ABSTRACT
Next-generation sequencing has been essential to the global response to the COVID-19 pandemic. As of January 2022, nearly 7 million severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) sequences are available to researchers in public databases. Sequence databases are an abundant resource from which to extract biologically relevant and clinically actionable information. As the pandemic has gone on, SARS-CoV-2 has rapidly evolved, involving complex genomic changes that challenge current approaches to classifying SARS-CoV-2 variants. Deep sequence learning could be a potentially powerful way to build complex sequence-to-phenotype models. Unfortunately, while they can be predictive, deep learning typically produces “black box” models that cannot directly provide biological and clinical insight. Researchers should therefore consider implementing emerging methods for visualizing and interpreting deep sequence models. Finally, researchers should address important data limitations, including (i) global sequencing disparities, (ii) insufficient sequence metadata, and (iii) screening artifacts due to poor sequence quality control.
KEYWORDS: COVID-19, SARS-CoV-2, viral genomics, bioinformatics, deep learning, explainable AI, genomics, machine learning
PERSPECTIVE
COVID-19 has been called the “first pandemic in the post-genomic era” (1). The first severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome was published on 12 January 2020, a week after the WHO first reported on the virus. Only 5 days later, the sequence was used to design the mRNA vaccines that have changed the course of the pandemic (2). Since then, next-generation sequencing technology has enabled an unprecedented view of genetic changes in the virus throughout both the duration of the pandemic and different parts of the world (1, 3). Global data sharing of sequence data has been equally critical, much to the credit of the GISAID EpiCoV database project (4). GISAID’s primary mission has been to share flu genomes, in part to help design the annual flu vaccine (its full name being the Global Initiative on Sharing All Influenza Data) (5). Now, at the beginning of 2022, the GISAID EpiCoV database has accumulated nearly 7 million SARS-CoV-2 genome sequences, and at present, around 800,000 sequences are being added each month. So much data has been generated and made available that it has spurred the development of computational tools for high-frequency sequence variant tracking (6) and even daily updates (7, 8).
Despite the surge in research efforts devoted to COVID-19 (9, 10), laboratory study of the virus remains a more specialized and time-consuming effort than sequencing. Clinical and epidemiological data are often superficial, measuring only a few variables, other than data sets specific to particular facilities or narrow populations. We need to fully capitalize on the abundant data that we do have to (i) anticipate how changes in the virus might affect health before we have time to gather empirical data and (ii) better design and interpret experiments to maximize our use of limited resources. So, how can we translate genome sequence data to as much biological understanding and actionable clinical insight as possible?
SARS-COV-2 IS RAPIDLY EVOLVING
SARS-CoV-2 has spent the first 2 years of the pandemic rapidly evolving in ways that have had a big impact on virulence, transmission, and ability to evade our immune responses (11). SARS-CoV-2 is an RNA virus, so its genome is prone to mutate—albeit at a rate mitigated by its large genome size and the proofreading function of its exoribonuclease (12). The most frequent mutations observed in coronaviruses are generally substitutions, although insertions and deletions are observed as well (13). In some cases, insertions from other viral genomes may occur, and, in fact, it appears as though the SARS-CoV-2 genome includes an insertion from human RNA (14). In other human coronaviruses, the estimated mutation rate is around 3 × 10−4 substitutions per site per year (15, 16).
The amount of mutation observed during the COVID-19 pandemic has been even more substantial than expected (17). An early estimate of SARS-CoV-2 mutation was 6 × 10−4 substitutions per site per year (18). But the disease has spread widely around the world since then, and novel variants transmit more quickly—increasing the opportunities for the virus to mutate (19, 20). The SARS-CoV-2 spike protein will continue to change in the future. Studies on another human coronavirus, HCoV-OC43, suggest that genetic drift plays a role in coronavirus adaptive evolution (21). One study estimates that as of July 2021, SARS-CoV-2 had only “explored” 31% of the potential space for spike gene variation, based on comparisons with related sarbecoviruses (22).
SEQUENCE ANALYSIS HAS STRUGGLED TO KEEP UP
The first widely used tool for tracking SARS-CoV-2 genomic variation was the Nextstrain project, https://nextstrain.org. Nextstrain, originally developed as a general tool for viruses, was adapted to offer clade definitions for SARS-CoV-2 based on phylogenetic analysis (23). Phylogenetic tree reconstruction has been effective in inferring viral origins and trace transmission changes but not as useful in classifying genomes because the virus can accumulate and drop mutations in parallel across clades and subclades (24). The Pango nomenclature (https://cov-lineages.org/), developed specifically for SARS-CoV-2, has largely supplanted Nextstrain clade definitions (25). New sequences are assigned to Pango classifications, called “lineages,” using the Random Forests classification algorithm. A new Pango lineage is defined when a sufficient number of viral sequences emerges with a phylogenetic dissimilarity from existing sequences above a set threshold (26). Particularly significant Pango lineages have been identified by the World Health Organization (WHO) as variants of concern (VOC), which are given Greek letter designations (27), such as Alpha (Pango lineage B.1.1.7), Beta (B.1.351), Delta (B.1.167.2), and, recently, Omicron (B.1.1.529).
