Abstract
YY1 is a sequence-specific DNA-binding transcription factor that has many important biological roles. It activates or represses many genes during cell growth and differentiation and is also required for the normal development of mammalian embryos. Previous studies have established that YY1 interacts with histone acetyltransferases p300 and CREB-binding protein (CBP) and histone deacetylase 1 (HDAC1), HDAC2, and HDAC3. Here, we present evidence that the activity of YY1 is regulated through acetylation by p300 and PCAF and through deacetylation by HDACs. YY1 was acetylated in two regions: both p300 and PCAF acetylated the central glycine-lysine-rich domain of residues 170 to 200, and PCAF also acetylated YY1 at the C-terminal DNA-binding zinc finger domain. Acetylation of the central region was required for the full transcriptional repressor activity of YY1 and targeted YY1 for active deacetylation by HDACs. However, the C-terminal region of YY1 could not be deacetylated. Rather, the acetylated C-terminal region interacted with HDACs, which resulted in stable HDAC activity associated with the YY1 protein. Finally, acetylation of the C-terminal zinc finger domain decreased the DNA-binding activity of YY1. Our findings suggest that in the natural context, YY1 activity is regulated through intricate mechanisms involving negative feedback loops, histone deacetylation, and recognition of the cognate DNA sequence affected by acetylation and deacetylation of the YY1 protein.
YY1, also known as δ, NF-E1, and UCRBP, is a multifunctional transcription factor. It binds, with its four C2H2 zinc fingers, to a specific DNA sequence (CGCCATNTT) located in many different promoters and either activates or represses transcription (for comprehensive reviews, see references 57 and 65). YY1 regulates the expression of both cellular and viral genes, including those encoding c-Myc, c-Fos, p53, α-actin, gamma interferon, P5 of adeno-associated virus, E6 and E7 of human papillomavirus (HPV), and a number of other viral long terminal repeats. Many of these gene products have important consequences for cell growth and differentiation (reviewed in reference 57). YY1 is highly conserved among human, mouse, and Xenopus laevis cells, and a Drosophila melanogaster homologue of YY1 exists (6, 14, 20, 47, 49, 58). Knockout studies show that deletion of YY1 results in peri-implantation lethality in mice (13), further demonstrating the importance of YY1 in fundamental biological processes such as development.
Various biochemical methods have been employed to dissect the functional domains of YY1 in order to understand how the activity of YY1 is regulated (1, 7, 8, 38–40, 58, 73). It can be summarized that YY1 contains two repression domains, one embedded within residues 170 to 200 and the other overlapping with the C-terminal zinc finger DNA-binding domain. YY1 might also contain an independent activation domain at the N terminus. This modular nature of YY1 supports the idea that YY1 is bifunctional, capable of both activating and repressing transcription. It is generally believed that whether YY1 behaves as a transcriptional activator or repressor depends on its relative concentration (7), other cell type-specific factors (4, 29, 36, 60, 74), and the promoter sequences surrounding the YY1 binding sites (59). However, the specific cues dictating when and how this decision is executed remain a mystery. It has been shown that YY1 is a stable phosphorylated protein expressed ubiquitously regardless of cell cycle position or the differentiation status of the cell (1), suggesting that the activity of YY1 is regulated at the posttranslational level, possibly through interactions with other proteins.
In support of the hypothesis that the actions of YY1 are controlled by protein-protein interactions, a wide variety of transcription factors have been shown to associate with YY1. These include proteins of the basal transcription machinery, such as TATA binding protein (1), TFIIB (67); sequence-specific DNA-binding transcriptional activators, such as SpI (37, 55), c-Myc (60), ATF/CREB (79), C/EBP (4); and various transcriptional coregulators, such as E1A (40), TAFII55 (11), p300, CREB-binding protein (CBP) (1, 36), and HDAC1, HDAC2, and HDAC3 (73, 75). The YY1-p300 and YY1-HDACs interactions are of particular interest. p300 and CBP are two closely related transcriptional coactivators (15) that have been shown to be histone acetyltransferases (HATs) (2, 46). Hyperacetylated histones have long been known to associate with activated genes (23, 54, 68), and recent studies further suggest a causal relationship between histone acetylation and gene activation (reviewed in references 30 and 33). It is thought that addition of acetyl groups to the lysine residues of histone tails facilitates access of transcription factors to DNA by disrupting higher-order packaging of the chromatin (34) and by neutralizing the positive charge of the histone proteins, which reduces the affinity of histones for DNA (24). In certain circumstances, the transcriptional activator activity of YY1 directly depends on its association with p300 (36). In contrast, HDACs have the opposite effects on transcription. HDAC1, HDAC2, and HDAC3 are class I HDACs (reviewed in reference 12) that have a high degree of homology to the Saccharomyces cerevisiae global transcriptional regulator Rpd3p. It has been shown that the transcriptional repression activity of YY1 is mediated by association of HDAC2 with residues 170 to 200 of YY1, which corresponds to one of the repression domains of YY1 (73). It is conceivable that by selectively associating with either HATs or HDACs, YY1 becomes an activator or a repressor. However, it has not been shown definitively that such an active selection system exists for YY1.
Recently p300, CBP, and another HAT, PCAF (p300-CBP associated factor) have been shown to acetylate transcription factors in addition to their histone substrates (reviewed in reference 63). Importantly, acetylation was a key regulatory mechanism for the regulation of these transcription factors. Because YY1 interacts with both p300 and HDACs, we explored the possibility that YY1 is also regulated by acetylation and deacetylation. Indeed, our results demonstrate that YY1 was acetylated by both p300 and PCAF and was deacetylated by HDAC1 and HDAC2. p300 acetylated the central region of YY1, amino acids (aa) 170 to 200, while PCAF acetylated both the central region and the C-terminal zinc finger domain of YY1. The acetylated central region of YY1 was a target of active deacetylation by HDAC1 and HDAC2, whereas the C-terminal zinc finger domain was not deacetylated. Acetylation and deacetylation, in turn, had intriguing influences on the sequence-specific DNA-binding and transcriptional activities of YY1.
MATERIALS AND METHODS
Plasmids.
Glutathione S-transferase (GST) was expressed from pGSTag (51). GST-YY1 (aa 1 to 414) [or, simply, GST-YY1(1–414)] was expressed from pGST-YY1 (73), and deletion constructs of GST-YY1 were generated by restriction enzyme digestion and religations of pGST-YY1. GST-p53 expression plasmid has been described (26). GST-p300 and GST-PCAF were expressed from plasmids pGEX2T-p300 (aa 1195 to 1810) and pGEX5X-PCAF (aa 352 to 832), respectively (9).
Gal4-YY1 was expressed from pM1-YY1, which was constructed by inserting the full-length YY1 cDNA in frame with the Gal4 DNA-binding domain in pM1 (52). pM1-YY1 (K170-R200) was prepared using adapter oligonucleotides containing AAG (lysine) to AGG (arginine) mutations within YY1 aa 170 to 200 of pM1-YY1. pG5CAT-Control was constructed by inserting five Gal4 DNA-binding sites into the BglII site of pCAT-Control (Promega), which contains the chloramphenicol acetyltransferase (CAT) gene downstream of the simian virus 40 promoter and enhancer sequences. Flag-tagged YY1 (F-YY1) was expressed from pCEP4F-YY1, which was generated by inserting the full-length YY1 cDNA into the pCEP4F vector (80). Serial Flag-YY1 deletion constructs were made by restriction enzyme digestion and religations of pCEP4F-YY1. pET15b-YY1, which expressed nontagged full-length YY1 in Escherichia coli in an inducible system, was constructed by cloning the full-length YY1 cDNA into pET15b (Novagen). pGEM7Zf3X-HD1, which was used to generate in vitro-translated HDAC1, was made by subcloning full-length HDAC1 into the pGEM7Zf3X vector, which is derived from pGEM7Zf(+) (Promega).
The following plasmids have been previously described: pBJ5-HD1F (64), which expresses HDAC1 C-terminally tagged with a Flag epitope; pBJ5.1-HD1F (H199F), HDAC1 point mutant (22); and pME18S-FLAG-HDAC2, which expresses Flag-tagged HDAC2 (35).
Recombinant proteins.
The GST fusion constructs of p300, PCAF, p53, and YY1 deletion mutants were expressed in E. coli DH5α, bound to glutathione-agarose beads (Sigma), washed extensively in phosphate-buffered saline (PBS), and eluted with 25 mM reduced glutathione. The eluate was then dialyzed against a buffer containing 20 mM Tris-HCl (pH 8), 0.5 mM EDTA, 100 mM KCl, 20% glycerol, 0.5 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonyl fluoride.
