Abstract
Unlike classical voltage-gated sodium (NaV) channels, NaX has been characterized as a voltage-insensitive, tetrodotoxin-resistant, sodium (Na+)-activated channel involved in regulating Na+ homeostasis. However, NaX remains refractory to functional characterization in traditional heterologous systems. Here, to gain insight into its atypical physiology, we determine structures of the human NaX channel in complex with the auxiliary β3-subunit. NaX reveals structural alterations within the selectivity filter, voltage sensor-like domains, and pore module. We do not identify an extracellular Na+-sensor or any evidence for a Na+-based activation mechanism in NaX. Instead, the S6-gate remains closed, membrane lipids fill the central cavity, and the domain III-IV linker restricts S6-dilation. We use protein engineering to identify three pore-wetting mutations targeting the hydrophobic S6-gate that unlock a robust voltage-insensitive leak conductance. This constitutively active NaX-QTT channel construct is non-selective among monovalent cations, inhibited by extracellular calcium, and sensitive to classical NaV channel blockers, including tetrodotoxin. Our findings highlight a functional diversity across the NaV channel scaffold, reshape our understanding of NaX physiology, and provide a template to demystify recalcitrant ion channels.
Subject terms: Electron microscopy, Physiology, Sodium channels
NaX is an atypical member of the voltage-gated sodium channel family that may contribute to Na+ homeostasis. Here, the authors describe the structural and functional attributes of the human NaX channel to reveal new insights into its physiology.
Introduction
Mammalian voltage-gated sodium (NaV) channel subtypes, NaV1.1–NaV1.9, perform essential roles in electrical signaling by initiating and propagating action potentials1. A tenth NaV channel-like gene (SCN7A) was cloned three decades ago, but atypical sequence features and the inability to record voltage-activated sodium (Na+)-currents raised speculation that this channel might have distinct physiological roles2–5. Evidence suggests that NaX may contribute to Na+ homeostasis5–10 and NaX shows restricted expression in a brain area that specializes in monitoring blood composition11–13. NaX-knockout mice ingest salt despite dehydration and are resistant to hypertension caused by elevated Na+ levels11,14. Autoimmunity against NaX is associated with chronic hypernatremia with impaired thirst perception and salt appetite in humans9. In the periphery, NaX regulates Na+ homeostasis upstream of the epithelial Na+ channel (ENaC) and may be a target to treat atopic dermatitis and hypertrophic scarring10,15.
Clues to NaX function derive mainly from in vivo studies and cells obtained from knockout mice5,16. Murine NaX is reportedly activated by extracellular Na+ concentrations above 150 mM5,17,18, where Na+-sensitivity can be shifted into the physiological range (135–145 mM) through a mechanism involving the endothelin receptor17. Na+ influx through NaX may activate the Na+/K+-ATPase19,20 leading to proton release and the activation of acid-sensing channels on nearby neurons14,16. However, because all attempts to record currents from traditional heterologous systems have failed3,4, the basic biophysical properties of NaX and the molecular determinants involved in Na+-sensing remain unknown.
NaX is the most sequence divergent member of the mammalian NaV channel family1, which has made structure-function correlations difficult to discern. Because murine NaX contains a reduced number of S4-gating charges and is voltage-insensitive2–4, the structure and function of the voltage-sensor-like domains (VSLDs) are unclear. Instead of the characteristic ion-selectivity filter DEKA-locus found in NaV channels, NaX contains a unique DENA-locus2–4 and is thought to be tetrodotoxin (TTX)-resistant5,12. NaX is reported to generate non-inactivating currents and has a divergent intracellular DIII–DIV linker sequence5,18. In this work, we determined the structure of the human NaX channel, which has provided new insights into its unusual biophysical properties and physiology.
Results
Evaluation of human NaX function
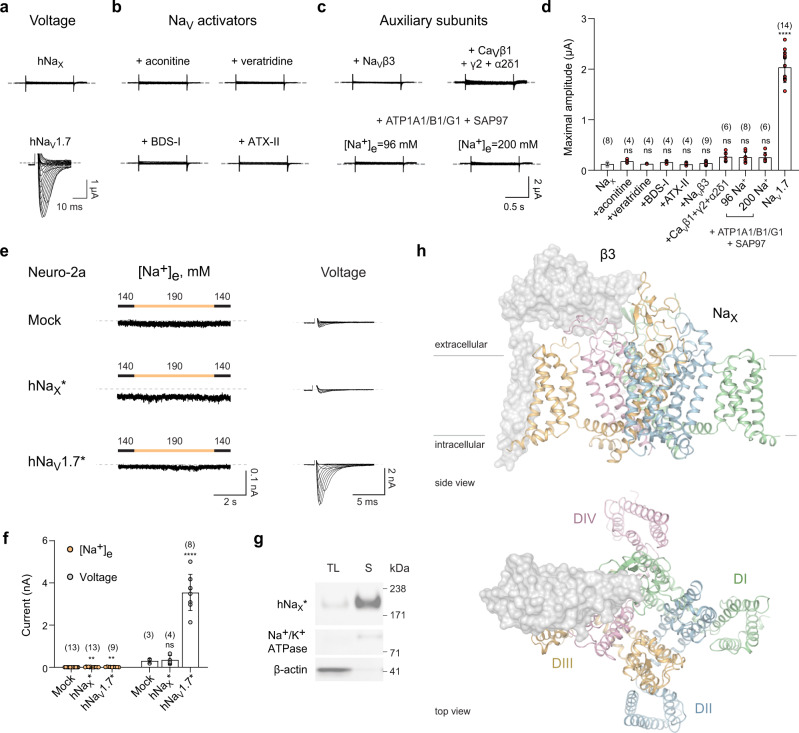
Reliable ionic currents were not detected for human NaX expressed in HEK293T cells or Xenopus laevis oocytes in response to changes in membrane voltage (Fig. 1a–d, Supplementary Fig. 1a–d). Application of NaV channel pharmacological activators like aconitine or veratridine did not invoke any clear NaX-mediated currents in oocytes (Fig. 1b, d, Supplementary Fig. 1g, h). Co-expression with NaV or voltage-gated calcium (CaV) channel auxiliary-subunits also failed to produce dependable currents (Fig. 1c, d, Supplementary Fig. 1i, j). Thus, as reported for the rodent proteins3,4, human NaX does not operate as a conventional voltage-gated channel.
Fig. 1. Characterization of and overall structure of human NaX.
a Representative currents from Xenopus laevis oocytes expressing human NaX or NaV1.7. NaX: steps between +80 to −100 mV, in 20 mV increments from a HP of 0 mV; NaV1.7: depolarizing steps between −80 and +65 mV, in 5 mV increments, from a HP of −100 mV. b Representative currents from oocytes expressing NaX in response to extracellular application of indicated compounds (BDS-I Blood depressing substance I, ATX-II Neurotoxin 2). Voltage protocols as above. See Methods for concentrations of compounds tested. c Representative currents from oocytes expressing NaX and co-expression of NaV or CaV channel auxiliary-subunits, Na+/K+-ATPase subunits and synapse-associated protein 97 (SAP97), and in the presence of the indicated extracellular Na+ concentration. Voltage protocols as above. d Data summary of independent experiments performed as in parts a–c. Data are shown as mean ± SD; ns not significant; ****p < 0.0001; one-way ANOVA with Dunnett’s test (against NaX). Exact p-values and statistical parameters are provided in Source Data. Numbers of biological replicates (n) are indicated. e Representative currents from murine Neuro-2a cells expressing human NaX or NaV1.7 in response to changes of the extracellular Na+ concentration (HP = −60 mV), as indicated, or voltage: depolarizing steps between −60 to +60 mV, in 10 mV increments from a HP of −100 mV. f Data summary of independent experiments performed as in parts e. Data are shown as mean ± SD; ns not significant; *p < 0.05; **p < 0.01; ****p < 0.0001; one-way ANOVA with Dunnett’s test (against mock-transfected cells). Exact p-values and statistical parameters are provided in Source Data. Numbers of biological replicates (n) are indicated. g Western blots of total lysate and surface fraction of proteins extracted from Neuro-2a cells probed for the indicated proteins. Data represent three independent biological replicates. h Side and extracellular view of the β3-NaX channel complex. Approximate membrane boundaries are indicated. DI, DII, DIII, and DIV are colored in green, blue, orange, and pink, respectively, with the β3-subunit in gray surface representation.