While Pango lineages appear clear and well-defined, the reality is that the genome is much more fluid. If we want to understand how genome affects viral function, we cannot rely on traditional taxonomic categorization. As mutations recur, revert, and proliferate, taxonomy hits its limits of utility (11). As an initial matter, changes to SARS-CoV-2 properties often implicate combinations of multiple mutations that emerge simultaneously—and then sometimes revert in whole or in part as the virus continues to evolve (28, 29). For example, one frequent spike protein amino acid substitution, N501Y, has appeared and reverted contemporaneously in multiple clades and lineages, with no evidence of recombination (30). Simultaneous mutations can also have unpredictable, nonlinear effects, i.e., they can be synergistic, antagonistic, or fully independent (31). This complicates classical and Bayesian logistic regression methods for predicting fitness or protein function from mutations, as they rely on assuming the independence between mutations of individual amino acids or bases (32).
SARS-CoV-2 evolution is also highly nonlinear. Widespread lineages, such as Delta, have spawned complex sublineages with distinct immune evasion and virulence properties, which often genetically share more in common with distantly related lineages than their most recent ancestor (33, 34). The increasingly complex evolutionary history of the virus stymies other proposed methods for genetically subtyping viral variants as well (35–37). Further complicating the picture, some immunocompromised individuals can have chronic infections lasting 6 months to a year (38). During long-term infection, a spike protein can emerge with multiple variations, which phylogenetic analysis identifies as “long branch” divergence from the phylogenetic tree (39). Some long-term patients may even be treated with convalescent plasma or antibodies, which may select for immune evasive mutations (40). The Omicron variant has such a long branch divergence, indicating that it may have emerged in an immunocompromised host or after incubating in a nonhuman host such as mice (41, 42).
CAN DEEP SEQUENCE LEARNING HELP?
How can we predict the virulence, fitness, antibody evasion, and other key properties of novel SARS-CoV-2 variants from complex, nonlinear changes in genetic sequence? Machine learning can tackle complex pattern recognition problems by training a model that can classify the organisms or genes by phylogeny or phenotype based on features of their genetic sequences. For example, we can extract k-mer (short subsequence) frequencies or other combinations of bases/amino acids and use them as features to train classifiers using naive Bayes classifier (NBC), support vector machines (SVM), decision tree-based methods, and neural networks (43–49). Machine learning with k-mer features has been used for SARS-CoV-2 to identify genetic fingerprints of specific infections (50), classify variants (51, 52), and train a model to predict the pathogenicity of unknown viruses (53). Another approach is to build profile hidden Markov models (HMMs), which can identify taxonomic lineages and variants of viruses. HMMs have been used to align SARS-CoV-2 sequences and compare its spike protein to that of other coronaviruses (22, 54, 55).
Deep learning has emerged as an even more powerful and flexible tool to find patterns in large and complicated data sets (56–59). Deep learning models use multiple layers of neural networks to automatically extract and transform features during training (56–58). We can borrow deep learning methods developed for natural language processing (NLP) to find patterns in sequence data, where bases and amino acids that make up genome and protein sequences are analogous to semantic relationships between the words that make up sentences (60–63). For example, one group of researchers has used concepts from semantic processing, e.g., the frequency of correlated words, to identify potential mutagenic sites in viruses including SARS-CoV-2 (64). An emerging approach to deep sequencing learning is to transform protein sequences to embeddings that reflect their semantic structure, using the BERT (bidirectional encoder representations from transformers) neural network architecture, which Google developed to handle natural language search (65–68). An example of this approach is k-means clustering of “ProtBERT” SARS-CoV-2 protein embeddings generated by pretraining a BERT model on millions of UniProt sequences, which can be used to identify mutational hot spots within the genome that may give rise to future variants (69).
A key goal for modeling is to predict the health risk of emerging variants before empirical data are available. To this end, our group has developed a deep learning model to predict patient outcomes for emerging sequence variants that takes into account patient demographics (70). Others are working to integrate sequence learning with computational protein structure models. For example, one project combines models of cell receptor binding and immune epitope alteration with transformer-based deep learning models to predict the fitness advantage of mutations (71). Deep learning has also been used to identify the relationship between protein sequence and function using data from deep mutational scanning, an experimental technique for massively parallel functional analysis of protein sequence site mutations (72, 73). Using this approach, another project predicts the risk for emerging variants by using a neural network to predict infectivity and vaccine breakthrough in combination with protein structure and binding prediction to model antibody resistance (74).
LOOKING INSIDE THE DEEP LEARNING BLACK BOX
Deep learning methods excel at identifying complex features within data that allow classification. But they have a major weakness. Deep learning relies on neural networks, and it is very hard to determine why a neural network makes a particular classification or prediction. Interpretable, or explainable, machine learning can fill this important gap (75, 76). Interpretable machine learning is particularly important in bioinformatics, since explaining a model’s predictions is critical to justify making high-stakes clinical or research decisions based on machine learning predictions (77, 78). Accordingly, developers of deep learning approaches to SARS-CoV-2 should consider providing some functionality to interpret or explain predictions.
Analytical tools for interpretability in deep learning include examining neural network structure through relevance propagation, activation difference propagation, sensitivity analysis, and saliency map methods (79–81). Integrated gradients have been used to analyze RNA splicing models (82). An increasingly popular approach is the “attention” mechanism originally developed for NLP (83, 84). Attention can highlight important features in text processed by deep learning models (85–87). The amount of “attention” at a position in a sequence correlates with the weight put on that position in a trained model, where high attention at a position implies potential significance. Architectures combining convolutional neural networks (CNNs) with attention have been used to identify sequence motifs for functional genomics, e.g., transcription factor binding site detection (88, 89). Another group generated predictive models of adverse drug reactions based on chemical structures by combining attention with a CNN for each chemical property and structural feature in the model (90). Our group has shown that attention in combination with a recurrent neural network-based sequence model can provide insight into taxonomic and phenotypic classification of microbial 16s rRNA sequences (91), as well as gene ontology classifications of protein sequences (92).
