 79 | 0.017 (0.001) | 21 | 0.42 (0.02) |
| Kinectin-GFP | 78 | 0.031 (0.002) | 22 | 0.43 (0.02) |
| GFP-RhoGT17N | 100 | 0.008 (0.001) | ||
| α-KiD1 | 95 | 0.01 (0.001) | 5 | 0.27 (0.03) |
| GFP-RacT17N | 77 | 0.015 (0.001) | 23 | 0.26 (0.02) |
DISCUSSION
We have previously shown that RhoG is specifically activated by the first Dbl-like RhoGEF domain of Trio (8), a multidomain protein recently implicated in axon pathfinding in Drosophila (4, 5, 30) and C. elegans (35). Once activated, RhoG produces biological effects dependent on Rac1 and Cdc42 activities, leading in neuronal cells to neurite outgrowth (18) and in fibroblastic cells to the formation of microvilli and peripheral and dorsal ruffles, as well as cell protrusions, through a microtubule-dependent process (14). However, the molecular mechanisms by which RhoG uses microtubules to exert its cytoskeletal effects remained unknown. We report here the characterization of kinectin as a major RhoG target. Kinectin has been reported to act as a kinesin anchor protein essential for vesicle movement along microtubules (21, 37). Although the precise role of kinectin remains a debated issue, our data unambiguously demonstrate that (i) kinectin contains two distinct domains that bind RhoG and RhoA, respectively; (ii) RhoG, kinectin, and kinesin show highly similar cell distributions, in particular in the ER; (iii) kinectin redistributes according to RhoG activity; (iv) kinectin and kinesin activities, as well as the microtubule network, are required for RhoG downstream signaling; (v) kinectin signals towards opposed pathways, leading to either peripheral effects or stress fiber assembly, depending on RhoG activity and microtubule integrity; and (vi) changes in both RhoG activity and the level of kinectin expression have a direct effect on kinesin-dependent vesicle velocity.
Kinectin displays all the biochemical and cellular hallmarks of a RhoG-interacting protein: in vitro, kinectin interacts directly and selectively with the GTP-bound active RhoG; in vivo, endogenous kinectin coprecipitates with RhoGG12V and colocalizes with endogenous RhoG. The kinectin domain responsible for binding to RhoG is located within a coiled-coil region of 300 aa, termed KiD1. Although KiD1 has been characterized previously in the yeast two-hybrid system as a putative target for RhoA, Rac1, and Cdc42 (17), our own data using yeast two-hybrid interaction showed that KiD1 binds RhoG twice as efficiently as Rac1 and at least 20-fold more than Cdc42 and RhoA. Furthermore, ectopic expression of KiD1 in REF-52 fibroblasts inhibits the morphogenic effects of RhoGG12V but not those of RhoAG14V, Rac1G12V, and Cdc42G12V. Thus, although RhoA, Rac1, and Cdc42 might interact with KiD1 in semipurified systems, only RhoG efficiently binds KiD1 in vivo. This is also supported by coprecipitation assays, which showed that endogenous kinectin does not bind RacG12V or Cdc42G12V (not shown). Thus, KiD1 appears to be a specific target for RhoG. In addition to KiD1, another region located at the C terminus of kinectin (KiD2) has the capacity to specifically bind GTP-bound RhoA. However, the level of interaction between KiD2 and RhoA is rather low, and although KiD2 expression specifically and efficiently inhibits the cellular effects of RhoAG14V, we were unable to coprecipitate GFP-RhoAG14V and kinectin from COS-7 cell extracts (not shown). Clearly, additional experiments are needed to confirm RhoA-kinectin interaction and to evaluate its biological significance.
What might be the cellular consequences of RhoG-kinectin interaction? As mentioned in the introduction, RhoG morphogenic activity requires the microtubule network (8, 14) and kinectin was first described as a receptor for kinesin (37), calling for a role of RhoG in kinesin-dependent vesicular transport. In vivo data presented here show that in REF-52 cells, RhoG, kinectin, and kinesin colocalize mainly in the ER, and in COS-7 cells, they also colocalize to other vesicles, including lysosomes. In addition, RhoG needs binding to kinectin and requires kinesin activity to establish its morphogenic activity, as demonstrated by antibody microinjection. Last, the frequency of kinesin-dependent fast movements of lysosomes appears directly linked to RhoG activity, supporting the notion that RhoG activates vesicle transport towards the cell periphery. All these observations suggest a model according to which RhoG would act on kinectin to facilitate kinesin-driven vesicular transport along microtubules. This is supported by a recent report which demonstrated in yeast and in vitro that the kinectin C terminus directly interacts with the kinesin cargo binding domain (33), leading to a significant increase in the microtubule-stimulated kinesin ATPase activity. Although the molecular mechanisms remain to be characterized, the binding of RhoG to the central coiled-coil region of kinectin might elicit structural changes in the whole complex, eventually enhancing kinesin motor activity. This would result in an activation of plus-end-directed microtubule-dependent vesicular transport. A similar effect has been reported for RhoD, which regulates early endosome dynamics as well as cell morphology (28).