Non-tagged YY1 was expressed in E. coli BL21(DE3), induced with 0.2 mM isoprophyl-β-d-thiogalactopyranoside (IPTG), and captured by Ni2+-immobilized metal affinity chromatography (Invitrogen). Unbound bacterial proteins were removed with 50 mM imidazole, and bound YY1 was eluted with 500 mM imidazole. F-YY1 used in electrophoretic mobility shift assays (EMSA) was purified on an anti-Flag column (Sigma) under stringent conditions following the manufacturer's suggestions.
In vitro acetylation reactions.
Purified GST-YY1 and the serial deletion proteins were incubated at 30°C for 30 min with 0.25 μCi of [3H]acetyl coenzyme A ([3H]acetyl-CoA) (Amersham) and either purified GST-p300 (0.2 μg) or GST-PCAF (1 μg) in 30 μl of acetylation buffer containing 50 mM Tris-HCl (pH 8.0), 5% glycerol, 0.1 mM EDTA, 50 mM KCl, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride and 10 mM sodium butyrate. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gels were fixed by Coomassie blue staining and subjected to signal amplification (Amplify; Amersham) prior to exposing to X-ray film.
For subsequent mass spectrometry analyses, 0.5 pmol of the YY1 peptide (GRVKKGGGKKSGKKSYLSGGAGAAGGRGADP) was acetylated in acetylation buffer for 2 h with CAT-assay grade acetyl-CoA (Amersham).
Cell culture, transfection, and CAT assays.
HeLa cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and penicillin-streptomycin. 106 HeLa cells were seeded into 60-mm-diameter tissue culture dishes. Sixteen hours later, 10 μg of plasmids (for CAT assays, 5 μg of pG5CAT-Control plus 5 μg of pM1 effector plasmids) were transfected into cells using the calcium phosphate coprecipitation method (17). Forty-eight hours after transfection, cells were harvested by scraping and lysed by repeated freeze-thawing, and extracts were assayed for CAT activity by thin-layer chromatography (16).
Immunoprecipitation and Western blot analysis.
Immunoprecipitation of Flag-YY1 deletion proteins was done using anti-Flag M2 affinity gel (Sigma) following the manufacturer's suggestions.
Western blot analyses were performed using standard protocols (21). Immunoprecipitated proteins were detected with diluted primary antibodies (1:1,000 dilution of anti-acetyl-lysine [Upstate]; 1:5,000 dilution of anti-Flag M2 [Sigma]; 1:1,000 dilution of anti-HDAC1, HDAC2, or HDAC3 rabbit anti-serum [35, 66, 72]) followed by 1:7,500-diluted alkaline phosphatase-conjugated secondary antibodies (Promega). The blots were subsequently developed with 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium (Promega).
In vitro protein-protein interaction assays.
35S-labeled HDAC1 was generated from pGEM7Zf3X-HD1 using T7 RNA polymerase and the TNT Reticulocyte Lysate System (Promega). GST-YY1 (aa 170 to 200) was either acetylated with cold acetyl-CoA or mock acetylated and was then captured onto glutathione-agarose beads. In vitro-translated HDAC1 (5 μl) was mixed with the beads in the presence of PBS plus 0.2% NP-40 at 4°C for 1 h. Beads were washed extensively in PBS plus 0.2% NP-40. Bound proteins were eluted by boiling in Laemmli sample buffer, separated by SDS-PAGE, and detected by Coomassie blue staining and autoradiography.
Chemical labeling of peptides with [3H]acetate.
Peptides were labeled chemically according to the protocol described (64) with minor modifications. The peptides (0.4 mg) were labeled with 5 mCi of [3H]sodium acetate (2 to 5 Ci/mmol; New England Nuclear) in the presence of 0.24 M benzotriazol-1-gloxytrif-(dimethylamino)-phosphonium hexafluorophosphate and 0.2 M triethylamine (Aldrich). Labeled peptides were purified on a Microcon-SCX column (Millipore).
Histone deacetylation assays.
HeLa cells were transfected with 10 μg of pCEP4F-YY1 deletion plasmids by the calcium phosphate coprecipitation method (17). Forty-eight hours after transfection, cells were harvested, lysed in PBS plus 0.1% NP-40, and immunoprecipitated with anti-Flag M2 affinity gel (Sigma). HDAC activities of the immunoprecipitated Flag-YY1 were determined using a peptide corresponding to residues 2 to 24 of histone H4 as described (64) except that incubation was performed at room temperature overnight. Trichostatin A (TSA) (400 nM [final concentration]; Sigma) or a five-column volume of Flag peptide (Sigma) was added to the immunoprecipitate 30 min prior to addition of the H4 substrate peptide, if appropriate.
Immunofluorescence analysis.
HeLa cells were grown on chamber slides (Nalge Nunc International) for about 24 h and transfected with 10 μg of F-YY1 deletion constructs. Two days later, cells were washed with ice-cold PBS, fixed with 4% paraformaldehyde for 10 min, rinsed again with PBS, covered with 400 μl of 1% bovine serum albumin in PBS for 1 h at room temperature, washed again in PBS, and then treated with 1:200 dilution of anti-Flag fluorescein isothiocyanate (FITC) conjugate antibody (Upstate) for 1 h at room temperature. Subsequently, cells were subjected to extensive washing with PBS and coverslips were applied with one drop of antifade mounting medium with DAPI (4′,6′-diamidino-2-phenylindole) (Vector) before analysis under a fluorescence microscope.
EMSAs.
Purified YY1 proteins and GST-p53 were first acetylated or mock acetylated. Single-stranded oligodeoxynucleotides corresponding to a consensus YY1-binding site (56) or a p53 cognate sequence (18) were labeled individually with [γ32-P]ATP and T4 polynucleotide kinase, heated together at 65°C, and allowed to anneal by slow cooling to room temperature. Binding reactions were performed in a 12-μl reaction volume containing 12 mM HEPES (pH 7.9), 10% glycerol, 5 mM MgCl2, 60 mM KCl, 1 mM DTT, 0.5 mM EDTA, bovine serum albumin (50 mg/ml), 0.05% NP-40, 0.1 mg of poly(dI-dC), approximately 1 ng of proteins, and 5 fmol of radiolabeled DNA. Reaction mixtures were incubated for 10 min at room temperature and separated on 4% nondenaturing polyacrylamide gels. The gels were then dried and exposed to film.
RESULTS
YY1 is acetylated by p300 and PCAF.
Acetylation has been shown to regulate the activity of many transcription factors, including sequence-specific DNA-binding factors p53 (18), GATA-1 (5, 27), E2F (43, 44), and MyoD (53). Analysis of the YY1 amino acid sequence reveals that YY1 contains multiple lysine residues that are potential substrates for acetylation. Interestingly, within YY1's HDAC interaction domain (residues 170 to 200), there are six lysines arranged in pairs (Fig. 1). As described previously, YY1 interacts with the HAT p300 (1, 36), which interacts with another HAT, PCAF (76). Remarkably, both p300 and PCAF catalyze the acetylation of transcription factors (reviewed in reference 63). To determine if p300 or PCAF acetylated YY1, we performed in vitro acetylation reactions using GST-tagged p300 and PCAF purified from E. coli as enzymes. YY1 serial deletion constructs were similarly purified from E. coli as GST fusion proteins and used as substrates. GST-PCAF acetylated various deletion constructs of YY1 (Fig. 2A, lanes 1 to 5, 8 to 11, 13, 15, and 16) but not GST alone (lanes 12 and 17). Unlike PCAF, GST-p300 efficiently acetylated GST-YY1(170–200) as well as GST-YY1(1–200) (Fig. 2B, lanes 3, 12, and 15). GST alone was not acetylated by p300, showing that the in vitro acetylation was specific to YY1 (Fig. 2B, lane 1). Moreover, acetylation of YY1 was dependent on p300 or PCAF and was not due to YY1 auto-acetylation (Fig. 2C).
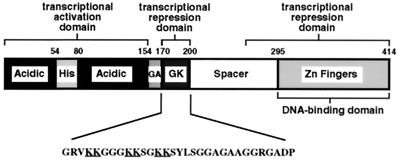
FIG. 1.
Multiple functional domains of transcription factor YY1. YY1 has one transcriptional activation domain at the N terminus and two repression domains, one encompassing residues 170 to 200 and the other one residing at the C terminus. The amino acid sequence of the central repression domain (residues 170 to 200) is given with lysine residues underlined. His, histidine-rich domain; GA, glycine-alanine-rich domain; GK, glycine-lysine-rich domain.