In a neuroblastoma (Neuro-2a) cell line expressing murine NaX, increases in the extracellular Na+ concentration ([Na+]e) reportedly produce a non-inactivating Na+ current17,18. When human NaX was expressed in Neuro-2a cells, small and flickery inward currents were observed upon changes in [Na+]e from 140 to 190 mM, but cells expressing the human NaV1.7 channel showed similar behavior (Fig. 1e, g, Supplementary Fig. 1e). A non-inactivating inward current was only observed when seal resistances were low (<1 GΩ) in both human NaX- and mock-transfected Neuro-2a cells (Supplementary Fig. 1f). Because sequence differences (Supplementary Fig. 2) or cellular context21 may account for these seemingly conflicting functional outcomes between murine and human NaX, we next considered if structure determination might provide insight into the function or regulation of NaX.
Structure of the β3-NaX channel complex in lipid nanodiscs
To facilitate cryogenic electron microscopy (cryo-EM) analysis, human NaX was co-expressed with the canonical auxiliary β3-subunit, purified in mild detergent (glyco-diosgenin), and reconstituted into phospholipid nanodiscs (Supplementary Fig. 3a–c). Prior to sample vitrification, the Na+ concentration was raised above the threshold reported to activate NaX (to 200 mM), and the resulting cryo-EM reconstruction of the β3-NaX complex extended to 3.2 Å resolution (Supplementary Fig. 4a–g, Supplementary Table 1).
The NaX channel resembles a four-leaf clover with VSLDs arranged in a domain-swapped organization (Fig. 1h). The β3-subunit is wedged between extracellular loops of the pore module and its single transmembrane helix is positioned against VSLD3 (Fig. 1h, Supplementary Fig. 5a). The intracellular DIII–DIV linker is bound alongside the pore (see below), and the carboxyl-terminal domain is not well-resolved (Supplementary Fig. 5a), establishing that the overall architecture of NaX is highly similar to available human NaV channel structures determined in detergent (Supplementary Fig. 5b–e)22–26. However, NaX does reveal structural deviations in regions implicated in NaV channel gating (Supplementary Fig. 5d, e), suggesting that these changes may be a consequence of sequence and functional divergence (Fig. 1a, b, Supplementary Fig. 2).
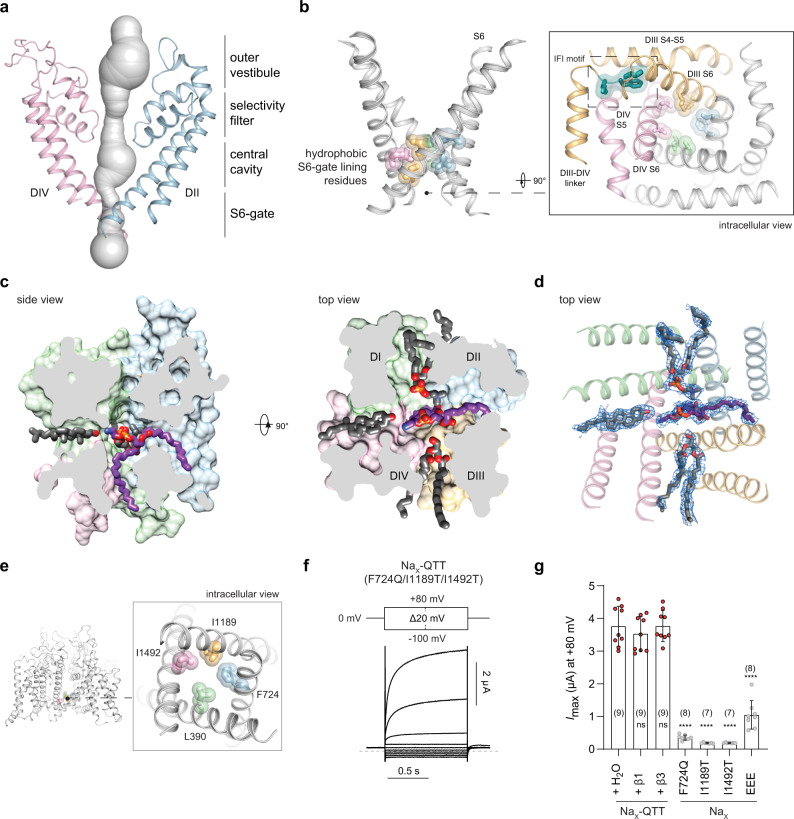
The NaX pore module is in a nonconductive state
The NaX pore module contains an outer vestibule, an ion-selectivity filter, a central cavity, and an S6-gate (Fig. 2a). Despite the high Na+ concentration in our sample, no distinguishing extracellular Na+ binding site was observed (Supplementary Fig. 5f), and residues lining the narrow S6-gate form a hydrophobic barrier to the passage of hydrated ions, defining a nonconductive state (Fig. 2b, Supplementary Fig. 5c)22–26. These structural observations align with our physiological results (Fig. 1a–f) and overturn expectations that human NaX alone gives rise to a Na+-activated, non-inactivating conductance17,18.
Fig. 2. NaX pore module structure reveals a nonconductive state.
a NaX pore volume shown as gray surface with DII and DIV in cartoon rendering (DI and DIII omitted for clarity). b View of the S6-helices with side-chains lining the activation gate shown. Orthogonal view provides a wider perspective with DIII and DIV colored orange and pink, and the IFI-motif (green) from the DIII–DIV linker shown in stick and semi-transparent surface representation. c Orthogonal views sliced through the pore module highlighting lateral fenestrations and bound lipids. The phosphatidylethanolamine that crosses the S6-gate is in purple stick representation. d Similar to middle panel c, but with cryo-EM map shown in blue mesh representation. e Location of S6-gate hydrophobic side-chains targeted by pore-wetting mutations. f Representative currents from Xenopus laevis oocytes expressing the NaX-QTT construct with voltage protocol indicated. g Data summary of independent experiments with indicated constructs (see Supplementary Fig. 8). Data are shown as mean ± SD; ns not significant; ****p < 0.0001; one-way ANOVA with Dunnett’s test (against NaX-QTT + H2O). Exact p-values and statistical parameters are provided in Source Data. Numbers of biological replicates (n) are indicated.
The NaX central cavity is similar to those in NaV channels, although the DIV-S6 phenylalanine targeted by pore-blocking drugs26,27 is replaced by DIV-Trp1484 (Supplementary Fig. 5g). NaX reveals four lateral fenestrations that penetrate the pore, suggesting that lipids or NaV and CaV channel antagonists might target the central cavity through membrane access pathways (Fig. 2c, d).
The NaX pore is infiltrated by membrane lipids
Four lipids penetrate the lateral pore fenestrations in NaX to enter the central cavity (Fig. 2c, d). These features can unambiguously be assigned as three phospholipids and one cholesterol, where one phospholipid even straddles and seals the intracellular S6-gate (Fig. 2c, d, Supplementary Fig. 5h). To evaluate if nanodisc reconstitution was responsible for lipid visualization, we prepared a β3-NaX sample in the detergent glyco-diosgenin without exogenous phospholipid supplementation and determined its structure in 200 mM Na+ to 2.9 Å resolution (Supplementary Fig. 6a–g, Supplementary Table 1). An essentially identical pore structure penetrated by four well-defined lipids was observed (Supplementary Fig. 6h). The presence of well-resolved, co-purifying lipids in NaX may reflect the hydrophobicity of the central cavity and S6-gate or the intrinsic stability of the nonconductive state.