Our data showing that RhoGT17N inhibits the cellular effects of kinectin overexpression seem to conflict with the notion that RhoG acts upstream of kinectin. Indeed, inhibition of the signaling emanating from a protein by a dominant-negative GTPase is generally interpreted as implicating the GTPase downstream of the signaling protein. As far as RhoG is concerned, two interpretations can be proposed. First, RhoG activity might facilitate the formation of kinectin/kinesin complex, thereby increasing microtubule-dependent transport. In this case, RhoG should be considered as acting upstream of kinectin. Alternatively, once GTP bound, RhoG might use kinectin only to travel across the cytoplasm and then exert its peripheral effects by interacting with other partners. Accordingly, RhoG should rather be considered as a downstream effector of kinectin. All of the experiments presented here that use F-actin staining as a readout can actually fit either scenario. However, the dramatic effects of RhoGG12V and RhoGT17N on lysosomal distribution can only be envisioned in light of the first scenario, since they show a direct implication of RhoG activity on microtubule-dependent transport. This scenario is also supported by the peripheral redistribution of kinectin upon TrioD1 expression and by the dramatic increase in actin stress fibers in cells coexpressing GFP-kinectin and RhoGT17N. On the other hand, since dominant-negative Cdc42 and, to a lesser extent, Rac1 simply revert the cellular effects of kinectin overexpression, these GTPases are more likely to act downstream of kinectin. Indeed, none of them coprecipitates with kinectin, and their activities appear independent of the presence of microtubules (14). The most probable scenario is therefore that RhoG acts on kinectin to facilitate kinesin- and microtubule-dependent transport, leading to the delivery of products at the cell periphery capable of activating pathways controlled by Rac1 and Cdc42. Whether the resulting translocation of RhoG elicits specific cellular events remains to be established.
The biological significance of the interaction between kinectin and RhoA remains unclear. On one hand, that kinectin is a RhoA target is not fully experimentally supported, as discussed above. On the other hand, a dramatic effect on stress fiber assembly is triggered by kinectin overexpression in the absence of RhoG activity or a microtubule network, a cellular effect probably ascribable to RhoA (19). This suggests that the potential RhoA-kinectin interaction may be not coincidental. For instance, should RhoG inhibition or microtubule disruption reduce the overall level of kinectin-kinesin interaction, this might make KiD2 available for RhoA, since KiD2 appears to be located close to the kinesin-binding domain (33). Whatever the underlying mechanisms, it is noteworthy that the apparent capacity of kinectin to mediate the RhoA pathway under particular physiological conditions is paralleled by the ability of Trio to activate RhoA through its second Dbl-like domain (6). Given the implication of Trio and microtubules in axon pathfinding (5, 23), as well as the antagonistic roles of RhoG and RhoA in neurite outgrowth (18, 20), one can speculate that kinectin might mediate two opposed pathways: one mediated by RhoG and microtubules (e.g., growth cone extension) and the other mediated by RhoA and local microtubule depolymerization (e.g., neurite retraction).
ACKNOWLEDGMENTS
We thank A. Debant for critical reading of the manuscript and C. Hüber, P. Roux, F. Comunale, and M. Puceat for fruitful discussion and constant support. We are indebted to J. Camonis, G. Gacon, and G. Zalcman for pLex vectors encoding Rho GTPases and to the Kazusa DNA Research Institute (Chiba, Japan) for kinectin cDNAs.
This work was supported by contracts mainly from the Ligue Nationale contre le Cancer (“Equipe labelisee”), by institutional grants from CNRS, and by the Association pour la Recherche contre le Cancer (contract no. 5284).