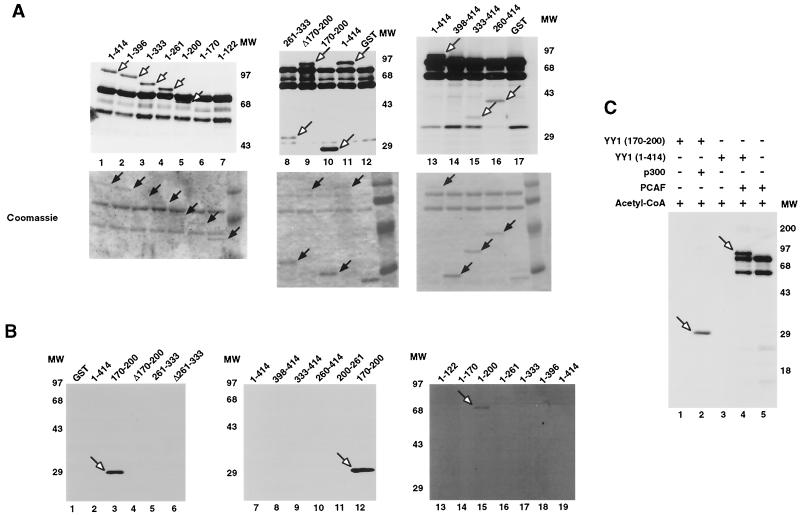
FIG. 2.
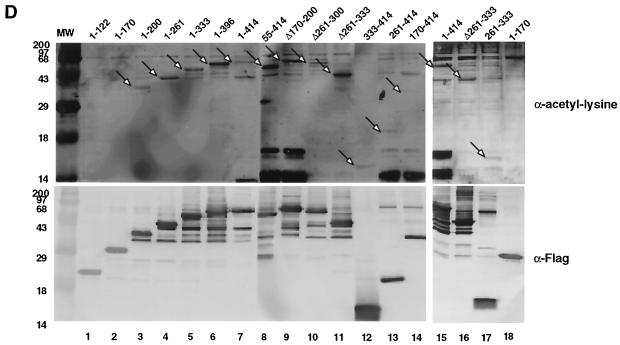
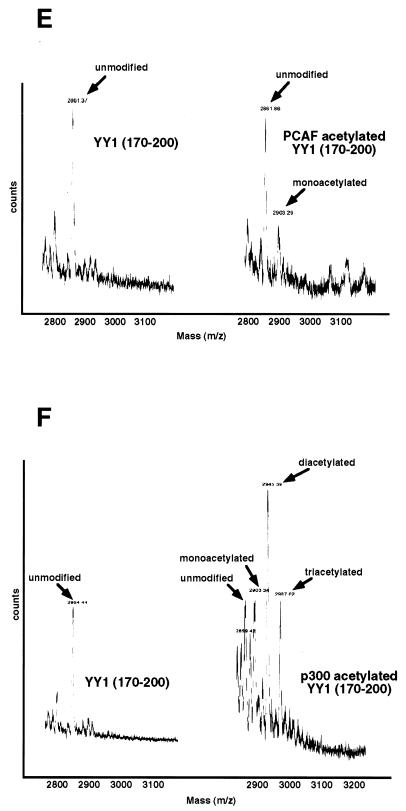
Acetylation of YY1 in vitro and in vivo. (A) Acetylation of YY1 by PCAF. Serial GST-YY1 deletion proteins were incubated with GST-PCAF and separated by SDS-PAGE. The gels were then exposed to X-ray film to detect acetylated proteins. Open arrows indicate acetylated YY1 proteins. Auto-acetylated forms of PCAF were detected as three bands of ∼68 kDa. Solid arrows indicate GST-YY1 deletion proteins. (B) Acetylation of YY1 by p300. Serial GST-YY1 deletion proteins were incubated with GST-p300. Arrows indicate YY1 proteins acetylated by p300. (C) Acetylation of YY1 is dependent on p300 or PCAF. In vitro acetylation reactions were performed in the presence or absence of p300 or PCAF. Arrows indicate that YY1 was acetylated only in the presence of p300 or PCAF. (D) Identification of regions of YY1 acetylated in vivo. F-YY1 serial deletion proteins were transiently expressed in HeLa cells, immunoprecipitated with anti-Flag antibody, and analyzed by Western blotting using anti-acetyl lysine antibody and anti-Flag antibody. Arrows indicate immunoprecipitated YY1 proteins that were acetylated. (E and F) Determination of the number of acetylated lysines by mass spectrometry. A YY1 peptide containing residues 170 to 200 was in vitro acetylated by PCAF or p300 and subjected to mass spectrometry analysis. Left panels are spectra from mock-acetylated peptides. Right panels contain spectra from acetylated as well as unacetylated peptides. Small peaks around 3100 m/z in Fig. 2E represent degraded GST-PCAF or background chemical noises.
The arrangement of the six lysines within the p300 acetylation domain of YY1 closely resembles the sequence of the p300-acetylated region of histone H2B (63), suggesting that YY1 residues 170 to 200 contain an authentic p300 acetylation domain. Furthermore, the PCAF-interacting domain of p300 overlaps with its YY1-interacting domain (1, 76), which suggests that acetylation of YY1 by PCAF versus by p300 might be either cooperative or mutually exclusive in vivo, depending on whether or not YY1 binding to p300 can be competed by PCAF. Our data that full-length GST-YY1 was acetylated by PCAF and not by p300 (Fig. 2A, lanes 1, 11, and 13, and Fig. 2B, lanes 2, 7, and 19) further strengthen the suggestion that the conformation of YY1, perhaps affected by selective interaction with p300 and PCAF, is important for YY1 acetylation. Taken together, we found that PCAF acetylated YY1 at residues 170 to 200 and at its C terminus. p300, in contrast, efficiently acetylated YY1 only at residues 170 to 200 and only when YY1 was in a C-terminal truncated form, implying that p300-mediated acetylation of YY1 is dependent on the conformation of YY1.
To determine which region of YY1 was acetylated in vivo, F-YY1 deletion constructs were transiently transfected into HeLa cells and immunoprecipitated with anti-Flag antibody. Acetylated forms of F-YY1 were detected by Western blotting with anti-acetyl lysine antibody (Fig. 2D, top panel). To ensure that all F-YY1 deletions were expressed, blots were also probed with anti-Flag antibody (Fig. 2D, bottom panel). The results of these experiments show that YY1 is acetylated in vivo at residues 170 to 200 as well as in the C-terminal residues 261 to 414. Taken together, our in vitro and in vivo acetylation results suggest that there are two acetylation domains on YY1: residues 170 to 200, which are acetylated by both p300 and PCAF, and the C terminus, which is acetylated by PCAF only.
Because both p300 and PCAF acetylate YY1 at residues 170 to 200, we were interested in determining the specificity of acetylation by these two enzymes. Using GST-p300 or GST-PCAF, we in vitro acetylated a synthetic peptide corresponding to YY1 residues 170 to 200. We then compared the mass spectra produced by mass spectrometry. Figure 2E shows that mock-acetylated YY1 peptide had an Mr of 2,861 (left, 2861 m/z). When this peptide was acetylated with PCAF, another peak emerged with an Mr of 2,903 (right, 2903 m/z). Compared to the mock-acetylated peptide, this additional peak had a mass corresponding to one additional acetyl group (2903 − 2861 = 42), suggesting that the YY1 peptide was acetylated once by PCAF. Interestingly, when we compared the spectrum from the p300-acetylated peptide with that of the mock-acetylated peptide, we found three additional peaks at 2903, 2945, and 2987 m/z, which contained one, two, and three additional acetyl groups, respectively (Fig. 2F). This result strongly suggests that YY1 can be acetylated by p300 at three different lysines between residues 170 and 200.
Lysine-to-arginine mutations within YY1 residues 170 to 200 significantly reduce the transcriptional repression activity of YY1.