The intracellular DIII–DIV linker restricts S6-gate dilation
The NaX DIII–DIV linker is bound to a pore module receptor site reminiscent of the fast-inactivated state in NaV channels (Fig. 2b, Supplementary Fig. 7a, b)22–26. Mutating the NaX IFI-motif to a NaV channel non-inactivating QQQ-motif28 was insufficient to generate currents in oocytes (Supplementary Fig. 7c, d), but disrupting the VSLD3 and DIII S4–S5 linker regions required to form the fast-inactivation receptor site in NaV channels produced robust leak currents in NaV1.7-NaX chimeric channels (Supplementary Fig. 7e–g)29. The DIII–DIV linker therefore appears to restrict S6-dilation in NaX and may stabilize the nonconductive state.
Pore-wetting mutations can activate human NaX
We hypothesized that targeted introduction of polar residues around the hydrophobic S6-gate might promote pore hydration, destabilization of bound lipids, displacement of the IFI-motif, and transition into a conductive state (Fig. 2a–d). A triple-mutant NaX channel construct containing polar substitutions at three S6-gate lining positions, NaX-QTT (F724Q-I1189T-I1492T), produced robust ionic currents when expressed alone in Xenopus oocytes (Fig. 2e, f; Supplementary Fig. 8a, b). Single-point mutant NaX channels failed to produce robust currents, indicating that a pore-wetting threshold likely exists (Fig. 2g, Supplementary Fig. 8c)30,31. The triple-mutant construct NaX-EEE (L390E-I1189E-I1492E) also displayed function, but with lower current amplitudes and higher current variability relative to NaX-QTT (Fig. 2g, Supplementary Fig. 8d).
NaX-QTT currents are outward-rectifying without signs of inactivation (Fig. 2f, Supplementary Fig. 8b). Current amplitudes or kinetics were not impacted when NaX-QTT was co-expressed with the β1- or β3-subunits (Fig. 2g, Supplementary Fig. 8e). NaX-QTT also behaved as a non-inactivating channel in patch-clamp experiments in HEK293T cells (see below), where minor differences in the kinetics of the outward currents are likely due to species differences between the expression systems, and the lack of control over intracellular ionic composition in Xenopus laevis oocytes. Because NaX-QTT contains only three mutations targeted to the S6-gate (Fig. 2e), we characterized this construct as a proxy to evaluate ion selectivity and pharmacology of the human NaX channel.
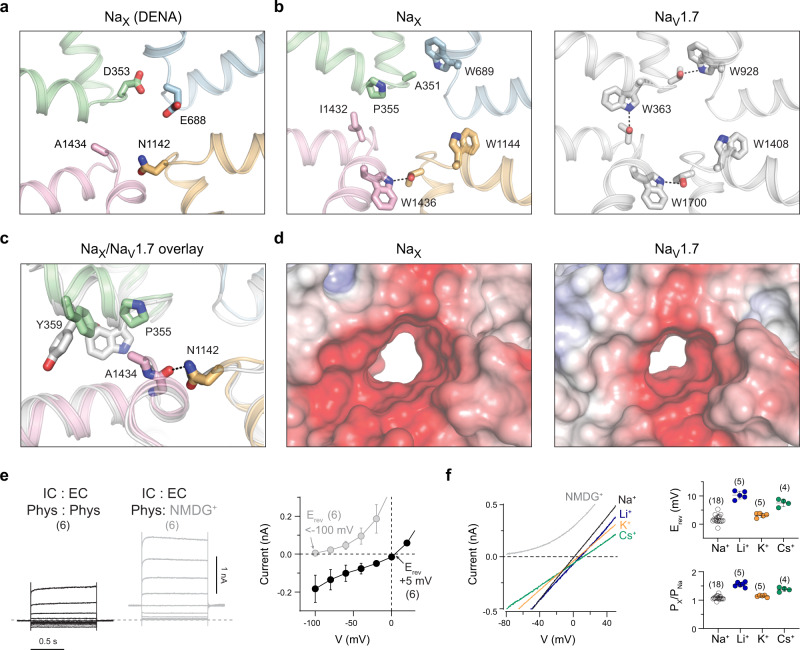
The NaX selectivity filter
The unique DENA-locus of NaX is supported by a local architecture that differs radically from the scaffold which supports the DEKA-locus in NaV channels (Fig. 3a–d). Consequently, the ion-selectivity filter of NaX is wider and more electronegative (Fig. 3d). At the DENA-locus, Asp353 (DI) bonds to the backbone of DII-Trp689, Glu688 (DII) points into the ion permeation pathway, and Asn1142 (DIII) bonds to the DIV-Phe1433 carbonyl to impose a ~1.5 Å radial shift onto Ala1434 (DIV) relative to the DIV alanine in NaV channels (Fig. 3c). Displacement of Ala1434 is accommodated by the distinct DI-Pro355 in NaX, which permits DI-Tyr359 to fill the volume vacated by the traditional DI tryptophan (Fig. 3b, c). This remodeling only slightly repositions side-chains within the NaX outer vestibule, suggesting that ion or toxin binding above the selectivity filter may not be substantially impacted (Supplementary Fig. 9a, b).
Fig. 3. Structure and characterization of the NaX selectivity filter.
a NaX DENA-motif side-chains shown as sticks. b Residues in NaX that form a conserved interaction network around the selectivity filter in NaV channels (NaV1.7, PDB 6J8J). c Superimposed view of the DI-DIV interface comparing NaX and NaV1.7 (gray, PDB 6J8J). d View as in b, NaX and NaV1.7 selectivity filter electrostatic surface rending. Note, central cavity and activation gate excluded for clarity. e Representative currents from HEK293T cells expressing human NaX-QTT with a C-terminal GFP-Flag tag in a physiological (left) or NMDG+-only extracellular solution (middle). See methods for composition of intracellular (IC) and extracellular (EC) solutions. Steps between +80 to −100 mV, in 20 mV increments, from a HP of 0 mV. Right, shows I–V curve data summary from n = 6 cells over two independent experiments. Data are shown as mean ± SD. Numbers of biological replicates (n) are indicated. f Representative currents from HEK293T cells expressing human NaX-QTT with indicated monovalent cations in the extracellular solution. Voltage ramp from −80 to +80 mV was applied. Right, summary of reversal potentials and permeability ratios measured from three independent experiments. Data are shown as mean ± SD. Numbers of biological replicates (n) are indicated.
NaX-QTT is non-selective among monovalent cations
The DIII-Asn1142 dramatically alters the structure and chemical profile of the NaX selectivity filter relative to the DIII-lysine that supports Na+-selectivity in NaV channels (Fig. 3d)32. Transferring a DEKA-locus into NaX produced no measurable currents (Supplementary Fig. 9c), establishing that features beyond the selectivity filter contribute to channel activation.
When cations in the extracellular solution were replaced with N-methyl-D-glucamine, the reversal potential for NaX-QTT in HEK293T cells shifted from +5 mV to below −100 mV (Fig. 3e), indicating that this construct behaves as a cation channel, and that large cations do not permeate. Replacement of Cl− ions with the large anion methansulfonate did not change the current profile (Supplementary Fig. 9d), establishing that NaX-QTT behaves as a cation-selective channel. Using a ramp protocol with a simple Na+ intracellular solution (150 mM), the reversal potential was close to 0 mV under symmetrical NaCl conditions, and only slight changes in the reversal potentials were measured upon exposure to different extracellular monovalent cations (150 mM; Fig. 3f). These results demonstrate that the NaX-QTT selectivity filter does not effectively discriminate between Na+, K+, Cs+, or Li+ ions.