Recently, transformer-based architectures have emerged, like the aforementioned BERT (93). Transformers are built on multiple attention modules (“heads”), which could be used for interpretability (94). For example, one recent paper demonstrated how different attention heads attended to different aspects of a learning task to identify nucleotide motifs for promoter sequences (95). However, attention cannot be inherently drawn out of transformers. Further processing steps are generally required to connect attention to specific linguistic features (96). Our group recently applied a self-attention layer after a transformer as a way to more readily extract and visualize attention across the sequence and applied it to SARS-CoV-2 (70). An important caveat is that, based on comparing attention to empirical evidence, attention does not necessarily imply explanation—at least in the sense of explaining precisely why a prediction took place (97). Attention can only highlight features that the attention layer of the deep learning model weighted most heavily during training, so it may only weakly indicate the complete set of important features for a classification problem.
SEQUENCING DISPARITIES AND DATA CHALLENGES
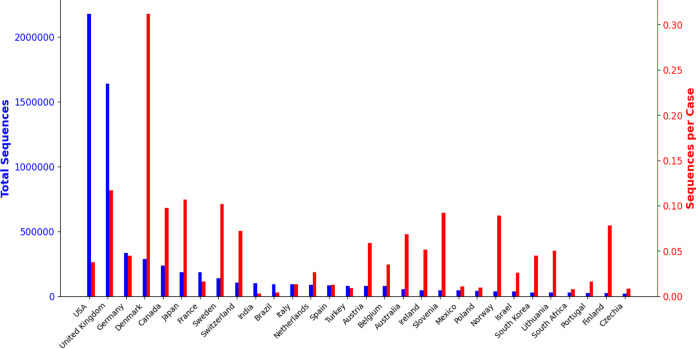
Finally, we highlight three important data limitations that researchers should address. First, as Fig. 1 shows, there are serious global inequities in sequencing data, with the overwhelming majority of sequences coming from Europe and North America. GISAID has encouraged data sharing from developing countries by trading restrictions on republishing sequence information for access to that information (98). But global sequencing resources are disparately available (99). Even within Europe and the United States, racial and regional disparities in sequencing found in other surveys (100) hamper SARS-CoV-2 sequencing as well. Second, the task of interpreting sequencing data is complicated by insufficient sample metadata, making it difficult to understand how SARS-CoV-2 sequences affect patient outcomes, for example. In GISAID, most sequences only have information about a patient’s age or gender (if available) and the location where the sample was collected. As of 7 January 2022, a little over 270,000 sequences (4%) of the nearly 6.9 million have any metadata for patient outcomes, and many metadata entries are unintelligible. Sequencing projects should be encouraged to collect and curate as much information as possible about the sample and meet minimum information standards for sequence metadata (101). Third, sequencing errors can lead to spurious results. Quality control is critical to make sure that low-frequency sequence variants are real (102). Sequences can pick up contaminants from other variants in the amplification process, leading to what appear to be recombinant variants but which are in fact simply artifacts (103).
FIG 1.
Total number of sequences submitted to GISAID (4) (left axis and bars, blue) and ratio of the number of submitted sequences to the total number of reported cases (104) as of 7 January 2022 for the 30 countries that have submitted the most sequences to GISAID. The overwhelming majority sequences in GISAID come from North America and Europe. Over half of all sequences are from the United States and United Kingdom alone. Sequencing rates show even greater disparities.
ACKNOWLEDGMENTS
G.L.R. received National Science Foundation (NSF) grants no. 1919691 and no. 2107108.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We gratefully acknowledge all data contributors, i.e., the authors and their originating laboratories responsible for obtaining the specimens and their submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative, from which Fig. 1 was generated.
Contributor Information
Gail L. Rosen, Email: glr26@drexel.edu.