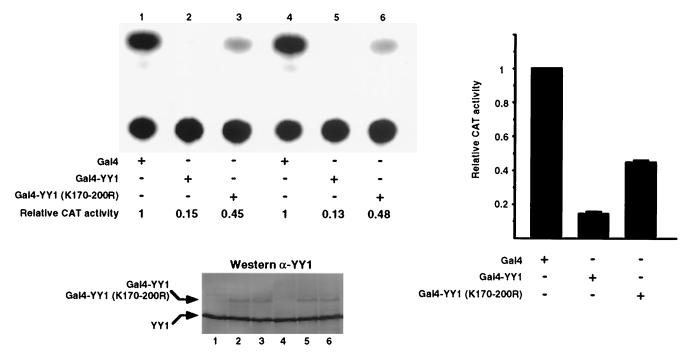
To understand the effect of acetylation on the transcriptional activity of YY1, we mutated the six lysines within YY1 residues 170 to 200 to arginines. Arginine substitutions preserve the charges of the affected amino acid residues but prevent acetylation in vivo by histone acetyltransferases. Both wild-type and mutant YY1 were fused to a Gal4 DNA-binding domain and transfected into HeLa cells in combination with a CAT reporter driven by the SV40 promoter containing five Gal4 binding sites. Consistent with previous findings (58, 73), wild-type Gal4-YY1 was a potent transcriptional repressor (Fig. 3, top panel, lanes 2 and 5; right panel). However, when the six lysines were mutated and no longer able to be acetylated, YY1 lost much of its repression activity (Fig. 3, top panel, lanes 3 and 6; right panel). This finding suggests that acetylation of YY1 is necessary for the maximum transcriptional repression activity of YY1. Western blot analysis showed that the loss of repression was not due to lack of expression of the mutant YY1 proteins (Fig. 3, bottom panel, compare lane 3 to lane 2 and lane 6 to lane 5). It is possible that lysine-to-arginine mutations result in a conformational change in YY1, which causes a lowered transcriptional repression activity. However, this would still suggest that the lysine residues within YY1 residues 170 to 200 are important for the full repression activity of YY1.
FIG. 3.
The effect of acetylation of YY1 residues 170 to 200 on the transcriptional repressor activity of YY1. HeLa cells were transfected with Ga14 DNA-binding domain alone (Gal4), Gal4-YY1 fusion construct (Gal4-YY1), or Gal4-YY1 mutant with the six lysines mutated to arginines [Gal4-YY1 (K170–200R)]. Transcriptional activities were analyzed by CAT assays using a CAT reporter containing five Gal4 binding sequences in tandem, and a representative autoradiogram is shown. Quantification of the relative CAT activities was performed using the PhosphoImager Storm system (model 860) and ImageQuant software (Molecular Dynamics). Western blot analyses were performed to verify the expression levels of the effector proteins.
Acetylation of YY1 residues 170 to 200 increases YY1 binding to HDACs.
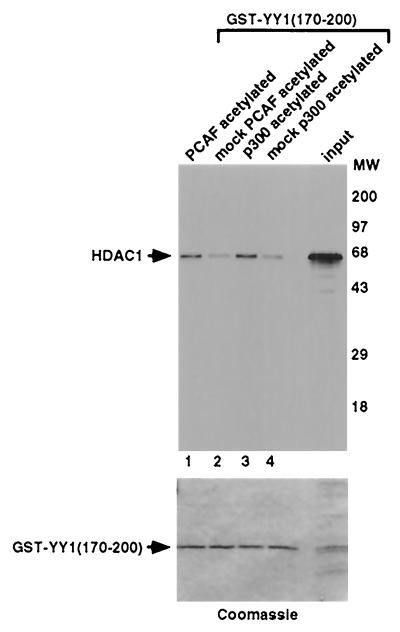
Using GST pull-down assays, we previously demonstrated that HDACs interact with YY1 residues 170 to 200 (73). Therefore, we asked if acetylation of YY1 in this region would affect YY1's interaction with HDACs. Figure 4 shows that [35S]methionine-labeled HDAC1 bound more efficiently to PCAF-acetylated and p300-acetylated GST-YY1(170–200) than to unacetylated GST-YY1(170–200) (compare lane 1 to lane 2 and lane 3 to lane 4). The same amount of GST-YY1(170–200) was used in all reactions as shown by Coomassie staining. Similar results were obtained using in vitro-transcribed and -translated HDAC2 (data not shown). In short, our data suggest that acetylation of YY1 at residues 170 to 200 significantly increases the binding of YY1 to HDACs.
FIG. 4.
Increased HDAC binding to YY1 residues 170 to 200 by acetylation. A representative autoradiogram of in vitro-translated HDAC1 captured by acetylated or mock-acetylated GST-YY1(170–200) is shown here. The input lane represents 1/10 the amount of HDAC1 used in each binding reaction. Reaction mixtures were separated by SDS-PAGE, and the gels were stained with Coomassie blue prior to exposure to film to confirm that equal amounts of GST-YY1(170–200) were used in the binding reactions.
HDAC1 and HDAC2 deacetylate YY1 residues 170 to 200 but not the C-terminal region of YY1.
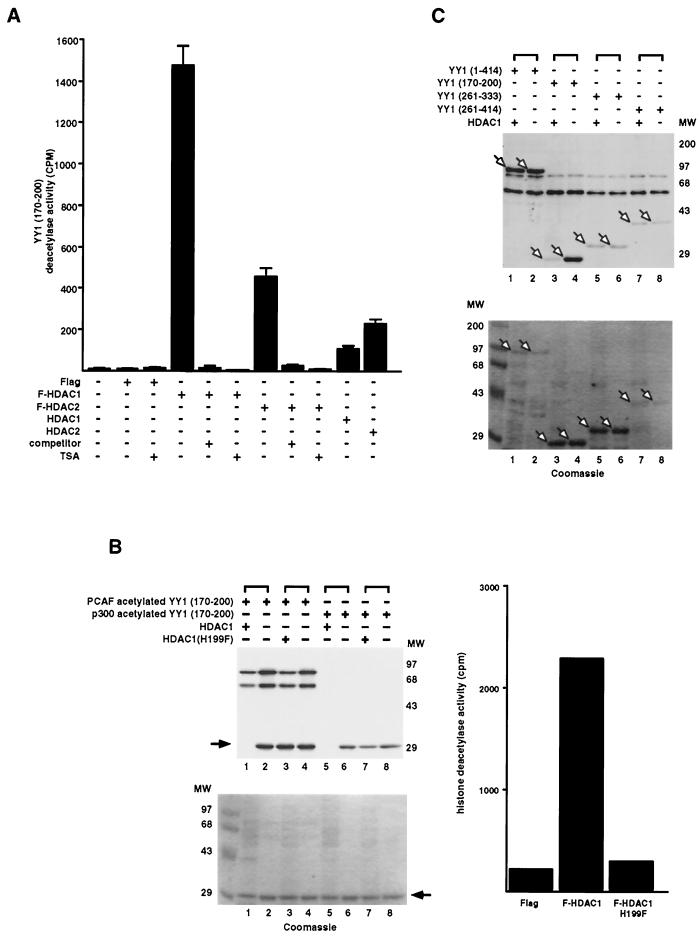
It is possible that the binding of HDACs to acetylated YY1(170–200) results in the subsequent deacetylation of YY1(170–200). To test this hypothesis, a synthetic peptide corresponding to YY1 residues 170 to 200 was chemically labeled with [3H]sodium acetate and used as a substrate in deacetylation assays. Figure 5A shows that immunopurified Flag-tagged HDAC1 and HDAC2 (F-HDAC1 and F-HDAC2) deacetylated the YY1(170–200) peptide. Deacetylation of YY1(170–200) by F-HDAC1 and F-HDAC2 was abolished by treatment with tricostatin A (TSA), a potent inhibitor of HDAC1 and HDAC2. Furthermore, if F-HDAC1 and F-HDAC2 were immunoprecipitated in the presence of an excess competitor, Flag peptide, deacetylase activity was not observed on the YY1 peptide. Similarly, endogenous HDAC1 and HDAC2 immunopurified from HeLa cells (HDAC1 and HDAC2) also deacetylated the YY1(170–200) peptide.
FIG. 5.
Identification of the region of YY1 deacetylated by HDACs. (A) YY1 peptide deacetylation by HDACs. Endogenous HDAC1 and HDAC2 and overexpressed F-HDAC1 and F-HDAC2 were immunoprecipitated from HeLa cells. A YY1 (residues 170 to 200) peptide was labeled with [3H]acetate, and 20,000 cpm of the labeled peptide was used in deacetylation reactions. (B) YY1 protein deacetylation by HDACs. (Left panels) GST-YY1(170–200) was in vitro acetylated by p300 or PCAF. Half of the acetylated GST-YY1(170–200) was mixed with protein G beads alone, and the other half was mixed with immunoprecipitated wild-type or mutant (H199F) HDAC1. Samples were then separated by SDS-PAGE, and the gels were subsequently stained with Coomassic blue and exposed to film. Arrows indicate the position of GST-YY1(170–200). (Right panel) Transiently expressed wild-type and H199F mutant HDAC1 were immunoprecipitated from HeLa cells and tested for their HDAC activity against an H4 peptide. (C) Deacetylation of YY1 serial deletion proteins by HDAC1. Serial deletion proteins of GST-YY1 were in vitro acetylated by PCAF. Half of the GST-YY1 proteins were mixed with protein G beads, and the other half were mixed with HDAC1 immunoprecipitate. Samples were then separated by SDS-PAGE, and the gels were subsequently stained with Coomassie blue and exposed to film. Arrows indicate the positions of GST-YY1 deletion proteins.