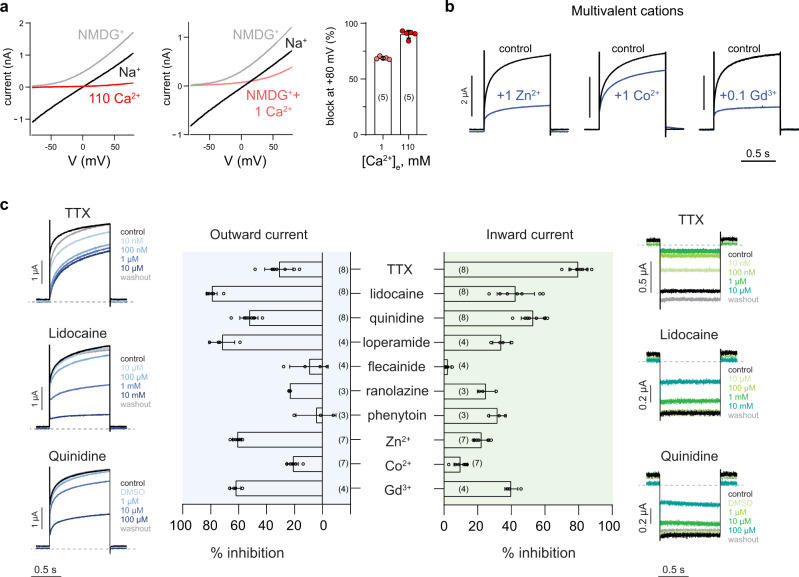
NaX-QTT is inhibited by Ca2+ and sensitive to NaV channel blockers
Ca2+ does not permeate through NaX-QTT, but 1 mM extracellular Ca2+ inhibited ~70% of the Na+ current (Fig. 4a). Zinc (Zn2+), cobalt (Co2+), and gadolinium (Gd3+) cations also inhibited NaX-QTT currents (Fig. 4b). Contrasting long-standing reports5,12, tetrodotoxin showed inhibitory effects on NaX-QTT, especially on the inward current (Fig. 4c). The high potency of TTX-block suggests that it occurs at the selectivity filter, where most side-chains implicated in TTX-block in NaV channels are structurally conserved (Supplementary Fig. 2, 9b)24. Lidocaine, quinidine, and loperamide blocked NaX-QTT currents in the µM to mM concentration range; however, strong inhibitory effects were not observed by all classical NaV channel pore-blocking drugs tested (Fig. 4c).
Fig. 4. Pharmacology of the human NaX-QTT channel.
a Representative I–V curves from HEK293T cells expressing human NaX-QTT with indicated extracellular monovalent cations with or without indicated amounts of CaCl2 in the extracellular solution. Voltage ramp from −80 to +80 mV was applied. Right, percentage of block of outward Na+ by indicated concentrations of Ca2+ at 80 mV. Data are shown as mean ± SD of n = 5 cells over three independent experiments. b Representative currents from Xenopus laevis oocytes expressing human NaX-QTT in standard extracellular solution with or without indicated divalent and trivalent cations (unit in mM), when stepping from 0 to +80 mV. c Representative currents from oocytes expressing human NaX-QTT in standard extracellular solution with or without indicated blockers added, when stepping from 0 to +80 mV (left) or from 0 to −100 mV (right). Middle, summary of currents measured from two independent experiments. Data are shown as mean ± SD. Numbers of biological replicates (n) are indicated. See Methods for concentrations of compounds tested.
NaX reveals atypical voltage-sensor-like domains
Conditions under which human NaX channel might produce voltage-activated currents have not been identified (Fig. 1a–f), although the VSLDs share a high structural similarity with the voltage-sensor domains (VSDs) of NaV and CaV channels (Supplementary Fig. 10a). Numerous features are implicated in noncanonical contributions to NaX channel function, for instance, VSLD4 is found in an unusual deactivated-like state (Supplementary Fig. 10a, b).
Discussion
The human NaX channel does not function as a conventional voltage-activated channel, but we find that the transfer of entire homologous domains or subdomains between NaX and NaV1.7 can produce channels with unusual gating characteristics (Supplementary Fig. 7d, e, 10c). These results suggest that the coupling interfaces or energetics of NaX gating have diverged substantially from NaV channels, underscoring the functional diversity that has evolved across this channel scaffold33. This does not exclude the possibility that NaX may function as a voltage-modulated channel, since binding of one or more putative auxiliary proteins may impart voltage sensitivity as described for the NALCN and TMEM16A channels33,34.
Murine NaX has been reported to produce non-inactivating inward currents in Neuro-2a cells upon increasing the extracellular Na+ concentration >150 mM5,17,18. We only observed qualitatively similar recordings with low seal resistance, and failed to reconstitute Na+-activated currents in cells expressing human NaX (Fig. 1d, Supplementary Fig. 1f). Moreover, our β3-NaX cryo-EM samples were prepared with NaCl concentrations above the reported threshold required for Na+-dependent gating5,16,17, but no clear Na+-sensing locus or Na+-based activation mechanism was observed (Fig. 2a, b, Supplementary Fig. 5f). The only map feature that we might assign as a Na+ ion is bound near the NaX selectivity filter at an extracellular site that is conserved in NaV channels (Supplementary Fig. 9a)23. We conclude that NaX lacks an intrinsic Na+-sensor, and speculate that the described Na+-sensing mechanism must be contributed by an exogenous factor or pathway, such as the Na+/K+ ATPase, the endothelin receptor, a yet to be identified system, auxiliary protein(s), or post-translational modification17,19,20.
Inspired by the concept of hydrophobic gating in channels30,31, mutation of only three residues around the S6-gate region was sufficient to produce robust ionic currents from NaX-QTT in Xenopus oocytes and HEK293T cells (Figs. 2e, f, 3e, f, 4a–c). Conceptually, the introduced polar side-chains help to hydrate the hydrophobic S6-gate, dislodge bound membrane lipids, and displace the IFI-motif to promote pore dilation and ion conduction (Fig. 2a–g, Supplementary Fig. 7a)28. NaX-QTT currents were non-inactivating and not modulated by voltage, and while NaX has been proposed to function as a leak channel in vivo13, future studies are required to define the biophysical properties of wild-type channels.
In the absence of extracellular Ca2+ ions, NaX-QTT gives rise to a near-Ohmic leak current that does not discriminate between the tested monovalent cations (Fig. 3f). This finding challenges prior suggestions that NaX is Na+-selective18 and confirms a key role for the DIII-lysine in NaV channels32. Extracellular Ca2+ block of NaX-QTT produces outward-rectifying currents with only a small inward leak component at negative membrane potentials (Fig. 4a). NaX may therefore function as a Na+-leak channel sensitive to modulation by extracellular Ca2+. Ca2+ block of NaX-QTT is ~10-fold more potent than a block on NaV channels (Fig. 4a)35, but similar to the Ca2+ sensitivity of the distantly related NALCN Na+-leak channel33, perhaps suggesting a shared mechanism to avoid excessive depolarizing Na+ influx into cells.
NaX-QTT has pharmacological sensitivities expected for a NaV-like channel, including a block by TTX, lidocaine, quinidine, and loperamide (Fig. 4b, c)1. However, flecainide, ranolazine, or phenytoin did not produce strong inhibition (Fig. 4c), possibly reflecting unique central cavity-lining residues in NaX (Supplementary Fig. 5g) or the absence of an inactivated state.
Lipids occlude the ion conduction pathway in our NaX channel structures (Fig. 2c, d, Supplementary Fig. 6h). The phospholipid which plugs the S6-gate is reminiscent of cholesterol-like molecules bound in NaV channels22,23, where the occupancy of these hydrophobic ligands appears to correlate with the up-conformation of a conserved DIV-S6-tyrosine side-chain implicated in drug block (Supplementary Fig. 6h, 10d)24,26. Drug binding can enforce a down-conformation of this S6-tyrosine, concurrent with the displacement of the lipid-like density at the S6-gate (Supplementary Fig. 10d)26. Thus, our NaX structures advance an emerging paradigm in channels whereby membrane lipids can directly modulate pore structure, drug binding, and ion conductance.
Under the conditions that we have examined, the human NaX channel does not function independently as a Na+-activated channel, nor as a voltage-activated channel. Using structure-guided engineering, we discovered a NaX-QTT channel construct that displays a non-selective cationic conductance and potent inhibition by extracellular Ca2+ and TTX. Overall, although the steps, factors, or pathways required for endogenous channel activation still need to be precisely defined, our study reveals that the NaX channel may operate as a Ca2+-modulated Na+ leak channel and provides new tools to further demystify this once recalcitrant channel.