Marta M. Gaglia, Tufts University
REFERENCES
- 1.van Dorp L, Houldcroft CJ, Richard D, Balloux F. 2021. COVID-19, the first pandemic in the post-genomic era. Curr Opin Virol 50:40–48. doi: 10.1016/j.coviro.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, Himansu S, Schäfer A, Ziwawo CT, DiPiazza AT, Dinnon KH, Elbashir SM, Shaw CA, Woods A, Fritch EJ, Martinez DR, Bock KW, Minai M, Nagata BM, Hutchinson GB, Wu K, Henry C, Bahl K, Garcia-Dominguez D, Ma L, Renzi I, Kong WP, Schmidt SD, Wang L, Zhang Y, Phung E, Chang LA, Loomis RJ, Altaras NE, Narayanan E, Metkar M, Presnyak V, Liu C, Louder MK, Shi W, Leung K, Yang ES, West A, Gully KL, Stevens LJ, Wang N, Wrapp D, Doria-Rose NA, Stewart-Jones G, Bennett H, et al. 2020. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiara M, D'Erchia AM, Gissi C, Manzari C, Parisi A, Resta N, Zambelli F, Picardi E, Pavesi G, Horner DS, Pesole G. 2021. Next generation sequencing of SARS-CoV-2 genomes: challenges, applications and opportunities. Brief Bioinform 22:616–630. doi: 10.1093/bib/bbaa297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khare S, Gurry C, Freitas L, Schultz MB, Bach G, Diallo A, Akite N, Ho J, Lee RT, Yeo W, Team GCC, Maurer-Stroh S. 2021. GISAID’s role in pandemic response. China CDC Wkly 3:1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shu Y, McCauley J. 2017. GISAID: global initiative on sharing all influenza data – from vision to reality. Eurosurveill 22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernasconi A, Mari L, Casagrandi R, Ceri S. 2021. Data-driven analysis of amino acid change dynamics timely reveals SARS-CoV-2 variant emergence. Sci Rep 11:21068. doi: 10.1038/s41598-021-00496-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McBroome J, Thornlow B, Hinrichs AS, Kramer A, De Maio N, Goldman N, Haussler D, Corbett-Detig R, Turakhia Y. 2021. A daily-updated database and tools for comprehensive SARS-CoV-2 mutation-annotated trees. Mol Biol Evol 12:5819–5824. doi: 10.1093/molbev/msab264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen C, Nadeau S, Yared M, Voinov P, Xie N, Roemer C, Stadler T. 2022. CoV-spectrum: analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics 38:1735–1737. doi: 10.1093/bioinformatics/btab856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rando HM, MacLean AL, Lee AJ, Lordan R, Ray S, Bansal V, Skelly AN, Sell E, Dziak JJ, Shinholster L, McGowan LD, Guebila MB, Wellhausen N, Knyazev S, Boca SM, Capone S, Qi Y, Park Y, Mai D, Sun Y, Boerckel JD, Brueffer C, Byrd JB, Kamil JP, Wang J, Velazquez R, Szeto GL, Barton JP, Goel RR, Mangul S, Lubiana T, Gitter A, Greene CS, COVID-19 Review Consortium . 2021. Pathogenesis, symptomatology, and transmission of SARS-CoV-2 through analysis of viral genomics and structure. mSystems 6:e00095-21. doi: 10.1128/msystems.00095-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rando HM, Wellhausen N, Ghosh S, Lee AJ, Dattoli AA, Hu F, Byrd JB, Rafizadeh DN, Lordan R, Qi Y, Sun Y, Brueffer C, Field JM, Guebila MB, Jadavji NM, Skelly AN, Ramsundar B, Wang J, Goel RR, Park Y, Boca SM, Gitter A, Greene CS, COVID-19 Review Consortium . 2021. Identification and development of therapeutics for COVID-19. mSystems 6:e00233-21. doi: 10.1128/mSystems.00233-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao K, Tzou PL, Nouhin J, Gupta RK, de Oliveira T, Kosakovsky PS, Fera D, Shafer RW. 2021. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet 22:1–17. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plante JA, Mitchell BM, Plante KS, Debbink K, Weaver SC, Menachery VD. 2021. The variant gambit: COVID-19’s next move. Cell Host Microbe 29:508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R. 2010. Viral mutation rates. J Virol 84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peacock TP, Bauer DLV, Barclay WS. 2021. Putative host origins of RNA insertions in SARS-CoV-2 genomes. https://virological.org/t/putative-host-origins-of-rna-insertions-in-sars-cov-2-genomes/761.
- 15.Mohammadi E, Shafiee F, Shahzamani K, Ranjbar MM, Alibakhshi A, Ahangarzadeh S, Beikmohammadi L, Shariati L, Hooshmandi S, Ataei B, Javanmard SH. 2021. Novel and emerging mutations of SARS-CoV-2: biomedical implications. Biomed Pharmacother 139:111599. doi: 10.1016/j.biopha.2021.111599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pyrc K, Dijkman R, Deng L, Jebbink MF, Ross HA, Berkhout B, van der Hoek L. 2006. Mosaic structure of human coronavirus NL63, one thousand years of evolution. J Mol Biol 364:964–973. doi: 10.1016/j.jmb.2006.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vankadari N. 2020. Overwhelming mutations or SNPs of SARS-CoV-2: a point of caution. Gene 752:144792. doi: 10.1016/j.gene.2020.144792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Dorp L, Acman M, Richard D, Shaw LP, Ford CE, Ormond L, Owen CJ, Pang J, Tan CCS, Boshier FAT, Ortiz AT, Balloux F. 2020. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet Evol 83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Egeren D, Novokhodko A, Stoddard M, Tran U, Zetter B, Rogers M, Pentelute BL, Carlson JM, Hixon M, Joseph-McCarthy D, Chakravarty A. 2021. Risk of rapid evolutionary escape from biomedical interventions targeting SARS-CoV-2 spike protein. PLoS One 16:e0250780. doi: 10.1371/journal.pone.0250780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, Russell TW, Tully DC, Washburne AD, Wenseleers T, Gimma A, Waites W, Wong KLM, van Zandvoort K, Silverman JD, Diaz-Ordaz K, Keogh R, Eggo RM, Funk S, Jit M, Atkins KE, Edmunds WJ, CMMID COVID-19 Working Group, COVID-19 Genomics UK (COG-UK) Consortium . 