Abstract
Decades of research have probed the interplay between chromatin (genomic DNA associated with proteins and RNAs) and transcription by RNA polymerase (RNAP) in all domains of life. In bacteria, chromatin is compacted into a membrane-free region known as the nucleoid that changes shape and composition depending on the bacterial state. Transcription plays a key role in both shaping the nucleoid and organizing it into domains. At the same time, chromatin impacts transcription by at least five distinct mechanisms: (i) occlusion of RNAP binding; (ii) roadblocking RNAP progression; (iii) constraining DNA topology; (iv) RNA-mediated interactions; and (v) macromolecular demixing and heterogeneity, which may generate phase-separated condensates. These mechanisms are not mutually exclusive and, in combination, mediate gene regulation. Here, we review the current understanding of these mechanisms with a focus on gene silencing by H-NS, transcription coordination by HU, and potential phase separation by Dps. The myriad questions about transcription of bacterial chromatin are increasingly answerable due to methodological advances, enabling a needed paradigm shift in the field of bacterial transcription to focus on regulation of genes in their native state. We can anticipate answers that will define how bacterial chromatin helps coordinate and dynamically regulate gene expression in changing environments.
Keywords: Nucleoid, RNA polymerase, supercoiling, topological stress, phase separation
Introduction
Compaction of DNA by DNA-binding proteins to form chromatin occurs in all forms of life. DNA compaction confines genomic DNA to an appropriate sub-cellular space, helps preserve the integrity of the genome, and helps determine the expression of genetic information. In eukaryotes and archaea, these functions are achieved by histones that assemble into regular, repeating structures [1–4]. The role of these structures in mediating gene expression is best understood in eukaryotes, where extensive post-translational modifications by chromatin regulators alter the properties of discrete, octa-histone nucleosomes to repress transcription when tightly packed or to allow transcription initiation in nucleosome-free promoter regions and elongation though modified nucleosomes. The comparable structuring, preserving, and expression-mediating functions of DNA compaction in bacteria, however, remain poorly understood. In part, the currently incomplete picture of bacterial chromatin reflects the near-bewildering variety of different DNA-binding proteins that associate with bacterial DNA in diverse and often poorly defined structures and patterns. This extreme heterogeneity of bacterial chromatin has made understanding its structure–function relationships an exceptional challenge despite much progress by a diligent community of researchers over many years.
Although many important questions remain about the molecular structure of bacterial chromatin, in this review we seek to summarize current understanding of a more narrowly defined topic: what basic mechanisms underlie the interplay between transcription of bacterial chromatin by RNA polymerase (RNAP) and the nucleoprotein structures that compact the bacterial genome. To review this topic from an RNAP-centric perspective, we will first briefly summarize relevant features of transcription and current models of bacterial chromatin structure from a transcriptional perspective. We refer the reader to the many recent and excellent reviews that cover bacterial chromatin structure more comprehensively [5–14]. We will then discuss five ways that these structures affect transcription: (1) occlusion of RNAP, (2) roadblocking RNAP progression, (3) changes in DNA supercoiling, (4) RNA-mediated interactions, and (5) macromolecular demixing (heterogeneity), condensation, and phase separation. Now is a particularly apt time to review and define key questions about transcription of bacterial chromatin because exciting recent advances provide new perspectives and because new approaches are now possible.
Multiple steps in transcription are important to understanding bacterial chromatin
Transcription is a highly regulated, multi-step process and thus bacterial chromatin may cause different effects at different steps. We will briefly highlight the relevant mechanisms, which are reviewed in detail elsewhere [15–17] and in this issue [18, 19]. Transcription is initiated when RNAP and an associated sigma (σ) factor recognize and bind an AT-rich promoter element containing canonical sequence elements (e.g., −10 and −35 hexamers) upstream of a transcription start site [20]. Once bound, the σ factor facilitates opening of the double helix at the transcription start site by unwinding ~1 helical repeat to form an open complex. RNAP then polymerizes templated ribonucleotides into an RNA chain. Once the chain is ≥~8 nt, RNAP escapes the promoter, releases σ, and continues RNA chain extension as an active elongation complex (EC) at 30–100 nt·s−1.
Elongation is punctuated by sequence-dependent pauses of ≥1 s every 100 base pairs (bp) on average [15, 16, 21–23]. Pausing, which occurs by multiple related mechanisms, helps regulate RNA synthesis by allowing transcription–translation coupling, properly timing the interaction of transcription factors (TFs) and small molecules, and aiding correct RNA folding [16]. Of particular relevance to interactions with bacterial chromatin, pausing can lead to and be prolonged by backtracking of RNA and DNA through RNAP so that the 3′ end of the RNA becomes disengaged from the enzyme’s active site [22, 24, 25]. Escape from a backtracked pause can be facilitated by cleavage factors (e.g., GreA and GreB), which stimulate cleavage of the displaced RNA to generate a new 3′ end in the active site [26].
Transcription terminates by one of two mechanisms that ultimately collapse the melted DNA bubble in an EC: intrinsic termination or rho (ρ)-dependent termination [17]. Intrinsic termination relies on specific sequence elements (a terminator RNA hairpin and a 3′ U-tract) that destabilize the EC and release the transcript and RNAP from DNA. ρ-dependent termination relies on the termination factor ρ, which binds to unstructured, C-rich RNA and then translocates RNA in a 3′-to-5′ direction to reach RNAP at a pause site where it dissociates the EC. After termination, the dissociated RNAP rebinds σ and is then available for the next round of transcription.
In vivo, transcription occurs on topologically constrained DNA that typically is negatively supercoiled in bacteria. Supercoiling is the over- or under-winding of the DNA helix relative to its naturally relaxed conformation in a short linear DNA (i.e., ~10.4 bp per helical turn). Over- or under-winding of DNA (supercoiling) can manifest either as a twist along the helical axis of DNA or as wrapping of the helix around itself in the form of plectonemes or toroids (Fig. 1). This wrapping is referred to as writhe. Twist (Tw) and writhe (Wr) can interconvert. Thus, the topological state of DNA is specified by the linking number (Lk; the number of times the helical axis crosses itself), which is a constant in a DNA segment between fixed points and follows the equation Lk = Tw + Wr, where changes from one Lk per 10.4 bp in relaxed B-form DNA are called supercoiling. Both twist and writhe can diffuse along bare or unconstrained DNA, but diffusion of supercoils can be blocked either by chromatin proteins or by RNAP (Fig. 1).