Methods
Protein expression and purification
Full-length human NaX containing tandem 2xStrepII and 2xFLAG tags at its N-terminus and untagged full-length human β3 were each cloned into a pRK vector behind a CMV promotor. Constructs were co-transfected with PEI into Expi293 cells and cultured for 48 h in SMM 293T-I medium under 5% CO2 at 37 °C. Cells were harvested by centrifugation at 800 × g for 10 min and resuspended and lysed in Buffer A (25 mM ADA pH 6.0, 200 mM NaCl, 1 mM PMSF, 1 μg/mL benzonase, and 1× Roche protease inhibitor cocktail) by dounce homogenization. To solubilize the cell membranes, glyco-diosgenin (GDN) and cholesterol hemisuccinate (CHS) were added to the sample to final concentrations of 2% and 0.3%, respectively, and the sample was gently stirred at 4 °C for two hours. Insoluble material was then collected by ultracentrifugation at 125,000 × g for 1 h at 4 °C. Supernatants were applied to anti-FLAG M2 agarose resin that had been pre-equilibrated in Buffer B (25 mM ADA pH 6.0, 200 mM NaCl, 0.01% GDN) and bound in batch at 4 °C for 1 h. The sample was then applied to a gravity column and the collected resin was washed with 10 CV Buffer B followed by 10 CV supplemented with 5 mM ATP and 10 mM MgCl2. Protein was eluted with 5 CV Buffer B supplemented with 300 μg/mL FLAG peptide. Eluates were applied directly to Strep-Tactin XT Superflow high capacity resin that had been pre-equilibrated in Buffer B and bound in batch for three hours, then washed with 10 CV Buffer B prior to sample elution in 5 CV supplemented with 50 mM biotin. For nanodisc incorporation, the sample was concentrated to 15 μM using an Amicon Ultra centrifugal filter device (100 kDa MWCO). For structural analysis in detergent (GDN), the eluate was instead concentrated to 100 μL and applied to a Superose 6 3.2/300 column that had been pre-equilibrated in Buffer B.
Mass spectrometry analysis
Following size exclusion chromatography, 10 µg of the NaX-containing peak fraction (co-expression with the β3-subunit) was denatured with 8 M guanidine (1:1 v/v), reduced with 1 M dithiothreitol (DTT, Sigma-Aldrich, St Louis, MO) to a final concentration of 100 mM DTT and incubated at 95 °C for 10 min, then centrifuged. 10 µL of 1 M Tris, pH 8 was added to the solution, and water was used to dilute the guanidine to a 2 M final concentration. Deglycosylation was performed with 2 µL of PNGaseF (15000 U, New England Biolabs) followed by overnight incubation at 37 °C. Samples were further reduced with 10 mM DTT at 60 °C (15 min) followed by alkylation with 20 mM iodoacetamide at room temperature. Proteins were digested with 0.2 µg trypsin (Promega) or chymotrypsin in 50 mM ammonium bicarbonate, pH 8 at 37 °C overnight. Digestions were quenched with formic acid and the supernatants were subjected to desalting on C18 PhyTips (PhyNexus), lyophilized, reconstituted in 0.1% formic acid containing 2% acetonitrile and analyzed without further processing by reversed-phase nano-LC/MS/MS on a Waters NanoAcquity HPLC system (Waters Corp.) interfaced to an Elite Orbitrap mass spectrometer (ThermoFisher Scientific). Peptides were loaded onto a Symmetry C18 column (1.7 mm BEH-130, 0.1 × 100 mm, Waters) and separated with a 60 min gradient from 2 to 25% solvent B (0.1% formic acid, 98% acetonitrile) at 1 µL/min flow rate. Peptides were eluted directly into the mass spectrometer with a spray voltage of 1.2 kV. Full MS data were acquired in FT for 350–1250 m/z with a 60,000-resolution. The most abundant ions from full MS scans were selected for MS/MS through a 2-Da isolation window.
Acquired tandem MS spectra were searched using the Mascot algorithm (Matrix Sciences) with trypsin or chymotrypsin enzyme specificity. Search criteria included a full MS tolerance of 50 ppm, MS/MS tolerance of 0.5 Da with oxidation of methionine (+15.9949 Da) as variable modification, and carbamidomethylation (+57.0215 Da) of cysteine as a static modification. Data were searched against the specific sequences of in-house constructs overlaid onto the Uniprot mammalian database, including reverse protein sequences. Peptide assignments were first filtered to a 2% false discovery rate (FDR) at the peptide level and subsequently to a 2% FDR at the protein level.
Reconstitution of β3-NaX into lipid nanodiscs
For reconstitution into nanodiscs, multiple 50 μL sample aliquots were each applied to 2 mL tubes with a 200-fold molar excess of a POPC:POPE:POPG lipid mix (3:1:1 ratio solubilized in a sonication bath at 10 mg/mL in a buffer containing 50 mM HEPES pH 7.5, 100 mM NaCl, 5 mM MgCl2, and 1% CHAPS) and incubated for 30 min at 4 °C. The membrane scaffold protein MSP1E3D1 (Sigma) was then applied to the samples in a 4-fold molar excess and incubated for an additional 30 min at 4 °C. Samples were then diluted to 1.5 mL and Bio-Beads were applied to a final concentration of 0.25 mg/mL. Samples were then nutated overnight at 4 °C prior to Bio-Bead removal. The samples were then pooled and passed over 1 mL Strep-Tactin XT resin in a column that had been pre-equilibrated in Buffer C (25 mM HEPES pH 7.0, 200 mM NaCl) to remove empty nanodiscs. The column was washed with 5 CV Buffer C prior to sample elution with 5 CV containing 50 mM biotin. The sample was then concentrated to 100 μL and applied to a Superose 6 3.2/300 column that had been pre-equilibrated in Buffer C. Peak fractions were pooled and concentrated to 3 mg/mL.
Cryo-EM sample preparation and data acquisition
Samples were cross-linked with a final concentration of 0.05% EM-grade glutaraldehyde at room temperature for 10 min. The crosslinking reactions were quenched by the addition of 1 M Tris pH 7.0. Ultrafoil R2/2 (200 mesh) cryo-EM grids (Quantifoil GMBH) were plasma cleaned for 5 s using the Solarus plasma cleaner (Gatan, Pleasanton, CA). Three microliters of the final samples at 2.25 mg/mL (nanodiscs) or 1.5 mg/mL (GDN) were applied to the cryo-EM grids and blotted for 2.5 s using a Vitribot Mark IV (ThermoFisher Scientific, Waltham, MA) using a blot force setting of 8 at 100% humidity, and plunged into liquid ethane. Grids were then imaged on a Titan Krios electron microscope (ThermoFisher Scientific, Waltham, MA) operated at 300 keV with a bioquantum energy filter using a K2 (nanodiscs) or K3 (GDN) Summit direct electron detector (Gatan, Pleasanton, CA). Images of the nanodisc were recorded at a magnification of ×165,000, which corresponded to 0.824 Å/pixel using a 20 eV energy slit. Image stacks contained 50 images recorded at 0.2 s intervals over 10 s, giving a total exposure of ~50 e−/Å2. Images of the GDN sample were recorded in super-resolution mode at ×105,000 magnification, corresponding to 0.419 Å/pixel, using a 20 eV energy slit. Image stacks contained 60 images recorded at 0.05 s intervals over 3 s, giving a total exposure of ~64 e−/Å2. All data collection was done using serialEM36.
Cryo-EM data processing
For the nanodisc sample, Cryo-EM data was processed using a combination of the WARP, RELION, and cisTEM software packages37–40. For the first dataset, 11,658 movies were corrected for frame movement using MotionCor241 in RELION. The resulting images were filtered to retain only those with an accumulated motion total below a value of 250 and contrast-transfer function (CTF) parameters were fit using the 30–4.5 Å band of the spectrum using CTFFIND4.142. Images were filtered to include only those with a detected fit resolution better than 5 Å, giving a total of 9988 good images for further processing. 1,669,107 particles were picked using a deep learning-based algorithm in WARP37. Particles were subjected to two rounds of 2D classification in cisTEM, and the best 30 classes were chosen (76,392 particles) and exported to RELION for 3D classification. A 20 Å low-pass filtered (LPF) KV1.2 reconstruction solved in nanodiscs (EMD-9024)43 for which all density outside of the nanodisc had been erased was used as the initial 3D reference. The best obtained 3D volume was then used as the reference for the second round of 3D classification using a broader selection of particles from the 2D classification in cisTEM (350,901 particles). The best 3D volume and its corresponding 91,435 particles were then imported back into cisTEM for iterative rounds of auto-refine without a mask and manual refinement with iteratively adjusted masks and 20 Å LPF outside the mask (outside weight of 0.8). The resulting 4.5 Å map was then used as the reference in a final round of unmasked auto-refinements and masked manual refinements in cisTEM using the particle stack from only a single round of 2D classification in cisTEM, giving the final 3.2 Å map.