2021. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372:eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren L, Zhang Y, Li J, Xiao Y, Zhang J, Wang Y, Chen L, Paranhos-Baccalà G, Wang J. 2015. Genetic drift of human coronavirus OC43 spike gene during adaptive evolution. Sci Rep 5:11451. doi: 10.1038/srep11451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cotten M, Robertson DL, Phan MVT. 2021. Unique protein features of SARS-CoV-2 relative to other sarbecoviruses. Virus Evol 7:veab067. doi: 10.1093/ve/veab067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin DP, Weaver S, Tegally H, San JE, Shank SD, Wilkinson E, Lucaci AG, Giandhari J, Naidoo S, Pillay Y, Singh L, Lessells RJ, Gupta RK, Wertheim JO, Nekturenko A, Murrell B, Harkins GW, Lemey P, MacLean OA, Robertson DL, de Oliveira T, Kosakovsky Pond SL, COVID-19 Genomics UK (COG-UK) . 2021. The emergence and ongoing convergent evolution of the SARS-CoV-2 N501Y lineages. Cell 184:5189–5200. doi: 10.1016/j.cell.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, Du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Toole Á, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, Colquhoun R, Ruis C, Abu-Dahab K, Taylor B, Yeats C, Du Plessis L, Maloney D, Medd N, Attwood SW, Aanensen DM, Holmes EC, Pybus OG, Rambaut A. 2021. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol 7:veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parums DV. 2021. Editorial: revised World Health Organization (WHO) terminology for variants of concern and variants of interest of SARS-CoV-2. Med Sci Monit 27:e933622. doi: 10.12659/MSM.933622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Le Guen J, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Péré H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Lorière E, Rey FA, Schwartz O. 2021. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 29.Tasakis RN, Samaras G, Jamison A, Lee M, Paulus A, Whitehouse G, Verkoczy L, Papavasiliou FN, Diaz M. 2021. SARS-CoV-2 variant evolution in the United States: high accumulation of viral mutations over time likely through serial founder events and mutational bursts. PLoS One 16:e0255169. doi: 10.1371/journal.pone.0255169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colson P, Levasseur A, Delerce J, Pinault L, Dudouet P, Devaux C, Fournier P-E, La Scola B, Lagier J-C, Raoult D. 2021. Spreading of a new SARS-CoV-2 N501Y spike variant in a new lineage. Clin Microbiol Infect 27:1352.e1–1352.e5. doi: 10.1016/j.cmi.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucas C, Vogels CBF, Yildirim I, Rothman JE, Lu P, Monteiro V, Gehlhausen JR, Campbell M, Silva J, Tabachnikova A, Peña-Hernandez MA, Muenker MC, Breban MI, Fauver JR, Mohanty S, Huang J, Pearson C, Muyombwe A, Downing R, Razeq J, Petrone M, Ott I, Watkins A, Kalinich C, Alpert T, Brito A, Earnest R, Murphy S, Neal C, Laszlo E, Altajar A, Tikhonova I, Castaldi C, Mane S, Bilguvar K, Kerantzas N, Ferguson D, Schulz W, Landry M, Peaper D, Shaw AC, Ko AI, Omer SB, Grubaugh ND, Iwasaki A, Yale SARS-CoV-2 Genomic Surveillance Initiative . 2021. Impact of circulating SARS-CoV-2 variants on mRNA vaccine-induced immunity. Nature 600:523–529. doi: 10.1038/s41586-021-04085-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obermeyer F, Schaffner SF, Jankowiak M, Barkas N, Pyle JD, Park DJ, MacInnis BL, Luban J, Sabeti PC, Lemieux JE. 2021. Analysis of 2.1 million SARS-CoV-2 genomes identifies mutations associated with transmissibility. medRxiv doi: 10.1101/2021.09.07.21263228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baj A, Novazzi F, Drago Ferrante F, Genoni A, Tettamanzi E, Catanoso G, Dalla Gasperina D, Dentali F, Focosi D, Maggi F. 2021. Spike protein evolution in the SARS-CoV-2 Delta variant of concern: a case series from Northern Lombardy. Emerg Microbes Infect 10:2010–2015. doi: 10.1080/22221751.2021.1994356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baj A, Novazzi F, Pasciuta R, Genoni A, Ferrante FD, Valli M, Partenope M, Tripiciano R, Ciserchia A, Catanoso G, Focosi D, Maggi F. 2021. Breakthrough infections of E484K-harboring SARS-CoV-2 Delta variant, Lombardy, Italy. Emerg Infect Dis 27:3180–3182. doi: 10.3201/eid2712.211792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Z, Sokhansanj BA, Malhotra C, Zheng K, Rosen GL. 2020. Genetic grouping of SARS-CoV-2 coronavirus sequences using informative subtype markers for pandemic spread visualization. PLoS Comput Biol 16:e1008269. doi: 10.1371/journal.pcbi.1008269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin L, Ding X, Li Y, Chen Q, Meng J, Jiang T. 2021. Co-mutation modules capture the evolution and transmission patterns of SARS-CoV-2. Briefings Bioinformatics 22:bbab222. doi: 10.1093/bib/bbab222. [DOI] [PubMed] [Google Scholar]