Figure 1. Bacterial chromatin.
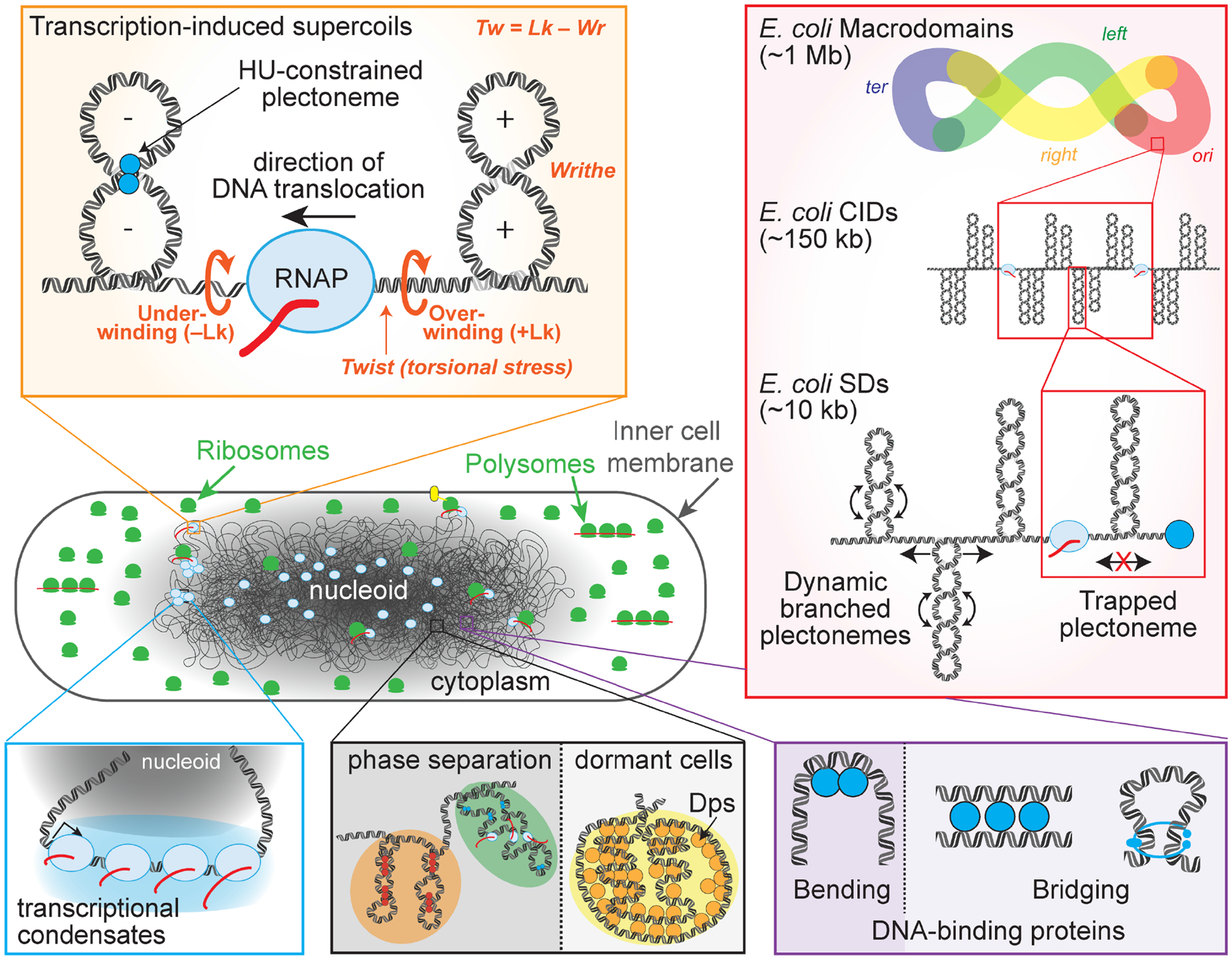
(Left center) The nucleoid in a growing bacterial cell shortly after cell division, which contains DNA (black), a set of abundant DNA-binding proteins, transcribing RNAP (blue), RNA (red), and ribosomes participating in co-transcriptional translation (green). The nucleoid is compacted into ~25% of the cell volume and is surrounded by cytoplasmic RNAs, proteins, polysomes, and membrane proteins, some of which are inserted co-translationally (transertion; yellow oval). In addition to chromatin proteins listed in Table 1, a freshly divided E. coli contains ~8,000 RNAPs (~5,000 ECs) and ~45,000 ribosomes (~10–15% of which are in the nucleoid). Various types of protein–DNA complexes help compact and organize the nucleoid, clockwise beginning top left as follows. (Orange box) Transcription supercoils DNA in a (+) direction (over-winding) downstream of RNAP and (−) direction (under-winding) upstream because neither transcribing RNAP nor DNA can rotate freely (called the ‘twin supercoiled domain model’). Supercoiling changes the linking number (Lk) of DNA (number of times DNA strands cross), which can be manifest as either twist (Tw) or writhe (Wr; Lk=Tw+Wr). Writhe generates either plectonemes or toroids and can be constrained (locked in place) by DNA-binding proteins like HU. (Red box) Nested bacterial chromatin domains. Four large domains with greater internal interactions have been proposed: ori, right, ter, and left. Within these macrodomains, more highly interacting chromosomal interaction domains (CIDs) and topologically isolated “supercoiled” domains (SDs) have been defined (see text). The global negatively supercoiled state of the bacterial chromosome generates plectonemes within these domains, which can be dynamic (double sided arrow) unless trapped by DNA-binding proteins or RNAP. (Purple box) DNA binding proteins (blue circles) can bend or bridge DNA to aid compaction and limit diffusion of supercoils. (Gray box) Some DNA-binding proteins can compact the DNA into an apparently phase-separated condensate (e.g., Dps in dormant cells). Other condensates may arise in growing cells in areas of high transcription or silenced transcription and be modulated by the cellular environment (e.g., the green phase with H-NS silencing and orange phase with transcription and HU binding). (blue box) Highly transcribed regions of the nucleoid may partition at the periphery of the nucleoid as a transcriptional condensate.
Neither DNA nor RNAP are free to rotate in cells because DNA is both continuous and bound by many proteins and because the nascent RNA is bound by ribosomes or other proteins. As a consequence, transcription generates positive (+) supercoils in front of the EC and negative (−) supercoils behind the EC (+1 and −1 Lk for every ~10 bp transcribed), known as the twin-supercoiled domain model (Fig. 1; [27–29]).
These topological constraints have widespread consequences for both transcription and the structure of bacterial chromatin [30–32]. Over- or under-twisting of DNA gives rise to torsion, which strongly affects RNAP at all steps of transcription. Because (−) supercoiling favors DNA unwinding and transcription initiation requires melting ~10 bp of DNA, (−) supercoiling (e.g., as generated upstream of a transcribing EC) favors transcription initiation (Fig. 2a). Conversely, (+) supercoiling inhibits initiation. Open complexes constrain ~1 (−) supercoil [33]. Because the bubble collapses during termination, the same effects mean (−) supercoiling inhibits and (+) supercoiling favors termination. During elongation, the EC also constrains ~1 (−) supercoil; however, the net (+) twist generated in front of RNAP and (−) twist generated behind RNAP oppose forward translocation and favor backtracking (Fig. 2a). RNAP can transcribe against a maximum of ~11 pN·nm of torque before it pauses and eventually backtracks [28]; GreB, which rescues backtracked ECs by transcript cleavage, increases the torque required to halt RNAP to ~18 pN·nm [34]. Other DNA-binding proteins can also modulate these torque effects either by constraining supercoils as writhe or blocking supercoil diffusion. Supercoils that are constrained into toroids or plectonemes by DNA-binding proteins (e.g. histones in nucleosomes or the bacterial chromatin protein HU) lessen twist and relieve torsion, which can aid transcription (see “DNA-binding proteins organize DNA” section). The recent discovery of a DNA-binding protein in Caulobacter that specifically targets (+) supercoiling, including in front of ECs, highlights the connection between the topological effects of transcription and the binding of chromatin proteins [35]. Topoisomerases, which can either relax (e.g., topoisomerase I) or increase (e.g., gyrase) supercoiling also aid transcription and maintain the overall (−) supercoiled state of the bacterial genomes [30, 32, 36, 37]. Overall, the dynamic interplay among RNAP, supercoiling, and DNA-binding proteins is integral to transcription of bacterial chromatin.
Figure 2. Supercoiling and DNA-binding proteins create domains that impact transcription.
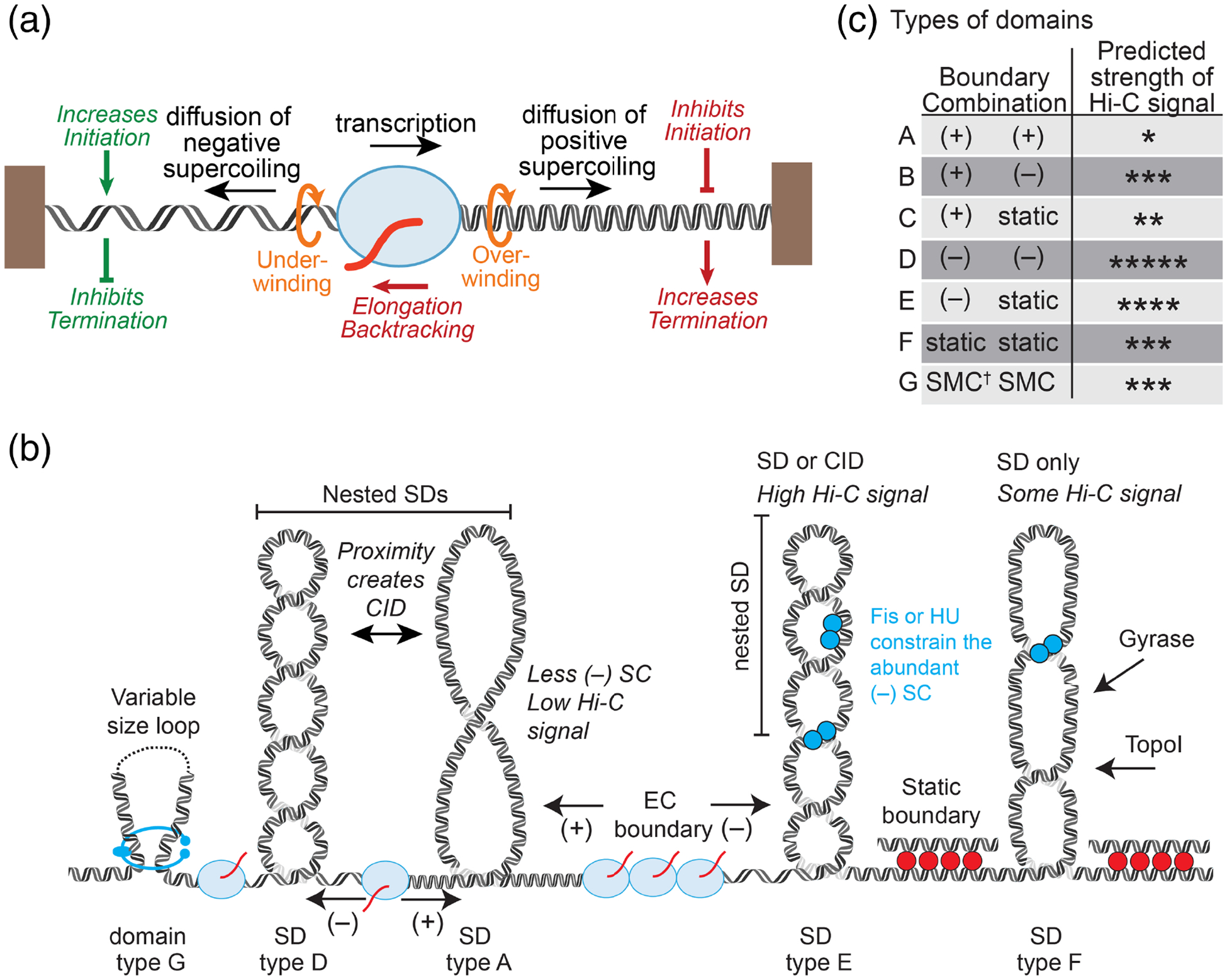
(a) Torsional stress generated by ECs directly impacts multiple steps in transcription. Both (+) supercoiling in front and (−) supercoiling behind ECs slow forward translocation, which slows elongation and favors backtracking. Upstream (−) supercoiling aids DNA melting and thus other transcription events by increasing initiation and inhibiting termination. Downstream (+) supercoiling inhibits DNA melting and thus other transcription events by inhibiting initiation and aiding termination. (b) Both CIDs and SDs are topologically isolated, but active versus static protein boundaries impact their properties differently. Both active (ECs; blue oval) and static (DNA-binding proteins; e.g., bridged H–NS filaments; red circles) block diffusion of supercoils, but active ECs will alter the supercoiling within a domain whereas static barriers will not generate supercoils. These differences in supercoiling could lead to a SD being classified as a CID if it is highly plectonemic or not if it is more relaxed. Nesting of SDs within CIDs may reflect interactions between SDs. Constraint of supercoils by Fis or HU could also create SDs. (c) Because ECs generate either (+) or (−) supercoiling within a domain depending on the direction of transcription, seven types of domains can be defined from four types of boundaries, leading to different probabilities that a domain will be highly interacting. Predicted strength of Hi-C signal (i.e., extent of inter-segment DNA interactions) are ranked from low (*) to high (*****). SMC boundaries differ in that they create isolated interaction domains without any apparent constrain of supercoils.