For the GDN detergent sample, Cryo-EM data were processed using a combination of the RELION, Gautomatch, and cisTEM software packages38–40,44. 10,850 movies were corrected for frame movement using MotionCor241 in RELION and binned to 1 Å/pixel. The resulting images were filtered to retain only those with an accumulated motion total below a value of 250 and contrast-transfer function (CTF) parameters were fit using the 30–4.5 Å band of the spectrum using CTFFIND4.142. Images were filtered to include only those with a detected fit resolution better than 5 Å, giving a total of 10,569 good images for further processing. A total of 2,780,663 particles were automatically picked using Gautomatch44 using 16 uniform projections of the reconstruction of NaV1.4 in complex with the β1 auxiliary subunit (EMD-9617) as templates. Particles were subjected to a single round of 2D classification into 250 classes in RELION and a particle stack containing 1,420,422 particles from the best 55 classes were extracted and exported to cisTEM. The structure of β3-NaX solved in nanodiscs was used as an initial reference in an initial round of auto-refine without a mask, followed by several rounds of manual refinement with iterative mask refinement and 20 Å LPF outside the mask (outside weight of 0.8). Manual refinement runs included defocus refinement and used a score threshold of 0.2. Iterative rounds of unmasked auto-refinements and masked manual refinements yielded a final reconstruction of 2.85 Å resolution.
Model building and structure analysis
The structure of NaV1.4 in complex with the β1 auxiliary subunit (PDB 6AGF)22 was used as a template to generate a NaX homology model using the Phyre2 server45. This model along with β1 was rigid body docked into the cryo-EM map for manual rebuilding in Coot46 to give an initial model of the β3-NaX complex. Multiple rounds of real-space refinement in Phenix47 and manual rebuilding in Coot were followed by molecular dynamics-assisted manual refinement in UCSF ChimeraX48 with ISOLDE49. After a final refinement in Phenix, the model was validated using the Phenix validation package. Structure figures were generated using PyMol50, UCSF Chimera51, and UCSF ChimeraX. Caver3.0 was used to analyze the channel pore52. Sequence alignments were performed using Clustal Omega53 and rendered with ESPript 3.054.
Molecular biology for biochemical and electrophysiological experiments
Complementary DNAs (cDNAs) of human NaX, NaX-eGFP-2 × FLAG, NaV1.7, NaX-NaV1.7 domain-swapped chimeras, ATP1A1, ATP1B1, and SAP97, codon optimized for Homo sapiens, were cloned into the pcDNA3.1/Hygro(+) vector. The human NaVβ1, NaVβ3, CaVβ1, CaVγ2, CaVα2δ1, ATP1A1, ATP1B1, ATP1G1, and SAP97 constructs were cloned into the pcDNA3.1(+) vector. For expression in Xenopus laevis oocytes, cDNAs were linearized using either NotI, BamHI, or XbaI restriction enzyme and then transcribed to capped RNAs with the T7 mMessage mMachine Kit (Ambion). For expression in HEK293T and Neuro-2a cells, plasmid DNAs purified with the NucleoBond Xtra Midi Plus kit (Macherey-Nagel) were used.
Two-electrode voltage-clamp electrophysiology
Oocytes were prepared as previously described33. Stage V/VI oocytes were obtained from ovaries of female Xenopus laevis frogs (obtained from African Reptile Park, South Africa). Animals were anaesthetized in 0.3% tricaine. Animal work and handling were carried out under license 2014–15-0201–00031, approved by the Danish Veterinary and Food Administration. Frogs were housed and cared for by an animal facility with ethical approval from the University of Copenhagen, Denmark. Healthy-looking stage V–VI oocytes were injected with 2.5–40 ng of RNA (in 5–41 nL) using a Nanoliter 2010 injector (World Precision Instruments). When excluding one or more constructs, an equivalent volume of nuclease-free water was added to maintain a constant RNA concentration across different construct combinations. Injected oocytes were kept at 18 °C, 140 rpm, in ND96 supplemented with 50 µg/mL gentamicin and 50 µg/mL tetracycline (in mM: 96 NaCl, 2 KCl, 1 MgCl2, 1.8 CaCl2, 2.5 sodium pyruvate, 0.5 theophylline, 5 HEPES; pH 7.4 with NaOH) for 2–5 days. Two-electrode voltage-clamp measurements were performed at room temperature using a Warner OC-725C Oocyte Clamp amplifier (Warner Instrument Corp, USA) and under constant perfusion with ND96 solution (in mM: 96 NaCl, 2 KCl, 1 MgCl2, 1.8 CaCl2, 5 HEPES; pH 7.4 with NaOH). For ion-selectivity experiments, external solutions contained 115 mM of test cations as chloride salts, 1.2 mM CaCl2, 2 mM MgCl2, 5 mM HEPES (pH 7.4 with the corresponding hydroxide). Data acquisition was performed using a Digidata 1550 digitizer (Molecular devices; sampled at 10 kHz) and pCLAMP 10 software (Molecular Devices). Microelectrodes from borosilicate glass capillaries (Harvard Apparatus) were prepared to have resistances around 0.2–1.0 MΩ using a P-1000 Flaming/Brown Micropipette Puller System (Sutter Instrument) and backfilled with 3 M KCl. The concentrations of compounds shown in Fig. 1b are as follows: aconitine (300 µM); veratridine (100 µM); blood depressing substance I (BDS-I, 300 nM); and neurotoxin 2 (ATX-II, 100 nM). Concentrations of compounds shown in Fig. 4c (middle panel) are as follows: tetrodotoxin (TTX, 10 µM); lidocaine hydrochloride (10 mM); quinidine (100 µM); loperamide hydrochloride (100 µM); flecainide acetate (300 µM); ranolazine dihydrochloride (300 µM); phenytoin sodium (300 µM); zinc chloride (ZnCl2; 1 mM); cobalt chloride (CoCl2; 1 mM); and gadolinium chloride (GdCl3; 100 µM).
Cell culture, cell surface biotinylation, and western blots
HEK293T and Neuro-2a cells (ATCC) were grown and maintained as described previously33. Approximately 800,000 HEK293T and 250,000 Neuro-2a cells were seeded in 35 mm cell culture dishes ~20 h before transient transfection with 1 µg of cDNA using LipoD293 ver. II (tebu-bio). For biochemical experiments, the cDNAs of NaX-eGFP-2 × FLAG and β3 were mixed in a mass ratio of 1:1. Equal amount of empty vector DNA was added to keep the total cDNA amount constant when β3 was excluded from transfection. Transfected HEK293T cells were used for biochemical experiments 24 h post-transfection. To induce differentiation, Neuro-2a cells were serum-starved 24 h post-transfection (cultured in DMEM for an additional 24–30 h) before they were used for biochemical and electrophysiological experiments. Cell surface biotinylation and western blots were performed as described previously33, with only slight modifications: (1) The quenching step was performed under gentle agitation on ice for 30 min, (2) Xenopus laevis oocytes were washed twice with tris-buffered saline before lysis, and (3) the total lysate fraction for oocytes was diluted 1:5 in SDS sample buffer before loading onto the gel due to excess protein. Antibodies were used as stated in ref. 33, except the mouse anti-Na+/K+-ATPase antibody was from Santa Cruz Biotechnology (sc-21712). Blots are representative of a minimum of three individual experiments.