REFERENCES
- 1.Alberts A S, Bouquin N, Johnston L H, Treisman R. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein, Skn7. J Biol Chem. 1998;273:8616–8622. doi: 10.1074/jbc.273.15.8616. [DOI] [PubMed] [Google Scholar]
- 2.Aspenström P. Effectors for the rho GTPases. Curr Opin Cell Biol. 1999;11:95–102. doi: 10.1016/s0955-0674(99)80011-8. [DOI] [PubMed] [Google Scholar]
- 3.Aspenström P. The Rho GTPases have multiple effects on the actin cytoskeleton. Exp Cell Res. 1999;246:20–25. doi: 10.1006/excr.1998.4300. [DOI] [PubMed] [Google Scholar]
- 4.Awasaki T, Saito M, Sone M, Suzuki E, Sakai R, Ito K, Hama C. The Drosophila trio plays an essential role in patterning of axons by regulating their directional extension. Neuron. 2000;26:119–131. doi: 10.1016/s0896-6273(00)81143-5. [DOI] [PubMed] [Google Scholar]
- 5.Bateman J, Shu H, Van Vactor D. The guanine nucleotide exchange factor trio mediates axonal development in the Drosophila embryo. Neuron. 2000;26:93–106. doi: 10.1016/s0896-6273(00)81141-1. [DOI] [PubMed] [Google Scholar]
- 6.Bellanger J M, Lazaro J B, Diriong S, Fernandez A, Lamb N, Debant A. The two guanine nucleotide exchange factor domains of Trio link the Rac1 and the RhoA pathways in vivo. Oncogene. 1998;16:147–152. doi: 10.1038/sj.onc.1201532. [DOI] [PubMed] [Google Scholar]
- 7.Benichou S, Bomsel M, Bodeus M, Durand H, Doute M, Letourneur F, Camonis J, Benarous R. Physical interaction of the HIV-1 Nef protein with beta-COP, a component of non-clathrin-coated vesicles essential for membrane traffic. J Biol Chem. 1994;269:30073–30076. [PubMed] [Google Scholar]
- 8.Blangy A, Vignal E, Schmidt S, Debant A, Gauthier-Rouvière C, Fort P. TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of RhoG. J Cell Sci. 2000;113:729–739. doi: 10.1242/jcs.113.4.729. [DOI] [PubMed] [Google Scholar]
- 9.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 10.Brady S T, Pfister K K, Bloom G S. A monoclonal antibody against kinesin inhibits both anterograde and retrograde fast axonal transport in squid axoplasm. Proc Natl Acad Sci USA. 1990;87:1061–1065. doi: 10.1073/pnas.87.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chardin P, Camonis J H, Gale N W, van Aelst L, Schlessinger J, Wigler M H, Bar-Sagi D. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science. 1993;260:1338–1343. doi: 10.1126/science.8493579. [DOI] [PubMed] [Google Scholar]
- 12.de Toledo M, Colombo K, Nagase T, Ohara O, Fort P, Blangy A. The yeast exchange assay, a new complementary method to screen for Dbl-like protein specificity: identification of a novel RhoA exchange factor. FEBS Lett. 2000;480:287–292. doi: 10.1016/s0014-5793(00)01953-0. [DOI] [PubMed] [Google Scholar]
- 13.Gallo G, Letourneau P C. Axon guidance: GTPases help axons reach their targets. Curr Biol. 1998;8:80–82. doi: 10.1016/s0960-9822(98)70051-x. [DOI] [PubMed] [Google Scholar]
- 14.Gauthier-Rouvière C, Vignal E, Mériane M, Roux P, Montcourier P, Fort P. RhoG GTPase controls a pathway that independently activates Rac1 and Cdc42Hs. Mol Biol Cell. 1998;9:1379–1394. doi: 10.1091/mbc.9.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 16.Hollenbeck P J, Swanson J A. Radial extension of macrophage tubular lysosomes supported by kinesin. Nature. 1990;346:864–866. doi: 10.1038/346864a0. [DOI] [PubMed] [Google Scholar]
- 17.Hotta K, Tanaka K, Mino A, Kohno H, Takai Y. Interaction of the Rho family small G proteins with kinectin, an anchoring protein of kinesin motor. Biochem Biophys Res Commun. 1996;225:69–74. doi: 10.1006/bbrc.1996.1132. [DOI] [PubMed] [Google Scholar]
- 18.Katoh H, Yasui H, Yamaguchi Y, Aoki J, Fujita H, Mori K, Negishi M. Small GTPase RhoG is a key regulator for neurite outgrowth in PC12 cells. Mol Cell Biol. 2000;20:7378–7387. doi: 10.1128/mcb.20.19.7378-7387.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 20.Kozma R, Sarner S, Ahmed S, Lim L. Rho family GTPases and neuronal growth cone remodelling: relationship between increased complexity induced by Cdc42Hs, Rac1, and acetylcholine and collapse induced by RhoA and lysophosphatidic acid. Mol Cell Biol. 1997;17:1201–1211. doi: 10.1128/mcb.17.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar J, Yu H, Sheetz M P. Kinectin, an essential anchor for kinesin-driven vesicle motility. Science. 1995;2674:1834–1837. doi: 10.1126/science.7892610. [DOI] [PubMed] [Google Scholar]