- 37.Pardo-Seco J, Gómez-Carballa A, Bello X, Martinón-Torres F, Salas A. 2021. Pitfalls of barcodes in the study of worldwide SARS-CoV-2 variation and phylodynamics. Zool Res 42:87–93. doi: 10.24272/j.issn.2095-8137.2020.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nussenblatt V, Roder AE, Das S, de Wit E, Youn JH, Banakis S, Mushegian A, Mederos C, Wang W, Chung M, Pérez-Pérez L, Palmore T, Brudno JN, Kochenderfer JN, Ghedin E. 2021. Year-long COVID-19 infection reveals within-host evolution of SARS-CoV-2 in a patient with B cell depletion. medRxiv doi: 10.1101/2021.10.02.21264267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voloch CM, da Silva Francisco R, de Almeida LGP, Brustolini OJ, Cardoso CC, Gerber AL, Guimarães APDC, Leitão IDC, Mariani D, Ota VA, Lima CX, Teixeira MM, Dias ACF, Galliez RM, Faffe DS, Pôrto LC, Aguiar RS, Castiñeira TMPP, Ferreira OC, Tanuri A, de Vasconcelos ATR. 2021. Intra-host evolution during SARS-CoV-2 prolonged infection. Virus Evol 7:veab078. doi: 10.1093/ve/veab078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L, Zody MC, Di Germanio C, Martinelli R, Mediavilla JR, Cunningham MH, Composto K, Chow KF, Kordalewska M, Corvelo A, Oschwald DM, Fennessey S, Zetkulic M, Dar S, Kramer Y, Mathema B, Germer S, Stone M, Simmons G, Busch MP, Maniatis T, Perlin DS, Kreiswirth BN. 2021. Emergence of multiple SARS-CoV-2 antibody escape variants in an immunocompromised host undergoing convalescent plasma treatment. mSphere 6:e00480-21. doi: 10.1128/mSphere.00480-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei C, Shan KJ, Wang W, Zhang S, Huan Q, Qian W. 2021. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J Genet Genom 48:1111–1121. doi: 10.1016/j.jgg.2021.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kupperschmidt K. 2021. Where did ‘weird’ Omicron come from? Science. 374:1179. doi: 10.1126/science.acx9738. [DOI] [PubMed] [Google Scholar]
- 43.Rosen G, Garbarine E, Caseiro D, Polikar R, Sokhansanj B. 2008. Metagenome fragment classification using N-mer frequency profiles. Adv Bioinformatics 2008:205969. doi: 10.1155/2008/205969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Libbrecht MW, Noble WS. 2015. Machine learning applications in genetics and genomics. Nat Rev Genet 16:321–332. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vervier K, Mahé P, Tournoud M, Veyrieras JB, Vert JP. 2016. Large-scale machine learning for metagenomics sequence classification. Bioinformatics 32:1023–1032. doi: 10.1093/bioinformatics/btv683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.I W, G P, Sc T. 2016. Correct machine learning on protein sequences: a peer-reviewing perspective. Briefings Bioinformatics 17:831–840. doi: 10.1093/bib/bbv082. [DOI] [PubMed] [Google Scholar]
- 47.Bzhalava Z, Tampuu A, Bała P, Vicente R, Dillner J. 2018. Machine learning for detection of viral sequences in human metagenomic datasets. BMC Bioinformatics 19:336. doi: 10.1186/s12859-018-2340-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alam MNU, Chowdhury UF. 2020. Short k-mer abundance profiles yield robust machine learning features and accurate classifiers for RNA viruses. PLoS One 15:e0239381. doi: 10.1371/journal.pone.0239381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Z, Yang W, Zhai Y, Liang Y, Zhao Y. 2022. Identify DNA-binding proteins through the extreme gradient boosting algorithm. Front Genet 12:821996. doi: 10.3389/fgene.2021.821996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lau BT, Pavlichin D, Hooker AC, Almeda A, Shin G, Chen J, Sahoo MK, Huang CH, Pinsky BA, Lee HJ, Ji HP. 2021. Profiling SARS-CoV-2 mutation fingerprints that range from the viral pangenome to individual infection quasispecies. Genome Med 13:62. doi: 10.1186/s13073-021-00882-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Basu S, Campbell RH. 2021. Classifying COVID-19 variants based on genetic sequences using deep learning models. bioRxiv doi: 10.1101/2021.06.29.450335. [DOI]
- 52.Ali S, Sahoo B, Ullah N, Zelikovskiy A, Patterson M, Khan I. 2021. A k-mer based approach for SARS-CoV-2 variant identification, p 153–164. In Wei Y, Li M, Skums P, Cai Z (ed), Bioinformatics research and applications lecture notes in computer science, Springer International Publishing, New York, NY. [Google Scholar]
- 53.Saha I, Ghosh N, Maity D, Seal A, Plewczynski D. 2021. COVID-DeepPredictor: recurrent neural network to predict SARS-CoV-2 and other pathogenic viruses. Front Genet 12:569120. doi: 10.3389/fgene.2021.569120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lemoine F, Blassel L, Voznica J, Gascuel O. 2021. COVID-align: accurate online alignment of hCoV-19 genomes using a profile HMM. Bioinformatics 37:1761–1762. doi: 10.1093/bioinformatics/btaa871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oliveira LS, Gruber A. 2021. Rational design of profile hidden Markov models for viral classification and discovery. In Helder IN (ed), Bioinformatics. Exon Publications, Brisbane, Australia. [PubMed] [Google Scholar]
- 56.LeCun Y, Bengio Y, Hinton G. 2015. Deep learning. Nature 521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 57.Zou J, Huss M, Abid A, Mohammadi P, Torkamani A, Telenti A. 2019. A primer on deep learning in genomics. Nat Genet 51:12–18. doi: 10.1038/s41588-018-0295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt B, Hildebrandt A. 2021. Deep learning in next-generation sequencing. Drug Discov Today 26:173–180. doi: 10.1016/j.drudis.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Villegas-Morcillo A, Makrodimitris S, van Ham RCHJ, Gomez AM, Sanchez V, Reinders MJT. 2021. Unsupervised protein embeddings outperform hand-crafted sequence and structure features at predicting molecular function. Bioinformatics 37:162–170. doi: 10.1093/bioinformatics/btaa701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eraslan G, Avsec Ž, Gagneur J, Theis FJ. 2019. Deep learning: new computational modelling techniques for genomics. Nat Rev Genet 20:389–403. doi: 10.1038/s41576-019-0122-6. [DOI] [PubMed] [Google Scholar]