To provide further evidence that HDACs deacetylated YY1, we used an F-HDAC1 immunoprecipitate to deacetylate full-length GST-YY1 and GST-YY1(170–200), both of which were in vitro acetylated with PCAF. As expected, YY1(170–200) was efficiently deacetylated by HDAC1 (Fig. 5B, top panel, compare lane 1 to lane 2). As a control, YY1(170–200) was not deacetylated by an HDAC1 mutant that was devoid of deacetylation activity (22) (Fig. 5B, right panel and top panel, lane 3). A Coomassie-stained gel (bottom panel) showed that the same amount of YY1(170–200) was present in each reaction. Similarly, YY1(170–200) acetylated by p300 was also deacetylated by HDAC1 (Fig. 5B, compare lane 5 to lane 6). To our surprise, and in contrast to YY1(170–200), full-length YY1 was not appreciably deacetylated by HDAC1 (Fig. 5C, lane 1). This observation suggests that deacetylases only target specific regions of YY1. In support of this argument, we also found that HDAC1 did not deacetylate two additional PCAF-acetylated GST-YY1 proteins, YY1(261–333) and YY1(261–414) (Fig. 5C, lanes 5 and 7). We conclude that only residues 170 to 200 of YY1, and not the C-terminal zinc finger region of YY1, can be deacetylated by HDAC1 and HDAC2.
HDACs bind YY1 at multiple regions.
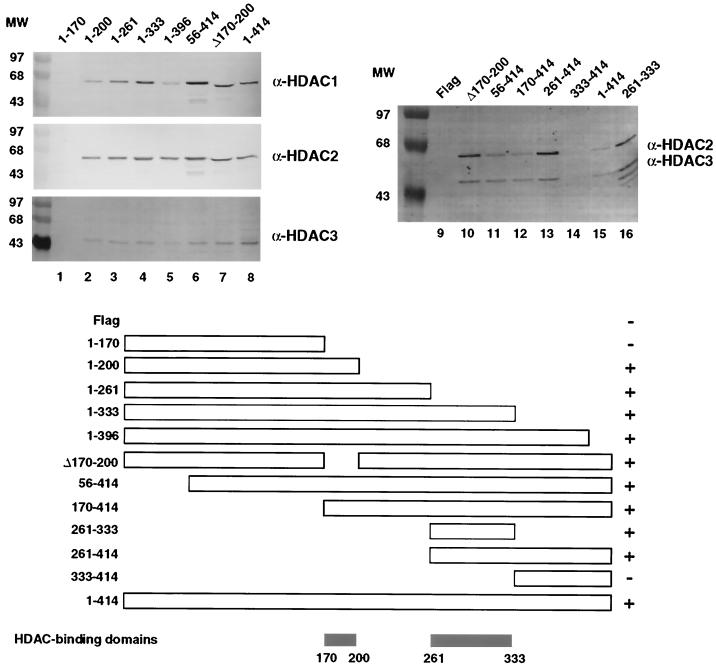
Based on our previous result that YY1 interacts with HDACs at residues 170 to 200 in vitro (73), the inability of HDAC1 to deacetylate the C-terminal zinc finger region of YY1 may arise from a general failure of HDACs to interact with YY1 in the zinc finger region. To test this possibility, we performed coimmunoprecipitation analyses to map the interaction domain of YY1 with HDACs in vivo (Fig. 6). Surprisingly, our results showed that in addition to residues 170–200, YY1 also interacted with HDACs at residues 261 to 333 in vivo. These results, together with those of our previous in vitro acetylation deacetylation experiment, suggest that YY1 interacts with HDACs at two domains: residues 170 to 200, where bound HDACs deacetylate YY1, and the C-terminal residues 261 to 333, where bound HDACs do not result in deacetylation of YY1.
FIG. 6.
Mapping of the HDAC interaction domains of YY1. Serial F-YY1 deletion constructs were transfected into HeLa cells and immunoprecipitated with anti-Flag antibody. Immunoprecipitated proteins were removed from the resin by competitive elution with excess Flag peptide and analyzed by Western blotting for the presence of HDAC1, HDAC2, and HDAC3 (lanes 1 to 8) or HDAC2 and HDAC3 only (lanes 9 to 16). The bottom panel summarizes the HDAC-binding domains of YY1. +, positive interactions between YY1 and HDACs; −, absence of YY1-HDAC interactions.
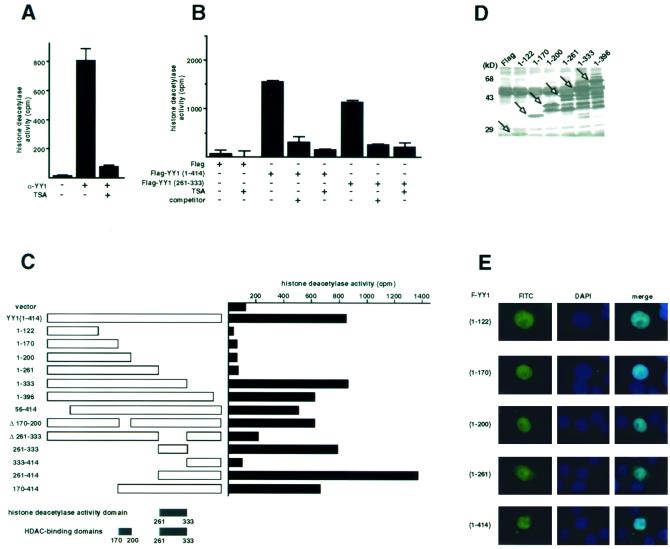
YY1 contains associated HDAC activity, which localizes to C-terminal residues 261 to 333 of YY1.
After we precisely mapped the HDAC-binding domains in YY1, we sought to determine the functional effect of this interaction. We found that immunoprecipitated endogenous YY1 from HeLa cells also contained histone deacetylase activity, which was inhibited by TSA (Fig. 7A). Using serial F-YY1 deletions, we determined the histone deacetylase activity domain of YY1, which localized to residues 261 to 333 (Fig. 7B and C). This region of YY1 was necessary and sufficient for the HDAC activity associated with YY1 (Fig. 7B and C). Most strikingly, the YY1 histone deacetylase activity domain completely overlapped with one of the HDAC-interacting domains of YY1. Furthermore, the HDAC activity associated with YY1 residues 261 to 333 was highly specific, because the activity was sensitive to TSA (Fig. 7B) and competed by excess Flag peptide (Fig. 7B). A representative Western blot shows that different F-YY1 deletion mutants expressed equally well (Fig. 7D). Immunofluorescence analysis confirmed that F-YY1 deletion proteins that did not exhibit HDAC activity localized to either the nucleus or both the nucleus and cytoplasm, ruling out the possibility that the lack of histone deacetylase activity was due to abnormal localization of the mutants (Fig. 7E). These results strongly suggest that stable interaction between HDACs and YY1 contributes to YY1's histone deacetylase activity.
FIG. 7.
Identification of the HDAC activity domain of YY1. (A) HDAC activity of endogenous YY1 in HeLa cells. Endogenous YY1 was immunoprecipitated from HeLa cells and assayed for deacetylase activity against the H4 peptide. Where indicated (+) TSA was added to 400 nM prior to addition of the peptide substrate. (B and C) Identification of aa 261 to 333 as the HDAC activity domain of YY1. F-YY1 deletion constructs were transfected into HeLa cells, immunoprecipitated with anti-Flag antibody, and assayed for deacetylase activity against the H4 peptide. Where indicated (+) TSA was added to 400 nM prior to addition of the peptide substrate or excess Flag peptides (competitor) were added prior to addition of the peptide substrate. The experiments in panel C were performed up to three times, with standard deviations less than or equal to 5%. (D) Expression of F-YY1 deletion mutants. Overexpressed F-YY1 deletion constructs and Flag alone from the parental vector were immunoprecipitated from HeLa cells using anti-Flag antibody, separated by SDS-PAGE, and analyzed by Western blotting using anti-Flag antibody. Arrows indicate the positions of F-YY1 deletion proteins. (E) Subcellular localization of F-YY1 deletion constructs. Various F-YY1 deletion constructs were transiently transfected into HeLa cells. Transfected HeLa cells were then fixed and probed with anti-Flag FITC conjugated antibody. The nuclei were stained with DAPI. Images were obtained from a fluorescence microscope. Merged images from FITC and DAPI stains indicate subcellular localization of F-YY1 deletion constructs.