Bacterial chromatin is structured by diverse regulators and transcription
Bacterial chromatin consists of DNA, RNA, and proteins that condense to form the nucleoid
Bacterial genomic DNA, which is typically ~2–6 Mbp despite outliers ranging from 0.1 Mbp in an obligate symbiont to 14 Mbp in an omnivorous myxobacterium [38, 39], must be compacted dramatically to fit inside a cell let alone the smaller nucleoid region. Uncompacted, a 5 Mbp DNA would form a random coil with a volume nearly 103 times larger than a typical cell volume, which is 0.4–3 femtoliters [8, 40]. Instead, the genome, associated proteins, and RNA compact into a central, membrane-free space called the nucleoid that takes up ~15–25% of the cell volume [8, 41]. The surrounding cell volume is occupied by cytoplasmic proteins and RNA, mostly in the form of ribosomes in an actively growing cell (Fig. 1; [6]). This ≥2,000-fold compaction (relative to an uncompacted, random coil) requires physical constraints in addition to the surrounding cytoplasmic and outer membranes to create the subcellular nucleoid. Both experiments and computational analyses suggest that chromatin organization is mediated by the combined effects of supercoiling [42], DNA-binding proteins [5], transcription [43], molecular crowding, electrostatics, and macromolecular demixing (heterogeneity) driven by weak differential chemical affinities that produce phase separation in the extreme form [44, 45]. Abundant cellular solutes, such as K+, Mg2+, and spermidine3+, aid nucleoid compaction [44], modulate protein–DNA–RNA interactions [46, 47], and alter RNAP activity directly [48]. Shifts in osmotic strength [49], solute concentrations [44], or temperature [50] can profoundly impact the nucleoid, but the underlying mechanisms of these effects are currently not well understood.
These different effects influence compaction via forces exerted in different dimensions (i.e., along the DNA axis or through space) and at different scales [10]. Supercoiling and protein binding events can act along the dimension of the DNA axis, whereas crowding, chemical interactions, electrostatic interactions, and phase separation operate in the three-dimensional (3D) environment of the entire cell. The complex and heterogeneous chromatin state generated by the balance of these different forces defines the substrate for transcription in living cells. Thus, understanding the types of chromatin structures that RNAP can encounter is crucial to understanding how transcription of chromatin occurs.
Many DNA-binding proteins organize DNA and are modulated by transcription
Compaction of DNA and constraint of DNA supercoiling in bacterial cells are achieved by diverse DNA-binding proteins that wrap, bend, or bridge the DNA [51]. Without these proteins, the chromosomal DNA itself would occupy a volume much larger than the cell, let alone the nucleoid, due to the intrinsic stiffness of DNA. Both DNA-binding proteins and solutes can aid DNA compaction and ensure cellular integrity [52, 53] by bending DNA or modulating DNA flexibility, respectively. In eukaryotes, DNA is compacted by the wrapping of the DNA around octa-histone nucleosomes, which constrain (−) supercoils, and further compacted by nucleosomal packing [2].
Bacteria possess abundant DNA-binding proteins that bind throughout the genome with variable specificity. Some of these proteins resemble histones in distribution and in some functions, but not in sequence or structure [54–59]. In E. coli, they include HU, IHF, Fis, H-NS, StpA, Dps, Lrp, CRP, MukBEF, and MatP, which differ greatly in abundance, effects, and extent of conservation [5, 10] (Table 1). Their DNA-binding modes include bending, wrapping, bridging, and some higher levels of compaction, illustrating the array of mechanisms of DNA organization in bacteria (Fig. 1; see “Organization of the nucleoid into domains” section below). Bridging brings distal dsDNA sequences together in 3D space, which creates a loop of non-bridged DNA. At least two modes of bridging dsDNA segments have been observed (Fig. 1): (i) by binding two DNA segments (e.g. H-NS [60]) or (ii) by encircling two DNA segments with a proteinaceous loop (e.g. structural maintenance of chromatin, SMC, proteins – MukBEF in Gram-negative bacteria [61]). Extensive wrapping, bridging (e.g., Dps [62] or StpA [63]), or constraint of plectonemes (e.g., hyperplectonemes stabilized by HU, H-NS, or Fis [64]) can also lead to higher-order DNA compaction. Due to the variability in abundance, structure, and DNA-binding mode, the precise roles of the most abundant bacterial chromatin proteins in organizing chromatin remain incompletely defined. Additionally, many of these proteins modulate gene expression, but the mechanisms of these effects remain incompletely characterized. For example, E. coli Lrp alters directly the expression of ~10% of genes and the expression of other genes indirectly possibly through different DNA-binding modes [65], with more studies needed to understand the mechanistic details. Conversely, transcription can influence binding of these proteins (e.g., HU and H-NS, discussed below). Although these proteins have been traditionally referred to as nucleoid-associated proteins, we refer to them here simply as DNA-binding proteins or chromatin proteins because no bright line distinguishes bacterial nucleoid-associated proteins, which structure the DNA and control gene expression globally, from site-specific DNA binding proteins like conventional TFs [5, 66]. Instead, a continuum of DNA-binding proteins exists in bacteria ranging from low-copy proteins that may bind only one site per genome (e.g., the E. coli MelR protein [67]) to highly abundant proteins that are present throughout the genome but nonetheless play key roles in transcription (e.g., HU [59]). Thus, chromatin or nucleoid-organizing protein might be a more meaningful descriptor than “nucleoid-associated.”
Table 1.
Prominent chromatin proteins in E. coli
| E. coli proteins | Fis | Dps | HU | H-NS | Hha |
|---|---|---|---|---|---|
| Conservation | γ-proteobacteria | In gram(–) & gram(+) | Highly conserved across species | Many gram(–) & some gram(+)* | Only in Enterobacteriaceae |
| Reference | [14] | [255] | [68] | [99, 271] | [272] |
| Monomer molecular weight | 11.2 kDa | 18.7 kDa | 9.5 kDa | 15.5 kDa | 8 kDa |
| Stoichiometry | dimer | homododecamer | dimers | dimers | |
| Abundance in exponential phase† | 6,000 – 30,000 dimers | 850 dodecamers | Avg. 70,000 dimers‡ | ~20,000 dimers†† | 200 monomers |
| Abundance in early stationary phase† | < 20 dimers | 6,500 – 15,000 dodecamers | Avg. 17,000 dimers‡ | ~13,000 dimers†† | 20 monomers |
| Binding sequence specificity | AT-rich; 15 bp consensus | Non-specific | Non-specific | AT-rich; 10 bp consensus | N/A |
| DNA-binding mode | Bending | Cooperative filament; Compaction | Cooperative filament; wrapping; bending; bridging | Cooperative filament; bending; bridging; compaction | |
| Effect on transcription | Occlusion; topological; stimulates | None | Topological; RNA-binding (?); phase separation (?) | Occlusion; Roadblock (?); topological; RNA binding (?); phase separation (?) | Enhances H-NS topological effect |
| E. coli proteins | IHF | Lrp | CRP | MukBEF° | MatP |
|---|---|---|---|---|---|
| Conservation | variety of proteobacteria | many bacteria (esp. γ-proteobacteria) to archaea | CRP-orthologs in almost all bacteria | multiple SMC paralogs in Euk; one SMC in bacteria | Enterobacteria |
| Reference | [273] | [65] | [274, 275] | [276, 277] | [278] |
| Monomer molecular weight | 11.3 kDa | 18 kDa | 23.6 kDa | 170 kDa | 17.6 kDa |
| Stoichiometry | dimer | octamer or hexadecamer | dimer | dimer | dimer |
| Abundance in exponential phase† | 4,000 – 30,000 dimers | 450 octamers | 1,700 dimers | 231 dimers | 20 dimers |
| Abundance in early stationary phase† | 7,000 – 13,000 dimers | 100–500 octamers | 430 dimers | 24 dimers | 10 dimers |
| Binding sequence specificity | AT-rich | AT-rich; 15 bp consensus | 22 bp symmetrical site | Non-specific; at ori | matS sites |
| DNA-binding mode | Bending; Bridging | Wrapping; Bending | Bending | Bridging | Bridging |
| Effect on transcription | Occlusion; Stimulate | Occlusion (?); Stimulate (?) | Stimulate | Unknown, but associates with highly transcribed genes | Unknown |
Well-studied H-NS paralogs and homologs: StpA, Rok, MvaT, Bv3F, Lsr2
Abundance varies based on method of detection, media, and strain. E. coli values presented here show a range of possible protein levels as reported by both [269, 270].