- 61.Iuchi H, Matsutani T, Yamada K, Iwano N, Sumi S, Hosoda S, Zhao S, Fukunaga T, Hamada M. 2021. Representation learning applications in biological sequence analysis. Comput Struct Biotechnol J 19:3198–3208. doi: 10.1016/j.csbj.2021.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Littmann M, Heinzinger M, Dallago C, Weissenow K, Rost B. 2021. Protein embeddings and deep learning predict binding residues for various ligand classes. Sci Rep 11:23916. doi: 10.1038/s41598-021-03431-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le NQK, Ho QT. 2021. Deep transformers and convolutional neural network in identifying DNA N6-methyladenine sites in cross-species genomes. Methods. S1046-2023(21)00274-7. doi: 10.1016/j.ymeth.2021.12.004. [DOI] [PubMed] [Google Scholar]
- 64.Hie B, Zhong ED, Berger B, Bryson B. 2021. Learning the language of viral evolution and escape. Science 371:284–288. doi: 10.1126/science.abd7331. [DOI] [PubMed] [Google Scholar]
- 65.Devlin J, Chang MW, Lee K, Toutanova K. 2019. BERT: pre-training of deep bidirectional transformers for language understanding, p 4171–4186. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers). Association for Computational Linguistics, Minneapolis, Minnesota. [Google Scholar]
- 66.Rives A, Meier J, Sercu T, Goyal S, Lin Z, Liu J, Guo D, Ott M, Zitnick CL, Ma J, Fergus R. 2021. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc Natl Acad Sci USA 118:e2016239118. doi: 10.1073/pnas.2016239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elnaggar A, Heinzinger M, Dallago C, Rehawi G, Wang Y, Jones L, Gibbs T, Feher T, Angerer C, Steinegger M, Bhowmik D, Rost B. 2021. ProtTrans: towards cracking the language of lifes code through self-supervised deep learning and high performance computing. IEEE Trans Pattern Anal Mach Intell doi: 10.1109/TPAMI.2021.3095381. [DOI] [PubMed] [Google Scholar]
- 68.Brandes N, Ofer D, Peleg Y, Rappoport N, Linial M. 2022. ProteinBERT: a universal deep-learning model of protein sequence and function. Bioinform doi: 10.1093/bioinformatics/btac020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mullick B, Magar R, Jhunjhunwala A, Barati FA. 2021. Understanding mutation hotspots for the SARS-CoV-2 spike protein using shannon entropy and K-means clustering. Comput Biol Med 138:104915. doi: 10.1016/j.compbiomed.2021.104915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sokhansanj BA, Zhao Z, Rosen GL. 2021. Interpretable and predictive deep modeling of the SARS-CoV-2 spike protein sequence. medRxiv doi: 10.1101/2021.12.26.21268414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beguir K, Skwark MJ, Fu Y, Pierrot T, Carranza NL, Laterre A, Kadri I, Lui BG, Sänger B, Liu Y, Poran A, Muik A, Sahin U. 2021. Early computational detection of potential high risk SARS-CoV-2 variants. bioRxiv doi: 10.1101/2021.12.24.474095. [DOI] [PMC free article] [PubMed]
- 72.Fowler DM, Fields S. 2014. Deep mutational scanning: a new style of protein science. Nat Methods 11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gelman S, Fahlberg SA, Heinzelman P, Romero PA, Gitter A. 2021. Neural networks to learn protein sequence–function relationships from deep mutational scanning data. Proc National Acad Sci USA 118:e2104878118. doi: 10.1073/pnas.2104878118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen J, Wang R, Gilby NB, Wei GW. 2021. Omicron (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. arXiv 2112.01318v1. [DOI] [PMC free article] [PubMed]
- 75.Linardatos P, Papastefanopoulos V, Kotsiantis SB. 2021. Explainable AI: a review of machine learning interpretability methods. Entropy (Basel) 23:18. doi: 10.3390/e23010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murdoch WJ, Singh C, Kumbier K, Abbasi-Asl R, Yu B. 2019. Definitions, methods, and applications in interpretable machine learning. Proc Natl Acad Sci USA 116:22071–22080. doi: 10.1073/pnas.1900654116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Auslander N, Gussow AB, Koonin EV. 2021. Incorporating machine learning into established bioinformatics frameworks. Int J Mol Sci 22:2903. doi: 10.3390/ijms22062903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watson DS. 2021. Interpretable machine learning for genomics. Hum Genet doi: 10.1007/s00439-021-02387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Montavon G, Samek W, Müller KR. 2018. Methods for interpreting and understanding deep neural networks. Digit Signal Process 73:1–15. doi: 10.1016/j.dsp.2017.10.011. [DOI] [Google Scholar]
- 80.Shrikumar A, Greenside P, Kundaje A. 2017. Learning important features through propagating activation differences, p 3145–3153. In International Conference on Machine Learning (PMLR). [Google Scholar]
- 81.Simonyan K, Vedaldi A, Zisserman A. 2014. Deep inside convolutional networks: visualising image classification models and saliency maps. arXiv 1312.6034.
- 82.Jha A, Aicher JK, Gazzara MR, Singh D, Barash Y. 2020. Enhanced integrated gradients: improving interpretability of deep learning models using splicing codes as a case study. Genome Biol 21:149. doi: 10.1186/s13059-020-02055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bahdanau D, Cho K, Bengio Y. 2014. Neural machine translation by jointly learning to align and translate. arXiv 1409.0473.