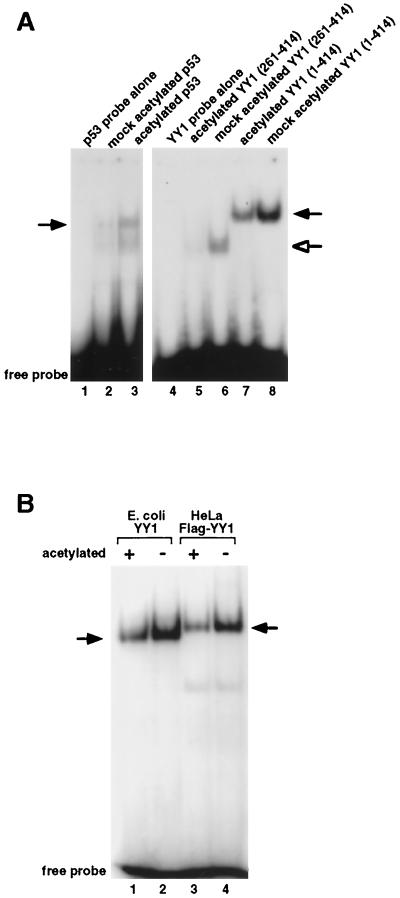
Acetylation of YY1 at the C-terminal zinc finger domain decreases the DNA-binding activity of YY1.
Because the C-terminal acetylation domain (residues 261 to 333) of YY1 overlaps with the zinc finger DNA-binding domain (residues 261 to 414) of YY1, we tested whether acetylation of YY1 at the zinc finger domain would affect the DNA-binding activity of YY1. Indeed, when in vitro acetylated by PCAF, GST-YY1 bound less avidly than did unacetylated GST-YY1 to the initiator element of the adeno-associated virus P5 promoter, which contains a consensus YY1 binding site (Fig. 8A, compare lane 7 to lane 8). Similar results were obtained when a GST-YY1 deletion construct that contained only the zinc finger domain of YY1 was tested for its DNA-binding properties (Fig. 8A, compare lane 5 to lane 6). In contrast, and consistent with earlier reports, acetylated GST-p53 bound to its recognition sequence better than unacetylated GST-p53 (Fig. 8A, compare lane 3 to lane 2). Thus, the decrease in DNA-binding activity caused by acetylation is specific to YY1. To rule out the possibility that this decrease in DNA-binding activity was associated with the GST fusion constructs, we tested nontagged YY1 purified from E. coli, as well as F-YY1 purified from HeLa cells. Both forms of YY1 exhibited decreased DNA-binding activity upon acetylation (Fig. 8B), proving that the DNA-binding activity of YY1 decreases when the zinc finger domain of YY1 is acetylated.
FIG. 8.
The effect of acetylation on YY1's sequence-specific DNA-binding activity. (A) Representative EMSA of GST-tagged YY1. Purified full-length GST-YY1(1–414), zinc finger domain of GST-YY1(261–414), and GST-p53 were in vitro acetylated with PCAF and mixed with 32P-labeled probes containing either a YY1 or p53-binding sequence. Mock-acetylated GST fusion proteins were treated identically as acetylated proteins with the exception that acetyl-CoA was omitted from the in vitro acetylation reactions. Protein-DNA complexes were resolved on nondenaturing polyacrylamide gels. Black arrows indicate the positions of full-length YY1- and p53-DNA complexes. The open arrow indicates the position of the complex between DNA and the zinc finger domain of YY1. (B) Representative EMSA of bacterially expressed nontagged YY1 and HeLa cell-expressed Flag-tagged YY1. Black arrows indicate the positions of the YY1-DNA complexes.
DISCUSSION
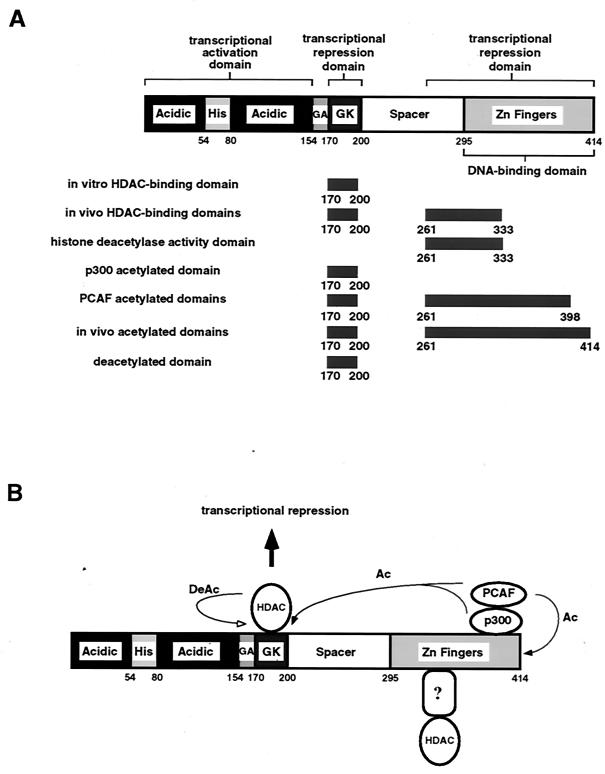
In this report, we demonstrate that the activity of the multifunctional transcription factor YY1 is regulated by acetylation and deacetylation. Acetylation and deacetylation of YY1 represent novel and complex means of regulating the activity of a DNA-binding transcription factor (Fig. 9). YY1 is acetylated in two regions, one at the previously identified HDAC-interacting domain of residues 170 to 200, and the other at the C-terminal DNA-binding zinc finger domain. Residues 170 to 200 of YY1 are acetylated by p300 and PCAF, while the C-terminal zinc finger domain is acetylated only by PCAF (Fig. 9A). Acetylation of these two regions results in dramatically different outcomes (Fig. 9). First, when residues 170 to 200 of YY1 are acetylated, YY1 becomes a more effective transcriptional repressor and binds HDACs more efficiently. However, upon binding to acetylated YY1 residues 170 to 200, HDACs also actively deacetylate this region, possibly resulting in a negative feedback loop. Second, YY1 possesses histone deacetylase activity toward histone H4 by associating with HDACs using the C-terminal zinc finger region. This is most likely a result of active targeting of acetylated YY1 zinc finger domains by HDACs. However, our data do not indicate that association of the YY1 C terminus with HDACs brings about deacetylation of YY1 in this region. In contrast, the interaction between YY1 and HDACs at residues 170–200 is likely to be a dynamic and highly regulated process and does not result in associated histone deacetylase activity stable enough to be detected in our experimental system. Finally, acetylation of YY1 zinc fingers decreases YY1's DNA-binding activity, which will have further impact on YY1's activity as a transcription factor.
FIG. 9.
Summary of regulation of YY1 by acetylation and deacetylation. (A) Summary of different domains of YY1 affected by acetylation and deacetylation. His, histidine-rich domain; GA, glycine-alanine-rich domain; GK, glycine-lysine-rich domain. (B) YY1 is acetylated in two regions: residues 170 to 200 and the C-terminal DNA-binding domain. Thin arrows represent YY1 protein modifications: arrows with solid heads represent acetylation (Ac), while an arrow with an empty head represents deacetylation (DeAc). Acetylation of YY1 at residues 170 to 200 by p300 and PCAF augments YY1's repressor activity (shown as a thick arrow leading to transcriptional repression), but acetylation of this region also targets YY1 for deacetylation, resulting in negative feedback regulation. Acetylation of the C terminus of YY1 results in stable association of histone deacetylase activity with YY1 as well as decreased DNA-binding activity. The in vivo association between YY1 and HDACs at the C-terminal region is probably mediated through an unidentified protein (depicted as a question mark in a rounded rectangle).