In exponential phase, measurements range from 30,000–140,000 HU dimers and in stationary phase, from 7,500–36,000 HU dimers [269, 270].
Plus, ~6,000 StpA monomers in exponential phase; ~3,000 StpA monomers in stationary phase [270].
indicates incompletely characterized, but still plausible effect
MukBEF is the condensin complex in E. coli and other γ-proteobacteria. Other bacteria and eukaryotes have condensin known as SMC-ScpAB. MukB and SMC are the core condensin complexes.
HU is the most abundant and highly conserved chromatin protein in growing bacteria, and is a key player both in structuring the nucleoid and in gene transcription (reviewed in [68]). HU exists as both heterodimers and homodimers of HUα and HUβ subunits, each an ~10 kDa DNA binding protein with little if any sequence-specificity. HU binds non-specifically to linear dsDNA with low affinity (μM range) as a dimer or a multimer [69, 70] but binds non-B-form DNA structures, such as DNA forks, sharp bends, kinks, or bulges, with higher affinity (nM range) [71, 72]. HU can also bind to RNA [73, 74]. HU plays multiple roles in condensing the nucleoid, including bending DNA [75, 76], wrapping DNA [69, 77], constraining (−) supercoils [78–80], putatively bridging DNA [69], and facilitating formation of RNA–DNA complexes [74, 81]. HU also stimulates topoisomerase I activity to remove excess (−) supercoiling [82]. This wide array of binding modes and dynamic interactions with DNA has left the precise roles of HU and underlying structures poorly defined. HU can constrain (−) supercoiling in plectonemes and recruit topoisomerase I upstream from ECs to reduce torsional stress (Fig. 1). HU exhibits a high intrinsic off-rate and can be displaced by 2 pN of force [83], suggesting HU would not impede transcribing RNAP. Further, similar to effects on eukaryotic nucleosomes [84], (+) supercoiling could help displace HU in front of RNAP if HU prefers to bind (−) supercoiled DNA [85]. Together, these properties of HU suggest that transcription is a key determinant of the genomic distribution of HU [80] and that HU helps modulate supercoiling throughout the genome (see “DNA topology mediates transcription-bacterial chromatin interplay”). Although incompletely substantiated, a mutant HU appears to bind (+) supercoiled DNA [79]. Further, the ability to constrain supercoils and the relative amounts in growing vs. non-growing cells varies among HU homo- and hetero-dimers [86, 87]. Possibly, some form of HU could constrain (+) supercoils in front of ECs in some conditions. Elucidation of the mechanistic details of HU-transcription interactions should be high priorities for studies of bacterial chromatin.
H-NS is an ~15 kDa basic protein containing a winged-helix DNA binding domain and two oligomerization interfaces that both helps organize the nucleoid and inhibits transcription. H-NS dimerizes in solution and is found in gram-negative bacteria, primarily γ-proteobacteria [88], with functional analogs in other bacteria (e.g., pseudomonal MvaT [89] and mycobacterial Lsr2 [90]). Multiple H-NS paralogs are often present in a single species, such as StpA in E. coli; H-NS and StpA form heterodimers [91]. Additionally, some enterobacteria contain Hha family proteins that associate with H-NS but do not bind DNA [92, 93]. Here, we refer to H-NS-like proteins and Hha family proteins as H-NS modulators because they likely modulate the structure and function of H-NS. H-NS binds to AT-rich DNA and forms nucleoprotein filaments that silence gene expression [94]. Two types of filaments have been observed in vitro: a linear (or “stiffened”) filament in which H-NS binds to one segment of DNA and a bridged filament in which H-NS binds two segments of DNA (Fig. 4a) [95, 96]. Silencing occurs when linear or bridged filaments block transcription initiation and when bridged, but not linear, filaments block elongation topologically by promoting backtrack pausing and ρ-dependent termination [97–99] (see “Occlusion” and “DNA topology” below). Both Hha and StpA enhance formation of bridged H-NS filaments in vitro [96, 100], but it remains unclear which conformation predominates in vivo.
Figure 4. Effect of H-NS filaments on transcription.
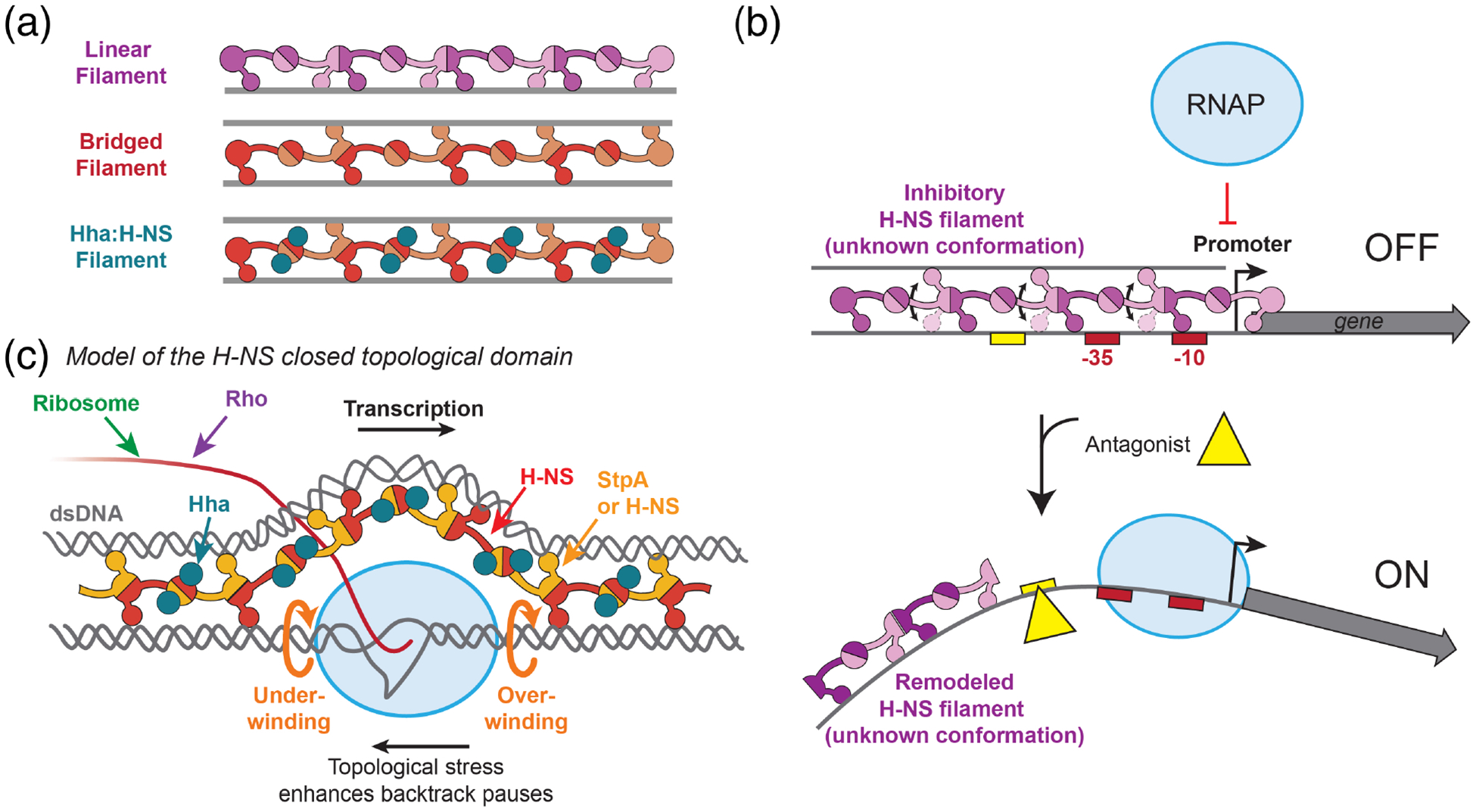
(a) H-NS can form either bridged (orange and red monomer) or linear (purple monomers) filaments at AT-rich sequences. StpA can substitute for H-NS monomers. Hha (blue circles) binds H-NS, but not DNA in an Hha:H-NS filament. (b) Bridged or linear filaments can be counter-silenced by antagonists (yellow triangle). Different conformations of the inhibitory filament are indicated by the double-sided arrow and alternative conformation of the DNA-binding domain. An antagonist could remodel a filament and aid transcription either by disrupting H-NS-DNA interactions (e.g., sterically blocking H-NS binding or distorting the DNA minor groove) or by perturbing multimerization (e.g., shortening the H-NS filament); either effect can aid RNAP binding to the promoter. (c) Bridged Hha:H-NS, StpA:H-NS, or H-NS filaments are proposed to form a closed topological domain around RNAP that highly constrains over-winding in front of and under-winding behind RNAP (orange arrows). This topological stress enhances backtracked pauses [98, 100], which can be relieved by GreB. Panel artwork adapted from [100].