Patch-clamp electrophysiology
Differentiated Neuro-2a cells expressing NaX-eGFP-2 × FLAG were seeded on glass coverslips an hour before patch-clamping experiments. Cells were voltage-clamped at −60 mV in whole-cell configuration (Fig. 1e–g; Supplementary Fig. 1f) using an Axopatch 200B amplifier (Molecular Devices). Digidata 1550 A digitizer (Molecular devices; sampled at 10 kHz) and pCLAMP 10 software (Molecular Devices). Glass pipettes for patch-clamp experiments were pulled from borosilicate glass capillaries (World Precision Instruments; Kwik-Fil 1.5/1.12; OD/ID), and fire polished to final resistances between 2.0 and 5.5 MΩ. Extracellular solutions contained (in mM): 140 or 190 NaCl, 5 KCl, 2.5 CaCl2, 1 MgCl2, 5 HEPES, 20 glucose (pH 7.4 with NaOH). The osmolarity values of these solutions were ~302 mOsm/L (for 140 NaCl) and ~395 mOsm/L (for 190 NaCl). The intracellular solution contained (in mM): 120 K-gluconate, 20 TEA-Cl, 2 MgCl2, 2Na2ATP, 1 EGTA and 10 HEPES, pH 7.3 with KOH (~281 mOsm/L).
HEK293T cells expressing NaX- or NaX-QTT-eGFP-2 × FLAG were seeded on glass coverslips an hour before whole-cell patch-clamping experiments. For a more physiological condition (Fig. 3d; Supplementary Fig. 1b), the extracellular solution contained 150 mM NaCl, 5 mM KCl, 0.5 mM CaCl2, 1.2 mM MgCl2, 10 mM HEPES, and 13 mM D-(+)-glucose (pH 7.4) with NaOH, ~320 mOsm/L, and the intracellular solution contained 140 mM CsCl, 10 mM CsF, 5 mM EGTA, 10 mM HEPES, and 2 mM Na2ATP (pH 7.2) with CsOH, ~304 mOsm/L. To determine if NaX-QTT was cation-permeable, we first substituted all extracellular cations with NMDG+ (Fig. 3e). Extracellular solution contained 150 mM NMDG and 10 mM HEPES (D-(+)-glucose was added accordingly to achieve osmolarity ~325 mOsm/L and pH was adjusted to 7.4 with hydrochloric acid). To determine if NaX-QTT was anion-permeable, we substituted all Cl− ions in both intra- and extracellular solutions with the large anion methanesulfonate (MS−) (Supplementary Fig. 9d). The extracellular solution contained 150 mM NaMS and 10 mM HEPES (D-(+)-glucose was added accordingly to achieve osmolarity ~325 mOsm/L, and pH was adjusted to 7.4 with NaOH). The intracellular solution contained 136 mM NaMS, 10 mM NaF, 5 mM EGTA, 10 mM HEPES, and 2 mM Na2ATP (pH 7.2 with NaOH, ~309 mOsm/L). The extracellular NMDG solution contained 150 NMDG and 10 mM HEPES (D-(+)-glucose was added accordingly to achieve osmolarity ~325 mOsm/L, and pH was adjusted to 7.4 with methanesulfonic acid).
For ion-selectivity experiments (Figs. 3e, f, 4a) (i) involving monovalent cations, the extracellular solution contained 150 mM XCl, 10 mM HEPES, and D-(+)-glucose was added accordingly to achieve osmolarity ~325 mOsm/L, and the pH was adjusted to 7.4 with XOH, where X indicates the cation of interest (Na+, K+, Li+, or Cs+); (ii) involving Ca2+, the extracellular solution contained 110 mM CaCl2, 10 mM HEPES, and D-(+)-glucose was added accordingly to achieve osmolarity ~325 mOsm/L, and the pH was adjusted to 7.4 with Ca(OH)2; (iii) the intracellular solution contained 136 mM NaCl, 10 mM NaF, 5 mM EGTA, 10 mM HEPES, and 2 mM Na2ATP (pH 7.2) with NaOH, ~309 mOsm/L.
Data analysis
For data analysis, raw current traces were typically filtered using 8-pole Bessel low-pass filter at 500–800 Hz. Current traces were subjected to factor 5 data reduction for display in figures. Data are presented as mean ± standard deviation (SD) from at least 3 cells from at least two batches of cells. For ion-selectivity experiments performed with Na+ intracellular solution (Figs. 3f, 4a), liquid junction potential (LJP) was measured with reference to the Na+ intracellular solution and corrected after recording. Relative ion permeabilities (PNa/PX) were calculated using the equation PX/PNa = [Na+]IC exp(ErevF/RT)/[X+]EC (X = Na, K, Li, or Cs), where F is Faraday’s constant, R is the gas constant, T = 274 K, and Erevs were measured from the −80 to +80 mV ramp (corrected for LJP). Data are presented as mean ± s.d. from at least two batches of cells. Statistical comparisons were performed using GraphPad Prism (v.8.1, GraphPad Software) and the tests are mentioned in text.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank members of the Pless laboratory and Genentech colleagues in the Research Materials, BioMolecular Resources, Microchemistry, Proteomics and Lipidomics, and Structural Biology departments for their support of this project. We appreciate advice from E. Green, A. Rohou, C. Koth, W. Sandoval and the encouragement of S. Hymowitz, V. Dixit, and A. Chan. We are grateful to Harold Zakon (University of Texas at Austin) for insightful discussions. Members of the Pless group acknowledge the Novo Nordisk Foundation (NNF20OC0064550), the Carlsberg Foundation (CF16-0504), and the Independent Research Fund Denmark (7025-00097 A) for financial support.
Source data
Author contributions
C.L.N. established protein purification and reconstitution methods. T.C and C.T. generated key protein expression reagents. C.L.N., M.K., and C.A. optimized cryo-EM sample preparation and data collection. C.L.N. and M.K. determined structures with guidance from C.C. H.C.C., S.A.H., and N.B. performed molecular biology, biochemistry, electrophysiology, and pharmacology experiments. With input from C.L.N., M.K., S.A.P., and J.P., H.C.C. characterized chimera, NaX-QTT and NaX-EEE constructs. J.T. performed mass spectrometry experiments. C.L.N., H.C.C., M.K., S.A.H., N.B., J.T., C.C., S.A.P., and J.P. analyzed the data. J.P., S.A.P., C.L.N., and H.C.C. prepared the manuscript with input from all authors. C.C., S.A.P., and J.P. supervised the project and are co-senior authors.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Data availability
The coordinates of the NaX-β3 structures deterimined in nanodiscs and GDN have been deposited in the Protein Data Bank under accession codes 7TJ8 and 7TJ9, respectively. Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession codes EMD-25919 and EMD-25920. Source data are provided with this paper.
Competing interests
C.L.N., M.K., T.C., C.T., J.T., C.A., C.C., and J.P. are or were employees of Genentech or Roche and own shares in the Genentech or Roche group. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Cameron L. Noland, Han Chow Chua, Marc Kschonsak.
Contributor Information
Claudio Ciferri, Email: ciferri.claudio@gene.com.
Stephan Alexander Pless, Email: stephan.pless@sund.ku.dk.
Jian Payandeh, Email: jpayandeh@interlinetx.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-28984-4.