This differential regulation of YY1 by acetylation and deacetylation suggests a complex regulatory system unlike that of any other transcription factor known to date. In cells where YY1 functions primarily as a transcriptional repressor, acetylation of YY1 at the central HDAC-binding region and the C-terminal DNA-binding region most likely will result in an intricate network of negative-feedback regulation (Fig. 9B): acetylation of YY1 at residues 170 to 200 augments YY1's repressor activity, but acetylation of this region also targets YY1 for deacetylation. In the meantime, acetylation of the C-terminal DNA-binding region of YY1 most likely stabilizes interaction between YY1 and HDACs, but acetylation of this region in turn decreases YY1's DNA-binding activity. To date, we have not been able to determine the relative levels of acetylation between the central region and the C terminus of YY1 under physiological conditions; therefore, the relative contribution of the acetylation of these two regions is unknown. Interestingly, we also found that TSA had little effect on the acetylation status of YY1 (data not shown), suggesting that regulation of YY1 by acetylation is more prominent than by deacetylation. This finding is also in agreement with our discovery that deacetylation of YY1 only occurs at residues 170 to 200, while acetylation of YY1 can happen at the C-terminal DNA-binding domain as well.
In many experimental systems, YY1 can also activate transcription, and it is still uncertain how this is accomplished. Different models, including bending of DNA, the relative distance between the YY1 binding site and the transcriptional initiation site, as well as protein-protein interactions, have been proposed (reviewed in references 57 and 65). Increasingly, more evidence shows that interactions with other proteins are probably the most important factors in YY1-mediated transcriptional activation. It has been suggested that interactions of YY1 with other cellular proteins or viral proteins can either disrupt the quenching activity of YY1 on other transcriptional activators or stimulate transcription with associated enzymatic activities such as HATs (36, 58, 65). Our findings here provide an additional speculation that on promoters activated by YY1, YY1-associated p300 and PCAF can activate transcription by both acetylating core histones and acetylating YY1 at the C-terminal zinc finger domain (PCAF only), which in turn decreases the overall histone deacetylation activity at the promoter.
In addition to histones and several nonhistone chromatin proteins, many transcription factors have been shown to be regulated by acetylation. These transcription factors include p53 (18), human immunodeficiency virus Tat (32), E1A (77), GATA-1 (5), EKLF (78), MyoD (50), E2F (43), TFIIE, TFIIF (28), CIITA (62), TCF (71), HNF-4 (61), UBF (48), TAL1 (also known as SCL) (25), and nuclear receptor coactivators ACTR, SRC-1, and TIF2 (10). Many of these factors are acetylated by both p300 and PCAF; therefore, it is not surprising that YY1 is also acetylated by both p300 and PCAF. The p300-interaction domain of YY1 has been mapped to the C-terminal 17 residues (36, 38), which were not acetylated by p300 in our study. This finding is reminiscent of acetylation of p53 by p300, in which the N terminus of p53 interacts with CBP (19, 41) while the region acetylated by p300 is at the C terminus of p53 (18). Moreover, in our in vitro acetylation studies, full-length GST-YY1 was acetylated by PCAF and not by p300, which suggests that the conformation of YY1, perhaps affected by selective interaction with p300 and PCAF, is important to YY1 acetylation. We also found that in HeLa cells, overexpression of PCAF, but not p300, partially alleviated the transcriptional repressor activity of a Gal4 DNA-binding domain-YY1 fusion protein. However, in NIH 3T3 cells, overexpression of p300, but not PCAF, relieved repression from Gal4-YY1 (data not shown). In this regard, it will be important to identify the in vivo triggers directing PCAF versus p300 acetylation. p53 is particularly interesting among acetylated transcription factors because it has been elegantly demonstrated that DNA damage (UV and ionizing radiation) causes p53 acetylation at two distinct lysines, one by p300 and the other by PCAF (42). To date, this is the only report linking specific environmental cues to acetylation of transcription factors by HATs, and yet it is still uncertain why acetylation would require two HATs. It is interesting that while an increase in DNA-binding activity has been observed for most acetylated DNA-binding transcription factors, HMG I(Y) binds DNA less when it is acetylated (45). The most striking observation is that only when HMG I(Y) is acetylated by CBP, not by PCAF, is there a decrease in its DNA-binding activity (45). This phenomenon is reminiscent of YY1 acetylation in its zinc finger domain by PCAF but not by p300 and the consequent reduction in the DNA-binding activity of YY1.
We were also interested in finding out if acetylation of YY1 also contributes to cellular events other than transcriptional control. So far, we have no evidence to suggest that acetylation changes the subcellular localization of YY1. YY1 has been shown to be a rather stable protein expressed at comparable levels in both growing and differentiating cells (1). Interestingly, acetylation affects the conformation of HNF-4 (61), the half-life of E2F (43), and promotes protein-protein interactions between Rch1 and importin-β (3). Therefore, it will be informative to test whether acetylation of YY1 may have similar consequences.
Our data demonstrating that YY1 interacts with HDACs at two different regions open the possibility that these two HDAC-interacting regions have distinct effects on YY1's role in transcriptional control. Recently a Drosophila homolog of YY1 was identified as PHO, which is encoded by pleiohomeotic, a member of the Polycomb group (PcG) genes (6). It has been proposed that the Drosophila YY1 homolog, PHO, binds to PcG response elements and interacts with other proteins to form a repressor complex with nucleosome remodeling activity (6, 31, 69, 70). YY1, PHO, and the Xenopus YY1 homolog FIII are almost completely identical in the zinc finger region in amino acid sequence, reinforcing the role of YY1 as a DNA-targeting factor in nucleating a repressor complex capable of modulating chromatin structures. The stable association of YY1 with HDACs at the C-terminal zinc finger region might represent an ancient mode of the functions of YY1, which is to form a repressor complex associated with the promoter. However, regulation of the affinity of the central region of YY1 for HDACs by acetylation might have evolved as a more sophisticated means of control, which defines a novel functional consequence of nonhistone factor acetylation.
ACKNOWLEDGMENTS
We thank Shelly Berger, Nobuo Horikoshi, and Stuart Schreiber for their generous gifts of plasmids; Nancy Olashaw, Rosalind Jackson, and Peter Neame for critical reading of the manuscript; and the core facilities at the Moffitt Cancer Center as well as the Protein Chemistry Core at the University of Florida for technical support. We also thank Jim Davie and his laboratory for their help in setting up preliminary experiments involving HDAC activity associated with YY1.
Y.-L.Y. was supported by a fellowship from the American Heart Association. This work was supported by a grant from the National Institutes of Health (GM58486) to E.S.