Although H-NS dramatically affects transcription, an elongating RNAP can also remodel an H-NS or H-NS:StpA filament [101, 102]. Elongating RNAP may encounter an H-NS filament in a coding region [54] or downstream of an antisense promoter [103], especially when ρ-dependent termination is suppressed [102]. Upon encountering certain filaments, ECs may disrupt or rearrange the filament; if ECs disrupt a filament covering a promoter, then H-NS silencing can be relieved depending on the number of ECs transcribing into the filament [101]. It remains unclear which interacting filaments (e.g., on different sequences, containing Hha or StpA [92, 100], or linear versus bridged) and ECs (e.g., with coding versus noncoding nascent RNAs) yield disrupted filaments versus halted ECs. Experiments designed to probe in vivo H-NS filament characteristics and interactions with RNAP will shed light on how ECs might affect H-NS silencing of nearby promoters.
Organization of the nucleoid into domains depends on an interplay between transcription and DNA-binding proteins
The overall structure of the nucleoid and its separation in distinct spatial, topological, and interaction domains is largely dictated by transcription itself, with modulation by DNA-binding proteins and their interplay with transcription. The organizing role of transcription is revealed when transcription is blocked by the RNAP inhibitor rifampicin. Upon addition of rifampicin to E. coli cells, the nucleoid first compacts due to loss of the contribution to expansion from coupled transcription–translation, but then eventually expands due to entropically driven intermixing of inactive ribosome subunits with the chromosome [6, 104]. However, deletion of abundant DNA-binding proteins like HU also decompacts the nucleoid [5, 68], showing that chromatin proteins also aid nucleoid organization.
The bacterial nucleoid is organized into sets of nested domain structures [10, 11] whose physical nature and functional definitions remain incompletely elucidated (Figs. 1&2). Large “macrodomains” [105, 106] are proposed to organize the chromosome into spatially distinct sections of the nucleoid based primarily on constraints imposed by the origin and terminus of DNA replication [11]. Topologically isolated “supercoil” domains were originally defined as segments of chromosomal DNA relaxed by the introduction of single- or double-strand nicks [32, 42, 107, 108]. With the advent of high-throughput in vivo DNA interaction assays (Hi-C; see below), segments of DNA exhibiting greater proximity as captured by formaldehyde-induced protein-DNA crosslinks were defined as chromosome interaction domains (CIDs) in bacteria [43, 106, 109–111] and topologically associated domains (TADs) in eukaryotes (reviewed in [112, 113]). The eukaryotic TAD refers to proximity of chromosome segments in three-dimensional space [114] rather than the mathematical definition of topology (properties of objects that are preserved when deformed as in stretched, bent, or twisted, but not when broken or rejoined) that underlies the classic definition of DNA topology by its linking number. This different use of topology in TAD confuses a rigorous description of chromatin domains. Thus, we will refer to domains demarcated by barriers to supercoil diffusion and within which supercoiling is connected as supercoil domains (SDs), although topologically isolated domain (TID) might otherwise be a better acronym. As we will explain below, CIDs and SDs are interrelated but distinct, can be nested, and can be further subdivided into several different subtypes depending on what forms the boundaries of the domains, transcription complexes being among the most important of these boundaries.
Large macrodomains (~1 Mbp each) have been reported in bacteria based on multiple methods (Fig. 1). Based on recombination frequencies between two λ att sites and microscopy in E. coli [115], Boccard and co-workers found four segments of the genome with higher levels of self-interaction than between-macrodomain interaction: ori, ter, left, and right with two nonstructured regions flanking ori [105, 116]. Macrodomains have also been reported in B. subtilis [106], but not in Caulobacter [11]. Macrodomains are proposed to play functional roles in replication and other cellular processes [13]. Their properties may depend in part on active transcription, at least during rapid growth [7, 117]. Recently, the Boccard group reported Hi-C data consistent with a robustly distinct ter region, long-range interactions within ori, left, and right at multiple scales, and strong interaction barriers defined by highly transcribed operons [110]. The distinct ori and ter segments of the chromosome are well-defined; they bind distinct chromatin proteins (DnaA [118] and MatP [106, 110], respectively, and membrane–ori-attachment proteins in some bacteria [11]) and have been found in all bacteria studied in detail. MukBEF modulates the long-range interactions in ori, left, and right and is excluded by MatP from ter. The relationships between the macrodomains, the CIDs and SDs nested within them, and transcription remain to be characterized definitively.
However, the organization of chromatin into CIDs and SDs unquestionably depends on transcription (Fig. 2b; see also “DNA topology”) [107, 108, 119]. CIDs are defined by chromosome segments with higher levels of internal interactions using Hi-C assays [43, 106, 110, 115, 119]. Hi-C assays report proximity between regions of the genome in 3D space using deep-sequencing of formaldehyde crosslinked and then ligated DNA fragments [115]. CIDs (~30–400 kb in E. coli) are smaller than macrodomains but larger than SDs (~2–70 kb; ~10–20 kb on average). About 30 CIDs have been identified in each species (E. coli [110], Caulobacter [43], and B. subtilis [106]) and some are nested; however, CIDs in E. coli appear less distinct and have proven difficult to capture [120]. In bacteria tested to date, inhibition of transcription by rifampicin causes a dramatic loss of the CID boundaries, which are typically located near highly transcribed genes (i.e., regions of high EC density); thus, active transcription appears to govern CID organization [43, 106, 110, 111]. Transcription creates domain boundaries by limiting supercoil diffusion for the same reasons it creates topological stress (see ‘twin supercoiled domain model’ above): the EC is unable to rotate when proteins bind its nascent RNA. A higher density of ECs may create a stronger topological barrier [32, 37]. Further, characterization of CID boundaries suggests that extensive transcription of longer operons (or possibly long bridged H-NS filaments) may favor physical separation of the domains on either side [119], which would appear as a strong CID boundary in Hi-C experiments.
Growing E. coli and Salmonella contain ~400 SDs, which were identified by assays that require two sites to be topologically connected (e.g., γδ or Tn3 resolvase action [32, 107, 108]) or by assays of transcription of supercoiling-sensitive genes near DNA nicks [42] (Fig. 1). Because genomic DNA is negatively supercoiled on average [36], plectonemes can form and move throughout the genome by slithering (dynamic plectonemes; Fig. 1). These plectonemes can be stabilized or trapped either by ECs or by DNA-binding proteins that limit diffusion of supercoils, thereby creating SDs (Figs. 1&2). Because ECs can’t rotate, they will both generate supercoils during transcription (twin-supercoiled domain) and prevent diffusion of supercoils [37]. Interestingly, the distribution of H-NS binding sites is consistent with a bridged H-NS filament creating ~11 kb loops in vivo [121], which matches the size of SDs.
Although CIDs and SDs have been separately defined experimentally, they are related because they can both be bounded by ECs. Given this relationship, why aren’t all SDs observed in Hi-C experiments? First, SDs may be more dynamic than CIDs, so they may be obscured in genome-scale experiments by averaging over a population of cells. Second, the physical separation between SDs may be small, allowing interactions between adjacent SDs. In other words, a low density of ECs may create a topological barrier and define an SD boundary, but still allow the domains on either side to interact and appear as a single CID in a Hi-C experiment [119] (Fig. 2b). This difference may explain in part why CIDs are larger than SDs on average. Modeling studies also suggest that SDs (i.e., plectonemes) can be nested within CIDs [43, 122] (Fig. 1). Thus, even though ECs can create boundaries for either CIDs or SDs, the interactions within and between these domains can differ. Differences may also arise from the extent of protein-constrained supercoiling within domains (e.g., by HU or Fis) or from contributions to boundaries or intersegment interaction by DNA-bridging proteins like H-NS or SMC [123].
ECs and DNA-binding proteins both form topological barriers that can demarcate CIDs and SDs, but these barriers differ in an important way (Fig. 2). ECs create active boundaries that generate additional supercoiling in both directions, whereas DNA-binding proteins that prevent diffusion of supercoiling (e.g., bridged H-NS filaments) create static barriers. In this context, we define an active barrier as one that generates supercoils (e.g., a barrier composed of ECs) and a static barrier as one that blocks supercoil diffusion without itself generating supercoils (e.g., bridged H-NS filaments; Fig. 2b); all domain barriers must rearrange at least once per cell cycle, but the lifetimes of barriers in general are not well-characterized. Further, abundant proteins like H-NS, HU, and Fis can either constrain supercoils within CIDs or SDs to relieve torsional stress and stabilize domains [43, 106, 110] or form bridged complexes between domains (e.g., hyperplectonemes [64]) and thereby form a larger CID with nested SDs. Finally, topoisomerases also affect both CIDs and SDs by modulating supercoiling. Together, active (EC) and static (DNA-binding protein) boundaries throughout the genome have the potential to form domain boundaries in different combinations that will have different predicted effects on the domain properties (Fig. 2c). For example, greater (−) supercoiling created by active EC boundaries and constrained as plectonemes may increase DNA-DNA interactions detected by Hi-C and form a CID, whereas ECs generating (+) supercoils or static boundaries may lessen DNA–DNA interactions within a domain. Targeted experiments coupled with computer modeling [43, 109, 122] to probe supercoiling, protein binding, and DNA-DNA interactions at domains with defined active (e.g., a highly transcribed gene) and static boundaries (e.g., bridged H-NS filaments) are needed to test the nested CID–SD model and to improve understanding of the interplay between supercoiling, transcription, chromatin proteins, and bacterial nucleoid substructure.