- 22.Lelias J M, Adra C N, Wulf G M, Guillemot J C, Khagad M, Caput D, Lim B. cDNA cloning of a human mRNA preferentially expressed in hematopoietic cells and with homology to a GDP-dissociation inhibitor for the rho GTP-binding proteins. Proc Natl Acad Sci USA. 1993;90:1479–1483. doi: 10.1073/pnas.90.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Letourneau P C. The cytoskeleton in nerve growth cone motility and axonal pathfinding. Perspect Dev Neurobiol. 1996;4:111–123. [PubMed] [Google Scholar]
- 24.Leung T, Manser E, Tan L, Lim L. A novel serine/threonine kinase binding the Ras-related RhoA GTPase which translocates the kinase to peripheral membranes. J Biol Chem. 1995;270:29051–29054. doi: 10.1074/jbc.270.49.29051. [DOI] [PubMed] [Google Scholar]
- 25.Liu B P, Chrzanowska-Wodnicka M, Burridge K. Microtubule depolymerization induces stress fibers, focal adhesions, and DNA synthesis via the GTP-binding protein Rho. Cell Adhes Commun. 1998;5:249–255. doi: 10.3109/15419069809040295. [DOI] [PubMed] [Google Scholar]
- 26.Machleidt T, Geller P, Schwandner R, Scherer G, Kronke M. Caspase 7-induced cleavage of kinectin in apoptotic cells. FEBS Lett. 1998;436:51–54. doi: 10.1016/s0014-5793(98)01095-3. [DOI] [PubMed] [Google Scholar]
- 27.Michiels F, Habets G G, Stam J C, van der Kammen R A, Collard J G. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375:338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- 28.Murphy C, Saffrich R, Grummt M, Gournier H, Rybin V, Rubino M, Auvinen P, Lutcke A, Parton R G, Zerial M. Endosome dynamics regulated by a Rho protein. Nature. 1996;384:427–432. doi: 10.1038/384427a0. [DOI] [PubMed] [Google Scholar]
- 29.Nakata T, Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newsome T P, Schmidt S, Dietzl G, Keleman K, Asling B, Debant A, Dickson B J. Trio combines with dock to regulate Pak activity during photoreceptor axon pathfinding in Drosophila. Cell. 2000;101:283–294. doi: 10.1016/s0092-8674(00)80838-7. [DOI] [PubMed] [Google Scholar]
- 31.Nobes C D, Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J Cell Biol. 1999;144:1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobes C D, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 33.Ong L L, Lim A P, Er C P, Kuznetsov S, Yu H. Kinectin-kinesin binding domains and their effects on organelle motility. J Biol Chem. 2000;275:32854–32860. doi: 10.1074/jbc.M005650200. [DOI] [PubMed] [Google Scholar]
- 34.Roux P, Gauthier-Rouvière C, Doucet-Brutin S, Fort P. The small GTPases Cdc42Hs, rac1 and RhoG delineate raf-independent pathways that cooperate to transform NIH3T3 cells. Curr Biol. 1997;7:629–637. doi: 10.1016/s0960-9822(06)00289-2. [DOI] [PubMed] [Google Scholar]
- 35.Steven R, Kubiseski T J, Zheng H, Kulkarni S, Mancillas J, Ruiz-Morales A, Hogue C W, Pawson T, Culotti J. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–795. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- 36.Symons M, Derry J M, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- 37.Toyoshima I, Yu H, Steuer E R, Sheetz M P. Kinectin, a major kinesin-binding protein on ER. J Cell Biol. 1992;118:1121–1131. doi: 10.1083/jcb.118.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tuma M C, Zill A, Le Bot N, Vernos I, Gelfand V. Heterotrimeric kinesin II is the microtubule motor protein responsible for pigment dispersion in Xenopus melanophores. J Cell Biol. 1998;143:1547–1558. doi: 10.1083/jcb.143.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Aelst L, Joneson T, Bar-Sagi D. Identification of a novel Rac1 interacting protein involved in membrane ruffling. EMBO J. 1996;15:3778–3786. [PMC free article] [PubMed] [Google Scholar]
- 40.Vaux D, Tooze J, Fuller S. Identification by anti-idiotype antibodies of an intracellular membrane protein that recognizes a mammalian endoplasmic reticulum retention signal. Nature. 1990;345:495–502. doi: 10.1038/345495a0. [DOI] [PubMed] [Google Scholar]
- 41.Vignal E, De Toledo M, Comunale F, Ladopoulou A, Gauthier-Rouviere C, Blangy A, Fort P. Characterization of TCL, a new GTPase of the Rho family related to TC10 and Cdc42. J Biol Chem. 2000;275:36457–36464. doi: 10.1074/jbc.M003487200. [DOI] [PubMed] [Google Scholar]
- 42.Vincent S, Jeanteur P, Fort P. Growth-regulated expression of rhoG, a new member of the ras homolog gene family. Mol Cell Biol. 1992;12:3138–3148. doi: 10.1128/mcb.12.7.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]