- 84.Xu K, Ba J, Kiros R, Cho K, Courville AC, Salakhutdinov R, Zemel RS, Bengio Y. 2015. Show, attend and tell: neural image caption generation with visual attention. arXiv 1502.03044.
- 85.Rush AM, Chopra S, Weston J. 2015. A neural attention model for abstractive sentence summarization. arXiv 1509.00685.
- 86.Yang Z, Yang D, Dyer C, He X, Smola A, Hovy E. 2016. Hierarchical attention networks for document classification, p 1480–1489. In Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. [Google Scholar]
- 87.Zhou P, Shi W, Tian J, Qi Z, Li B, Hao H, Xu B. 2016. Attention-based bidirectional long short-term memory networks for relation classification, p 207–212. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics. [Google Scholar]
- 88.Deming L, Targ S, Sauder N, Almeida D, Ye CJ. 2016. Genetic architect: discovering genomic structure with learned neural architectures. arXiv 1605.07156.
- 89.Lanchantin J, Singh R, Lin Z, Qi Y. 2016. Deep motif: visualizing genomic sequence classifications. arXiv 1605.01133.
- 90.Dey S, Luo H, Fokoue A, Hu J, Zhang P. 2018. Predicting adverse drug reactions through interpretable deep learning framework. BMC Bioinformatics 19:476. doi: 10.1186/s12859-018-2544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao Z, Woloszynek S, Agbavor F, Mell JC, Sokhansanj BA, Rosen GL. 2021. Learning, visualizing and exploring 16S rRNA structure using an attention-based deep neural network. PLoS Comput Biol 17:e1009345. doi: 10.1371/journal.pcbi.1009345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao Z, Rosen G. 2020. Visualizing and annotating protein sequences using a deep neural network, p 506–510. In 2020 54th Asilomar Conference on Signals, Systems, and Computers. [Google Scholar]
- 93.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. 2017. Attention is all you need, p 6000–6010. In Proceedings of the 31st International Conference on Neural Information Processing Systems NIPS’17, Curran Associates Inc., Red Hook, NY, USA. [Google Scholar]
- 94.Vig J. 2019. BertViz: a tool for visualizing multihead self-attention in the BERT model. In ICLR Workshop: Debugging Machine Learning Models. [Google Scholar]
- 95.Clauwaert J, Menschaert G, Waegeman W. 2021. Explainability in transformer models for functional genomics. Briefings Bioinform 22:bbab060. doi: 10.1093/bib/bbab060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kobayashi G, Kuribayashi T, Yokoi S, Inui K. 2020. Attention is not only a weight: analyzing transformers with vector norms. arXiv 2004.10102 [cs].
- 97.Jain S, Wallace BC. 2019. Attention is not explanation. arXiv 1902.10186.
- 98.Van Noorden R. 2021. Scientists call for fully open sharing of coronavirus genome data. Nature 590:195–196. doi: 10.1038/d41586-021-00305-7. [DOI] [PubMed] [Google Scholar]
- 99.Brito AF, Semenova E, Dudas G, Hassler GW, Kalinich CC, Kraemer MUG, Hill SC, Sabino EC, Pybus OG, Dye C, Bhatt S, Flaxamn S, Suchard MA, Grubaugh ND, Baele G, Faria NR, Danish Covid-19 Genome Consortium . 2021. Global disparities in SARS-CoV-2 genomic surveillance. medRxiv doi: 10.1101/2021.08.21.21262393. [DOI] [Google Scholar]
- 100.Spratt DE, Chan T, Waldron L, Speers C, Feng FY, Ogunwobi OO, Osborne JR. 2016. Racial/ethnic disparities in genomic sequencing. JAMA Oncol 2:1070–1074. doi: 10.1001/jamaoncol.2016.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schriml LM, Chuvochina M, Davies N, Eloe-Fadrosh EA, Finn RD, Hugenholtz P, Hunter CI, Hurwitz BL, Kyrpides NC, Meyer F, Mizrachi IK, Sansone SA, Sutton G, Tighe S, Walls R. 2020. COVID-19 pandemic reveals the peril of ignoring metadata standards. Sci Data 7:188. doi: 10.1038/s41597-020-0524-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jacot D, Pillonel T, Greub G, Bertelli C. 2021. Assessment of SARS-CoV-2 genome sequencing: quality criteria and low-frequency variants. J Clin Microbiol 59:e00944-21. doi: 10.1128/JCM.00944-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lagerborg KA, Normandin E, Bauer MR, Adams G, Figueroa K, Loreth C, Gladden-Young A, Shaw BM, Pearlman LR, Berenzy D, Dewey HB, Kales S, Dobbins ST, Shenoy ES, Hooper D, Pierce VM, Zachary KC, Park DJ, MacInnis BL, Tewhey R, Lemieux JE, Sabeti PC, Reilly SK, Siddle KJ. 2022. Synthetic DNA spike-ins (SDSIs) enable sample tracking and detection of inter-sample contamination in SARS-CoV-2 sequencing workflows. Nat Microbiol 7:108–119. doi: 10.1038/s41564-021-01019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hasell J, Mathieu E, Beltekian D, Macdonald B, Giattino C, Ortiz-Ospina E, Roser M, Ritchie H. 2020. A cross-country database of COVID-19 testing. Sci Data 7:345. doi: 10.1038/s41597-020-00688-8. [DOI] [PMC free article] [PubMed] [Google Scholar]