Macromolecular demixing may contribute to nucleoid and chromatin substructuring in bacteria
Macromolecular demixing arises because solutions can sometimes increase entropy by partitioning into heterogeneous sub-volumes to increase favorable and decrease unfavorable molecular interactions [124]. The molecular environment of the bacterial cell is crowded (200–300 mg protein/mL, 100 mg RNA/mL, and 11–18 mg DNA/mL, plus high mM concentrations of many solutes including nucleic acid compactors like spermidine; [125, 126]). Water molecules interact with each other and with macromolecules differently in the crowded nucleoid than in dilute solution [46]. In the cellular environment, molecular interactions both create the nucleoid and lead to substructuring within it and in the surrounding cytoplasm. These membraneless substructures are variously called macromolecular condensates, droplets, granules, speckles, bodies, densities, or clusters that in the extreme constitute a physical phase separation. These substructures are dynamic and aid rapid regulation of intracellular reactions [127]. The formation, properties, and impacts of substructures on cellular processes, including transcription, are a rapidly growing focus in eukaryotic cell and molecular biology research [127–131].
In bacteria, the nucleoid itself is a consequence of these demixing interactions. Computational analyses suggest that the electrostatic repulsion between DNA and ribosomes is a key factor driving nucleoid formation with a surrounding rich in ribosomes and mRNAs [6, 44, 45, 132]. The nucleoid remains liquid; thus, structural rearrangements and diffusion occur throughout it [8, 41, 44, 45, 133]. For example, an increase in transcription creates more mRNA, which favors demixing and further compaction of the nucleoid; in turn, compaction can decrease transcription. This feedback loop reflects the dynamic nature of the nucleoid and the physical properties driving demixing [132], and illustrates another way that transcription organizes the nucleoid.
In bacteria, macromolecular condensates are also thought to form within the nucleoid in subregions of highly transcribed DNA (Fig. 1; e.g., rrn ribosomal RNA operons; [7, 117, 134–137]). These transcription-driven condensates may arise because the high levels of nascent RNAs and associated proteins (e.g., ribosomes on mRNAs or ribosomal proteins assembling on rRNAs) generate a demixed heterogeneity in the nucleoid [117]. Evidence exists for a nucleolus-like structure formed when six of the seven ribosomal RNA operons in E. coli cluster together [134], and clusters of RNAP at the rrn operons have been observed during fast growth conditions [7, 117, 135, 136]. Clustering of non-rrn genes has also been reported [109, 138–140]. These transcription-driven clusters may provide substructure that play a role in organizing the nucleoid in conjunction with CIDs and SDs, and may facilitate gene regulation by localizing extensive transcription to the periphery of the nucleoid (Fig. 1; [117]). Formation of these transcription clusters could also be facilitated by the supercoiling effects discussed above.
Evidence for other condensates in bacteria, including the FtsZ, SlmA, and DNA complex formed during cell division [141] and the α-proteobacterial RNA degradasome [142], suggests that macromolecular demixing may play roles in many cellular processes including gene expression and regulation. The understanding of nucleoid substructuring remains in its infancy relative to studies of macromolecular demixing in eukaryotes owing principally to the much smaller size of bacteria. New computational, microscopic, and HT-sequencing methods are needed to overcome the size barrier and provide a better understanding of the interplay between transcription, nucleoid substructuring, and macromolecular demixing (i.e., phase separation) in bacteria.
Five types of effects of bacterial chromatin on transcription
As discussed above, transcription is a primary determinant of bacterial chromatin structure, but the opposite is equally true: bacterial chromatin structures strongly impact transcription of DNA. These effects of chromatin on transcription can be organized into five types of effects on RNAP (Fig. 3): occlusion, roadblocking, topological effects, RNA-mediated effects, and segregating effects of nucleoid substructuring. Some chromatin proteins also activate genes via direct effects on RNAP (e.g., recruitment to promoters), but these direct effects are beyond our scope; we refer the reader to several relevant reviews that cover this topic [5, 14, 66, 143, 144].
Figure 3. Five possible effects of chromatin on transcription.
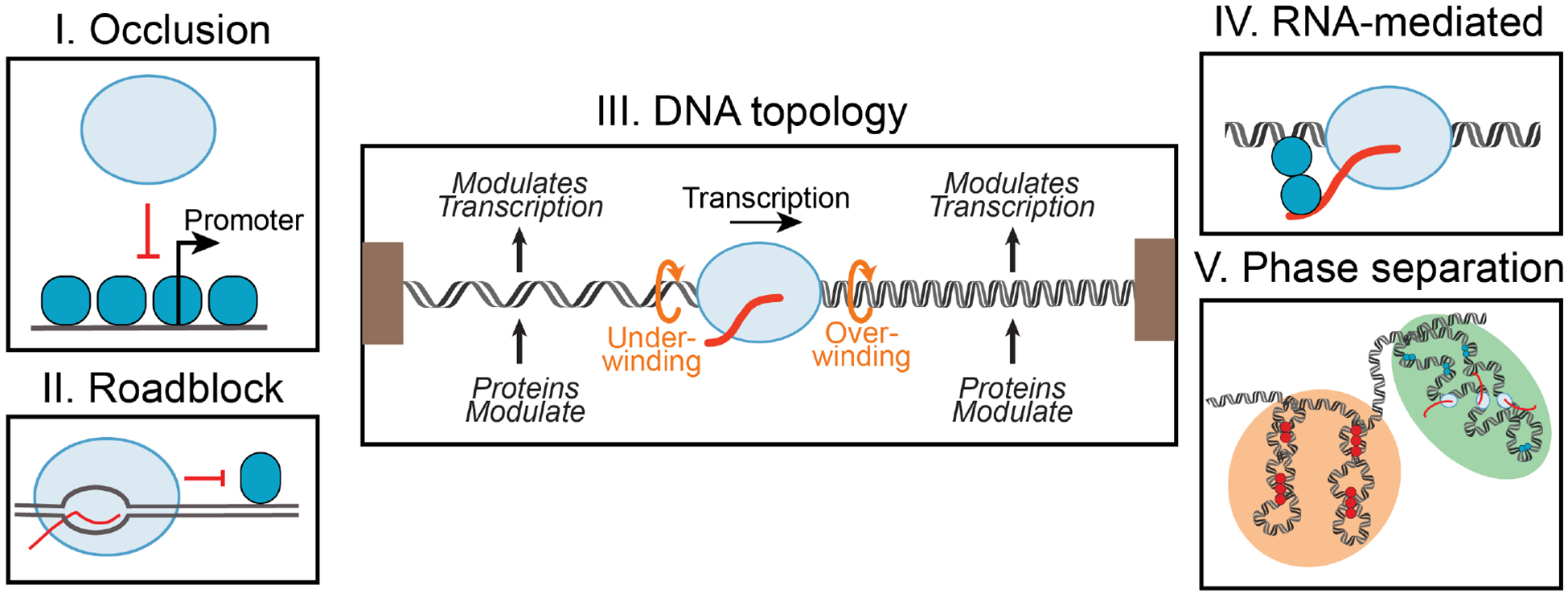
(I) Occlusion of RNAP (blue oval) by proteins (dark blue circle) bound at the promoter. (II) Roadblocks to RNAP progression by DNA-binding proteins. (III) DNA topology generated by transcription affects expression of nearby genes. (IV) RNA-mediated effects of chromatin on transcription. (V) Phase separation aided by chromatin proteins can organize transcription.
I. Occlusion of RNAP plays a role in virulence and stress responses
The simplest and most obvious way that bacterial chromatin affects transcription is by occluding promoters so that RNAP cannot bind (Fig. 3i). In E. coli and related bacteria, this competition may be accentuated at the −10 element of promoters in the absence of activators because the consensus −10 sequence resembles the binding motif of chromatin proteins like H-NS [99]. Alternatively, chromatin proteins may prevent binding of TFs to keep genes silenced. Fis and H-NS are among the most well-characterized chromatin proteins that occlude RNAP; they play notable roles in regulating genes required for virulence or stress responses.