References
- 1.Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol. Rev. 2005;57:397–409. doi: 10.1124/pr.57.4.4. [DOI] [PubMed] [Google Scholar]
- 2.George AL, Jr., Knittle TJ, Tamkun MM. Molecular cloning of an atypical voltage-gated sodium channel expressed in human heart and uterus: evidence for a distinct gene family. Proc. Natl Acad. Sci. USA. 1992;89:4893–4897. doi: 10.1073/pnas.89.11.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felipe A, Knittle TJ, Doyle KL, Tamkun MM. Primary structure and differential expression during development and pregnancy of a novel voltage-gated sodium channel in the mouse. J. Biol. Chem. 1994;269:30125–30131. [PubMed] [Google Scholar]
- 4.Akopian AN, Souslova V, Sivilotti L, Wood JN. Structure and distribution of a broadly expressed atypical sodium channel. FEBS Lett. 1997;400:183–187. doi: 10.1016/s0014-5793(96)01389-0. [DOI] [PubMed] [Google Scholar]
- 5.Hiyama TY, et al. Na(x) channel involved in CNS sodium-level sensing. Nat. Neurosci. 2002;5:511–512. doi: 10.1038/nn0602-856. [DOI] [PubMed] [Google Scholar]
- 6.Noda M, Hiyama TY. Sodium sensing in the brain. Pflug. Arch. 2015;467:465–474. doi: 10.1007/s00424-014-1662-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe U, et al. A comparison of voluntary salt-intake behavior in Nax-gene deficient and wild-type mice with reference to peripheral taste inputs. Brain Res. 2003;967:247–256. doi: 10.1016/s0006-8993(03)02247-9. [DOI] [PubMed] [Google Scholar]
- 8.Hiyama TY, Watanabe E, Okado H, Noda M. The subfornical organ is the primary locus of sodium-level sensing by Na(x) sodium channels for the control of salt-intake behavior. J. Neurosci. 2004;24:9276–9281. doi: 10.1523/JNEUROSCI.2795-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiyama TY, et al. Autoimmunity to the sodium-level sensor in the brain causes essential hypernatremia. Neuron. 2010;66:508–522. doi: 10.1016/j.neuron.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 10.Xu W, et al. Sodium channel Nax is a regulator in epithelial sodium homeostasis. Sci. Transl. Med. 2015;7:312ra177. doi: 10.1126/scitranslmed.aad0286. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe E, et al. Nav2/NaG channel is involved in control of salt-intake behavior in the CNS. J. Neurosci. 2000;20:7743–7751. doi: 10.1523/JNEUROSCI.20-20-07743.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grob M, Drolet G, Mouginot D. Specific Na+ sensors are functionally expressed in a neuronal population of the median preoptic nucleus of the rat. J. Neurosci. 2004;24:3974–3984. doi: 10.1523/JNEUROSCI.3720-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tremblay C, et al. Neuronal sodium leak channel is responsible for the detection of sodium in the rat median preoptic nucleus. J. Neurophysiol. 2011;105:650–660. doi: 10.1152/jn.00417.2010. [DOI] [PubMed] [Google Scholar]
- 14.Nomura K, et al. [Na(+)] increases in body fluids sensed by central Nax induce sympathetically mediated blood pressure elevations via H(+)-dependent activation of ASIC1a. Neuron. 2019;101:60–75 e66. doi: 10.1016/j.neuron.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Hou C, et al. Knockout of sodium channel Nax delays re-epithelializathion of splinted murine excisional wounds. Wound Repair Regen. 2021;29:306–315. doi: 10.1111/wrr.12885. [DOI] [PubMed] [Google Scholar]
- 16.Shimizu H, et al. Glial Nax channels control lactate signaling to neurons for brain [Na+] sensing. Neuron. 2007;54:59–72. doi: 10.1016/j.neuron.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Hiyama TY, et al. Endothelin-3 expression in the subfornical organ enhances the sensitivity of Na(x), the brain sodium-level sensor, to suppress salt intake. Cell Metab. 2013;17:507–519. doi: 10.1016/j.cmet.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto M, et al. Channel properties of Nax expressed in neurons. PLoS ONE. 2015;10:e0126109. doi: 10.1371/journal.pone.0126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berret E, et al. Regulation of central Na+ detection requires the cooperative action of the NaX channel and alpha1 Isoform of Na+/K+-ATPase in the Na+-sensor neuronal population. J. Neurosci. 2013;33:3067–3078. doi: 10.1523/JNEUROSCI.4801-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berret E, et al. Extracellular Na(+) levels regulate formation and activity of the NaX/alpha1-Na(+)/K(+)-ATPase complex in neuronal cells. Front. Cell Neurosci. 2014;8:413. doi: 10.3389/fncel.2014.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nehme B, Henry M, Mouginot D, Drolet G. The expression pattern of the Na(+) sensor, Na(X) in the hydromineral homeostatic network: a comparative study between the rat and mouse. Front. Neuroanat. 2012;6:26. doi: 10.3389/fnana.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan, X. et al. Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta1. Science10.1126/science.aau2486 (2018). [DOI] [PubMed]
- 23.Pan X, et al. Molecular basis for pore blockade of human Na(+) channel Nav1.2 by the mu-conotoxin KIIIA. Science. 2019;363:1309–1313. doi: 10.1126/science.aaw2999. [DOI] [PubMed] [Google Scholar]
- 24.Shen H, Liu D, Wu K, Lei J, Yan N. Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins. Science. 2019;363:1303–1308. doi: 10.1126/science.aaw2493. [DOI] [PubMed] [Google Scholar]
- 25.Pan, X. et al. Comparative structural analysis of human Nav1.1 and Nav1.5 reveals mutational hotspots for sodium channelopathies. Proc. Natl Acad. Sci. USA10.1073/pnas.2100066118 (2021). [DOI] [PMC free article] [PubMed]
- 26.Li Z, et al. Structural basis for pore blockade of the human cardiac sodium channel Nav 1.5 by the antiarrhythmic drug quinidine. Angew. Chem. Int Ed. Engl. 2021;60:11474–11480. doi: 10.1002/anie.202102196. [DOI] [PubMed] [Google Scholar]
- 27.Pless SA, Galpin JD, Frankel A, Ahern CA. Molecular basis for class Ib anti-arrhythmic inhibition of cardiac sodium channels. Nat. Commun. 2011;2:351. doi: 10.1038/ncomms1351. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, D. et al. Open-state structure and pore gating mechanism of the cardiac sodium channel. Cell10.1016/j.cell.2021.08.021 (2021). [DOI] [PMC free article] [PubMed]
- 29.Angsutararux, P., Kang, P. W., Zhu, W. & Silva, J. R. Conformations of voltage-sensing domain III differentially define NaV channel closed- and open-state inactivation. J. Gen. Physiol.10.1085/jgp.202112891 (2021). [DOI] [PMC free article] [PubMed]
- 30.Jensen MO, et al. Principles of conduction and hydrophobic gating in K+ channels. Proc. Natl Acad. Sci. USA. 2010;107:5833–5838. doi: 10.1073/pnas.0911691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aryal P, Sansom MS, Tucker SJ. Hydrophobic gating in ion channels. J. Mol. Biol. 2015;427:121–130. doi: 10.1016/j.jmb.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun YM, Favre I, Schild L, Moczydlowski E. On the structural basis for size-selective permeation of organic cations through the voltage-gated sodium channel. Effect of alanine mutations at the DEKA locus on selectivity, inhibition by Ca2+ and H+, and molecular sieving. J. Gen. Physiol. 1997;110:693–715. doi: 10.1085/jgp.110.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chua CH, Wulf M, Weidling C, Rasmussen LP, Pless SA. The NALCN channel complex is voltage sensitive and directly modulated by extracellular calcium. Sci. Adv. 2020;6:eaaz3154. doi: 10.1126/sciadv.aaz3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avalos Prado P, et al. KCNE1 is an auxiliary subunit of two distinct ion channel superfamilies. Cell. 2021;184:534–544 e511. doi: 10.1016/j.cell.2020.11.047. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto D, Yeh JZ, Narahashi T. Voltage-dependent calcium block of normal and tetramethrin-modified single sodium channels. Biophys. J. 1984;45:337–344. doi: 10.1016/S0006-3495(84)84159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 37.Tegunov D, Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. elife10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed]
- 39.Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. eLife10.7554/eLife.35383 (2018). [DOI] [PMC free article] [PubMed]
- 40.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matthies, D. et al. Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs. elife10.7554/eLife.37558 (2018). [DOI] [PMC free article] [PubMed]
- 44.Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D. Struct. Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goddard TD, et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Croll TI. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D. Struct. Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.The PyMOL Molecular Graphics System, V. r. p.https://pymol.org/2/support.html (Schrödinger, LLC).
- 51.Pettersen EF, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 52.Chovancova E, et al. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of the NaX-β3 structures deterimined in nanodiscs and GDN have been deposited in the Protein Data Bank under accession codes 7TJ8 and 7TJ9, respectively. Cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession codes EMD-25919 and EMD-25920. Source data are provided with this paper.