REFERENCES
- 1.Austen M, Luscher B, Luscher F J. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J Biol Chem. 1997;272:1709–1717. doi: 10.1074/jbc.272.3.1709. [DOI] [PubMed] [Google Scholar]
- 2.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 3.Bannister A J, Miska E A, Gorlich D, Kouzarides T. Acetylation of importin-alpha nuclear import factors by CBP/p300. Curr Biol. 2000;10:467–470. doi: 10.1016/s0960-9822(00)00445-0. [DOI] [PubMed] [Google Scholar]
- 4.Bauknecht T, See R H, Shi Y. A novel C/EBP beta-YY1 complex controls the cell-type-specific activity of the human papillomavirus type 18 upstream regulatory region. J Virol. 1996;70:7695–7705. doi: 10.1128/jvi.70.11.7695-7705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 6.Brown J L, Mucci D, Whiteley M, Dirksen M L, Kassis J A. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 7.Bushmeyer S, Park K, Atchison M L. Characterization of functional domains within the multifunctional transcription factor, YY1. J Biol Chem. 1995;270:30213–30220. doi: 10.1074/jbc.270.50.30213. [DOI] [PubMed] [Google Scholar]
- 8.Bushmeyer S M, Atchison M L. Identification of YY1 sequences necessary for association with the nuclear matrix and for transcriptional repression functions. J Cell Biochem. 1998;68:484–499. [PubMed] [Google Scholar]
- 9.Candau R, Zhou J X, Allis C D, Berger S L. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 11.Chiang C M, Roeder R G. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- 12.Cress W D, Seto E. Histone deacetylases, transcriptional control, and cancer. J Cell Physiol. 2000;184:1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 13.Donohoe M E, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flanagan J R, Becker K G, Ennist D L, Gleason S L, Driggers P H, Levi B Z, Appella E, Ozato K. Cloning of a negative transcription factor that binds to the upstream conserved region of Moloney murine leukemia virus. Mol Cell Biol. 1992;12:38–44. doi: 10.1128/mcb.12.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giordano A, Avantaggiati M L. p300 and CBP: partners for life and death. J Cell Physiol. 1999;181:218–230. doi: 10.1002/(SICI)1097-4652(199911)181:2<218::AID-JCP4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 16.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 18.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 19.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 20.Hariharan N, Kelley D E, Perry R P. Delta, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc Natl Acad Sci USA. 1991;88:9799–9803. doi: 10.1073/pnas.88.21.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harlow E, Lane D. Using antibodies: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 22.Hassig C A, Tong J K, Fleischer T C, Owa T, Grable P G, Ayer D E, Schreiber S L. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci USA. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebbes T R, Thorne A W, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong L, Schroth G P, Matthews H R, Yau P, Bradbury E M. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J Biol Chem. 1993;268:305–314. [PubMed] [Google Scholar]
- 25.Huang S, Qiu Y, Shi Y, Xu Z, Brandt S J. P/CAF-mediated acetylation regulates the function of the basic helix-loop-helix transcription factor TAL1/SCL. EMBO J. 2000;19:6792–6803. doi: 10.1093/emboj/19.24.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huibregtse J M, Scheffner M, Howley P M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung H L, Lau J, Kim A Y, Weiss M J, Blobel G A. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol Cell Biol. 1999;19:3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 29.Inouye C J, Seto E. Relief of YY1-induced transcriptional repression by protein-protein interaction with the nucleolar phosphoprotein B23. J Biol Chem. 1994;269:6506–6510. [PubMed] [Google Scholar]
- 30.Jones K A, Kadonaga J T. Exploring the transcription-chromatin interface. Genes Dev. 2000;14:1992–1996. [PubMed] [Google Scholar]
- 31.Kehle J, Beuchle D, Treuheit S, Christen B, Kennison J A, Bienz M, Muller J. dMi-2, a hunchback-interacting protein that functions in polycomb repression. Science. 1998;282:1897–1900. doi: 10.1126/science.282.5395.1897. [DOI] [PubMed] [Google Scholar]
- 32.Kiernan R E, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang K T, Benkirane M, Van Lint C. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J. 1999;18:6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 34.Kornberg R D, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 35.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 36.Lee J S, Galvin K M, See R H, Eckner R, Livingston D, Moran E, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 37.Lee J S, Galvin K M, Shi Y. Evidence for physical interaction between the zinc-finger transcription factors YY1 and Spl. Proc Natl Acad Sci USA. 1993;90:6145–6149. doi: 10.1073/pnas.90.13.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J S, See R H, Galvin K M, Wang J, Shi Y. Functional interactions between YY1 and adenovirus E1A. Nucleic Acids Res. 1995;23:925–931. doi: 10.1093/nar/23.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee T C, Zhang Y, Schwartz R J. Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene. 1994;9:1047–1052. [PubMed] [Google Scholar]
- 40.Lewis B A, Tullis G, Seto E, Horikoshi N, Weinmann R, Shenk T. Adenovirus E1A proteins interact with the cellular YY1 transcription factor. J Virol. 1995;69:1628–1636. doi: 10.1128/jvi.69.3.1628-1636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 42.Liu L, Scolnick D M, Trievel R C, Zhang H B, Marmorstein R, Halazonetis T D, Berger S L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez-Balbas M A, Bauer U M, Nielsen S J, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marzio G, Wagener C, Gutierrez M I, Cartwright P, Helin K, Giacca M. E2F family members are differentially regulated by reversible acetylation. J Biol Chem. 2000;275:10887–10892. doi: 10.1074/jbc.275.15.10887. [DOI] [PubMed] [Google Scholar]
- 45.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 46.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 47.Park K, Atchison M L. Isolation of a candidate repressor/activator, NF-E1 (YY-1, delta), that binds to the immunoglobulin kappa 3′ enhancer and the immunoglobulin heavy-chain mu E1 site. Proc Natl Acad Sci USA. 1991;88:9804–9808. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pelletier G, Stefanovsky V Y, Faubladier M, Hirschler-Laszkiewicz I, Savard J, Rothblum L I, Cote J, Moss T. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol Cell. 2000;6:1059–1066. doi: 10.1016/s1097-2765(00)00104-0. [DOI] [PubMed] [Google Scholar]
- 49.Pisaneschi G, Ceccotti S, Falchetti M L, Fiumicino S, Carnevali F, Beccari E. Characterization of FIII/YY1, a Xenopus laevis conserved zinc-finger protein binding to the first exon of L1 and L14 ribosomal protein genes. Biochem Biophys Res Commun. 1994;205:1236–1242. doi: 10.1006/bbrc.1994.2797. [DOI] [PubMed] [Google Scholar]
- 50.Polesskaya A, Duquet A, Naguibneva I, Weise C, Vervisch A, Bengal E, Hucho F, Robin P, Harel-Bellan A. CREB-binding protein/p300 activates MyoD by acetylation. J Biol Chem. 2000;275:34359–34364. doi: 10.1074/jbc.M003815200. [DOI] [PubMed] [Google Scholar]
- 51.Ron D, Dressler H. pGSTag—a versatile bacterial expression plasmid for enzymatic labeling of recombinant proteins. BioTechniques. 1992;13:866–869. [PubMed] [Google Scholar]
- 52.Sadowski I, Bell B, Broad P, Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 53.Sartorelli V, Puri P L, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang J Y, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 54.Sealy L, Chalkley R. DNA associated with hyperacetylated histone is preferentially digested by DNase I. Nucleic Acids Res. 1978;5:1863–1876. doi: 10.1093/nar/5.6.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seto E, Lewis B, Shenk T. Interaction between transcription factors Sp1 and YY1. Nature. 1993;365:462–464. doi: 10.1038/365462a0. [DOI] [PubMed] [Google Scholar]
- 56.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 57.Shi Y, Lee J S, Galvin K M. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 58.Shi Y, Seto E, Chang L S, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 59.Shrivastava A, Calame K. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shrivastava A, Saleque S, Kalpana G V, Artandi S, Goff S P, Calame K. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science. 1993;262:1889–1892. doi: 10.1126/science.8266081. [DOI] [PubMed] [Google Scholar]
- 61.Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 62.Spilianakis C, Papamatheakis J, Kretsovali A. Acetylation by PCAF enhances CIITA nuclear accumulation and transactivation of major histocompatibility complex class II genes. Mol Cell Biol. 2000;20:8489–8498. doi: 10.1128/mcb.20.22.8489-8498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sterner D E, Berger S L. Acetylation of histories and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 65.Thomas M J, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 66.Tsai S C, Valkov N, Yang W M, Gump J, Sullivan D, Seto E. Histone deacetylase interacts directly with DNA topoisomerase II. Nat Genet. 2000;26:349–353. doi: 10.1038/81671. [DOI] [PubMed] [Google Scholar]
- 67.Usheva A, Shenk T. TATA-binding protein-independent initiation: YY1, TFIIB, and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell. 1994;76:1115–1121. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]
- 68.Vidali G, Boffa L C, Bradbury E M, Allfrey V G. Butyrate suppression of histone deacetylation leads to accumulation of multiacetylated forms of histones H3 and H4 and increased DNase I sensitivity of the associated DNA sequences. Proc Natl Acad Sci USA. 1978;75:2239–2243. doi: 10.1073/pnas.75.5.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wade P A, Jones P L, Vermaak D, Veenstra G J, Imhof A, Sera T, Tse C, Ge H, Shi Y B, Hansen J C, Wolffe A P. Histone deacetylase directs the dominant silencing of transcription in chromatin: association with MeCP2 and the Mi-2 chromodomain SWI/SNF ATPase. Cold Spring Harbor Symp Quant Biol. 1998;63:435–445. doi: 10.1101/sqb.1998.63.435. [DOI] [PubMed] [Google Scholar]
- 70.Wade P A, Jones P L, Vermaak D, Wolffe A P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 71.Waltzer L, Bienz M. Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature. 1998;395:521–525. doi: 10.1038/26785. [DOI] [PubMed] [Google Scholar]
- 72.Wen Y D, Perissi V, Staszewski L M, Yang W M, Krones A, Glass C K, Rosenfeld M G, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci USA. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang W M, Inouye C J, Seto E. Cyclophilin A and FKBP12 interact with YY1 and alter its transcriptional activity. J Biol Chem. 1995;270:15187–15193. doi: 10.1074/jbc.270.25.15187. [DOI] [PubMed] [Google Scholar]
- 75.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 76.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Q, Yao H, Vo N, Goodman R H. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc Natl Acad Sci USA. 2000;97:14323–14328. doi: 10.1073/pnas.011283598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang W, Bieker J J. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou Q, Gedrich R W, Engel D A. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J Virol. 1995;69:4323–4330. doi: 10.1128/jvi.69.7.4323-4330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu X, Mancini M A, Chang K H, Liu C Y, Chen C F, Shan B, Jones D, Yang-Feng T L, Lee W H. Characterization of a novel 350-kilodalton nuclear phosphoprotein that is specifically involved in mitotic-phase progression. Mol Cell Biol. 1995;15:5017–5029. doi: 10.1128/mcb.15.9.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]