Fis and H-NS connect gene expression to environment signals
Fis silences genes in E. coli and related bacteria by occluding either TF or RNAP binding to the promoter region (reviewed in [14]). Many AT-rich, Fis-binding motifs are found throughout the genome in intragenic regions where promoter elements are frequent [57]. Fis is highly expressed during exponential phase and decreases to undetectable levels in stationary phase (Table 1); Fis regulation coordinates gene expression, especially of virulence genes, with growth phase. Recent work on Dickeya dadantii (a γ-proteobacterial plant pathogen closely related to E. coli) highlights this role of Fis in virulence regulation [145]. Virulence is activated in D. dadantii by expression of the major virulence regulator, pelD, when cell density is high (i.e., stationary phase for D. dadantii in a plant). pelD is silenced during a rapid growth phase by Fis, which both blocks RNAP binding to the promoter, and blocks its activator CRP (the catabolite response or cAMP receptor protein) from binding an adjacent site. Thus, Fis silencing keeps pelD off when cell density is low but allows activation of the virulence program when cells reach high density in coordination with a decline in Fis levels. H-NS also binds to the pelD promoter, creating a second mechanism to occlude RNAP during non-virulent conditions [146]. H-NS silencing is relieved by changes in DNA supercoiling at the promoter that destabilize the H-NS filament [147]. pelD provides one example of many gene regulatory circuits that respond to growth conditions using Fis [148] or H-NS [149] – a paradigm by which growth-dependent changes in chromatin coordinate changes in gene expression.
H-NS occlusion of RNAP occurs by formation of inhibitory nucleoprotein filaments across promoter regions. These H-NS filaments typically form on horizontally-acquired genes, pathogenic operons, and antisense transcripts, all of which are silenced by H-NS (Fig. 4b) [54, 94, 97, 150]. Environmental factors, most notably increased temperature [151] or changes in osmolarity [152], may contribute to relief of H-NS occlusion at these genes. For DNA from enteropathogenic operons, the H-NS filament forms in vitro at lower temperatures (~20 °C), consistent with stronger effects on RNAP binding and gene silencing at lower temperatures (Fig. 4b) [98, 151, 153]. At higher temperatures (~37 °C, e.g., the temperature encountered inside mammals), occlusion by H-NS is reduced in vitro [153] by either temperature-dependent changes in the DNA structure (bending or changing supercoiling [153, 154]) or in H-NS conformation (unfolding of H-NS dimerization domains [155]) that could prevent formation of an inhibitory filament [151, 153]. The structural details of this derepression remain unclear in part because conflicting evidence exists as to whether bridged or linear H-NS filaments are responsible for gene silencing in vivo. H-NS mutants deficient in linear filament formation in vitro do not silence genes in vivo [156], but silencing of some genes requires the H-NS modulators, Hha and StpA [55, 157, 158], known to stimulate bridging by H-NS in vitro [96, 100]. Despite the clear role of environmental factors and H-NS modulators in H-NS-mediated occlusion, the changes to H-NS and the DNA that mediate occlusion in vivo remain unclear and in need of structural elucidation.
Although most H-NS occlusion inhibits transcription initiation, in some cases it may aid gene expression by facilitating RNAP access to a specific promoter. For example, in the E. coli ehxCABD operon, which encodes genes allowing secretion of the virulence factor hemolysin, H-NS filaments occlude promoters and promoter-like sequences that are adjacent to the primary promoter; occlusion of these other promoters appears to direct RNAP to the primary promoter and thus ensure that the correct mRNA is produced [159]. H-NS may also increase transcription of σS-dependent promoters in stationary phase by occluding promoters for the housekeeping sigma factor, σ70 [160]. This function of H-NS adds yet another layer to the complex repertoire of gene regulation by H-NS.
Counter-silencing of H-NS filaments
In addition to temperature-induced derepression, promoter occlusion by H-NS filaments can be relieved by binding of H-NS antagonists in a process known as “counter-silencing” (Fig. 4b). H-NS antagonists typically do not activate transcription when H-NS is absent, suggesting that they do not directly contact RNAP but simply prevent the formation of repressive H-NS filaments (although some antagonists can also recruit RNAP) [161]. Conventional TFs can act as H-NS antagonists even when binding outside the typical distance of TF action (−60 to +20 relative to a transcription start site; e.g., PhoP in Salmonella, LeuO in enterohemorrhagic E. coli, VirB in Shigella [162], and IHF in E. coli [161, 163]). The ubiquity of antagonists among bacteria that utilize H-NS suggests that counter-silencing is a widely used mechanism of transcription regulation [164, 165]. Counter-silencing may involve one of three non-exclusive events: (i) displacement of the H-NS DNA-binding domain; (ii) remodeling of the H-NS filament into a non-inhibitory conformation; or (iii) binding of an antagonist to the H-NS oligomerization domain. In Salmonella, remodeling of the H-NS filament bound at the curli operon, which encodes curli fimbriae required for virulence and cell survival, was recently reported [166]. In vitro DNaseI footprinting suggests that the antagonist, CsgD, bends the DNA and remodels the H-NS filament without preventing H-NS binding to DNA [166]. The remodeled filament allows RNAP binding and gene expression, but the conformation of the remodeled filament is unknown (Fig. 4b).
In enterohemorrhagic E. coli (EHEC), expression of the LEE operon, which encodes virulence factors required to create attaching and effacing lesions, is induced when the H-NS antagonists, Pch and Ler, remodel or prevent binding of an H-NS:StpA:Hha filament, respectively [55, 167]. In Vibrio, the H-NS antagonist ToxT, along with the DNA-binding protein IHF, relieve H-NS repression of ctxAB, which encodes a potent Vibrio toxin, by displacing H-NS binding at a high-affinity site [149, 162, 168].
Some H-NS antagonists appear to interact with the oligomerization domain of H-NS to derepress genes. Both a phage protein, gp5.5 [169], and an EHEC transcription factor, Aar [170], can bind the oligomerization domain of H-NS to disrupt multimerization, but not DNA binding. In enteropathogenic E. coli, the truncated H-NS protein (H-NST) can form heterodimers with full-length H-NS, which disrupts bridged filament formation [96] and results in derepression of genes [171]. Evidence also exists that H-NST antagonizes H-NS DNA binding and induces expression of the LEE operon [172], suggesting some H-NS antagonists act by more than one mechanism. It is unclear if antagonism by H-NS binding proteins is gene-specific or if it affects H-NS silencing globally. One possibility is that the effect of H-NS binding antagonists may depend on the stability of H-NS filaments, which is determined by DNA sequence.
Overall, counter-silencing likely depends on a combination of interactions between H-NS and its modulators, antagonists, the DNA path, RNAP, and physio-chemical conditions. Some evidence suggests that inhibitory filaments adopt a linear conformation [173] and that remodeling of this filament may reduce filament length or perturb multimerization (Fig. 4b), both of which could make the filament too weak to compete with RNAP for DNA binding. Further studies of filament structure in the context of counter-silencing should contribute to our broader understanding of H-NS filament structure–function.
II. Chromatin roadblocks can inhibit DNA translocation through RNAP
Transcription of chromatin necessitates that RNAP encounters DNA-binding proteins during transcript elongation. Despite the high probability that RNAP will encounter such roadblocks, only a subset of DNA-binding proteins dwell on DNA long enough to strongly inhibit transcript elongation (Fig. 3ii; Fig. 5). Examples of proteins capable of roadblocking include catalytically inactivate CRISPR Cas9 [174], E. coli Lac repressor (LacI [175, 176]), E. coli GalR [177], B. subtilis CodY [178], and B. subtilis CcpA [179, 180]. Biochemical studies using these and other roadblocks have defined requirements for effective transcription roadblocking and mechanisms of roadblock bypass. RNAP can move past a roadblock by transcribing around the protein (e.g., as happens for eukaryotic nucleosomes [181]), by actively dislodging the roadblocking protein, or by translocating forward when the roadblock transiently dissociates from the DNA. Each mechanism can be modulated by additional factors (Fig. 5) including, (i) the concentration and binding kinetics of the roadblocking protein [175]; (ii) the number of RNAPs simultaneously transcribing a gene because RNAPs can cooperatively translocate DNA [182]; (iii) the stability of the backtracked or arrested RNAP at the roadblock [177]; (iv) the presence of ribosomes on the nascent RNA, which inhibit backtracking and aid elongation [183]; (v) the action of proteins that relieve topological stress (see next section); and (vi) the presence of transcription rescue proteins like Mfd or Gre factors.
Figure 5. Mechanisms to overcome chromatin roadblocks.
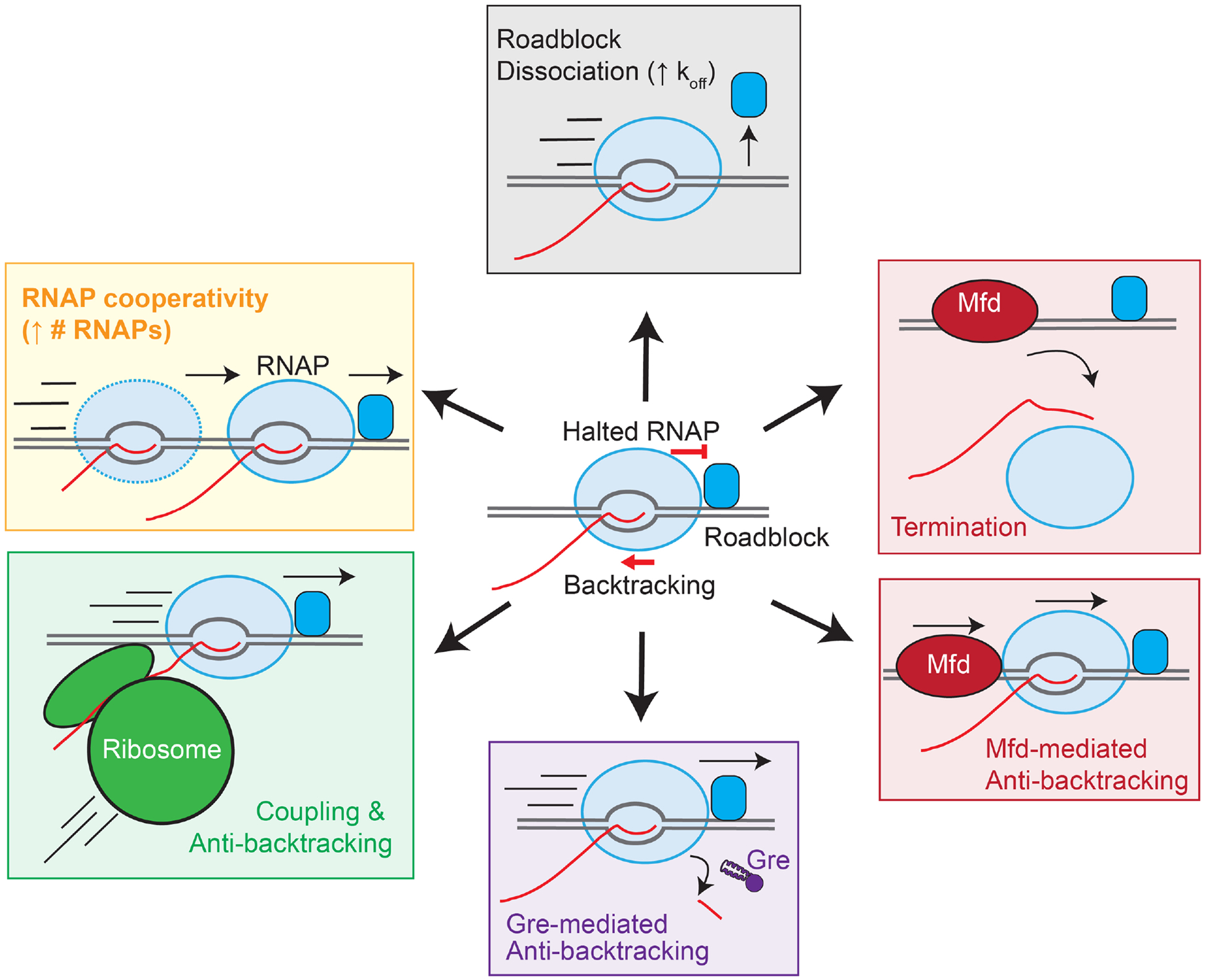
Strong protein roadblocks (blue rounded rectangle) can induce RNAP pausing, leading to backtracking (center; red “T” barrier). At least five mechanisms exist to help RNAP escape backtracking and transcribe past the roadblock (black arrows): roadblock protein dissociation, high EC density at the roadblock, transcription– translation coupling, Gre factors, and Mfd. A higher DNA-binding protein off-rate will decrease the roadblock strength (black box). Trailing ECs can help the leading EC transcribe through a DNA-binding protein (yellow box). During coupling, ribosomes inhibit EC backtracking (green box). Gre factors stimulate cleavage of the backtracked RNA to restore an active EC (purple box). Mfd translocates on DNA and binds RNAP. Mfd can either help ECs through a roadblock by preventing backtracking or, if the roadblock is strong, can dissociate RNAP from DNA (i.e., terminate transcription; red boxes).
Interplay between kinetics of RNAP and protein binding determines roadblock efficiency
As originally shown by Steege and co-workers [184, 185], a protein must bind DNA tightly to pose an effective roadblock to RNAP, although the kinetics of transcript elongation, specifically the sequence-dependent propensity for pausing and backtracking, also dictates roadblocking efficiency. The primary determinant of roadblocking efficiency is the dwell time of a specifically bound DNA-binding protein (assuming a protein level sufficient to occupy the site). To affect RNAP, a roadblock must remain on DNA considerably longer than the step time of RNAP (20–50 ms at non-pause sites in vivo; slower at pause sites) because RNAP can advance after this step time if the protein releases. This dwell time corresponds to relatively tight binding; assuming near-diffusion-limited association, even proteins with Kds in the micromolar to nanomolar range, where many DNA-binding proteins lie [186, 187], have little effect on transcription because their dwell times are in the 50 ms to few sec range. Demonstrated roadblocks, which include non-cleaving EcoRI [185], noncleaving Cas9 [174], LacI [184, 188], and biotin-streptavidin [189] all bind with dwell times in the several minute to multi-hour range [190–193]. Thus, even though the bacterial chromosome may be littered with DNA-binding proteins, in many cases transcription may easily prevail. This includes bacterial chromatin proteins; HU binds with micromolar affinity [71] and even H-NS does not slow RNAP in vitro unless it forms topologically entangled, bridged filaments [98].
As a case in point, the most-studied roadblocking barrier [184, 194–197], LacI, binds its operator in physiological solutes within a long DNA with a Kd of ~10−12 M and dwell time of ~10 min [191]. Translocation through the LacI roadblock can occur with an increase in cooperating RNAPs transcribing the DNA [182], weakening the LacI Kd by mutation, or reducing the LacI concentration [175], whereas formation of a DNA loop by tetrameric LacI increases roadblocking [176]. Overcoming a tightly bound LacI roadblock is difficult in poorly transcribed regions but may become possible in regions of high transcription (Fig. 5) [175].
A more recent model for roadblocking, CRISPR interference (CRISPRi), supports these foundational mechanisms. CRISPRi silences genes by targeting the catalytically dead Cas9 (dCas9) to a gene downstream of the promoter with a complementary single guide RNA (sgRNA) [174]. The efficiency of the CRISPRi roadblock is influenced by dCas9 binding affinity (Kds in the low nM range; [198]), the length of the RNA–DNA hybrid (≥10 bp of complementarity is required for gene repression; [199]), and the level of transcription in subsaturating dCas9 conditions. Additionally, dCas9 binding to the non-template strand nearer to the promoter appears to increase roadblock efficiency [174], although conflicting evidence exists about the importance of these parameters [200].
We note that extrapolation of in vitro DNA-binding protein behaviors (e.g., roadblocking) to behaviors in the nucleoid is fraught with unknowns. Diffusion rates, and thus the off-rates and dwell times, of DNA binding proteins in vivo will be strongly impacted by molecular crowding. GFP, for instance, diffuses in the E. coli cytoplasm about ten times more slowly than in dilute solution [201], but diffusion rates and dwell times in the nucleoid are reported for very few DNA-binding proteins and are difficult to predict. Besides crowding, at least two factors complicate such predictions. First, the effective on- and off-rates are likely to be dominated by electrostatics [202], so even slower diffusion of a model protein may not reflect the behaviors of DNA-binding proteins. Second, both classical and single-molecule experiments for the limited cases studied (e.g., HU, LacI [203, 204], RNAP [117, 205]) suggest DNA-binding proteins move by a combination of 1D sliding and 3D hopping among non-specific DNA locations rather than by free diffusion while searching for a specific binding site. Current and nascent strategies to study protein movements in vivo using fluorophores should be able to assess specifically bound DNA-binding protein dwell times, including with and without active transcription (e.g., downstream of a tightly regulated promoter). Such studies are needed now to gain better insight into transcriptional roadblocking in vivo.
RNAP backtracking modulates roadblock strength
In addition to protein binding kinetics and the number of transcribing RNAPs, the propensity for RNAP to pause and backtrack at a roadblock influences the strength of the roadblock. Pause sequences can trigger RNAP to reverse translocate RNA and DNA (backtrack) [206]; conversely, backtracking by roadblocked RNAP frequently occurs [182]. As first shown using nucleosomes [207, 208], the precise locations of halting and even whether RNAP halts appreciably, depend on the locations of pause sequences relative to a protein roadblock. Additionally, the upstream RNA structures [197], R-loop formation [177, 209, 210], and Gre factors [196, 206] all can influence the partition between active and backtracked RNAP [207, 208]. At the GalR roadblock, for example, base-pairing in the RNA–DNA hybrid within the roadblocked EC is thermodynamically unstable relative to a hybrid formed in a backtracked complex; this difference favors EC backtracking at the roadblock [177]. Introducing more stable DNA base-pairing upstream from the EC prevents backtracking and thus increases transcription through the GalR roadblock. At other backtracked pauses induced by roadblocks, like LacI, the addition of Gre factors to shift RNAP toward active elongation also can increase transcription through the roadblock [196]. The synergy between pause site locations, backtracking propensity, and DNA-binding site roadblocks in vivo remains unmapped. Further, it is difficult to separate effects of steric occlusion of transcription by a protein roadblock from topological stress that may itself induce delay, halting, or backtracking of RNAP and be exacerbated by inability of positive supercoils generated in front of ECs to diffuse through the roadblock. We address this topological effect of chromatin on transcription in the next section.
Genome-wide, transcription–translation coupling between the pioneering ribosome and RNAP also decreases backtracking. Specific interactions between RNAP and the ribosome are thought to facilitate coupling [211], which may aid RNA–DNA translocation, inhibit backtracking, and facilitate RNAP movement past roadblocks (Fig. 5) [183]. Thus, coupling aids in expression of genes bound by putative roadblocking DNA-binding proteins. For example, lipopolysaccharide biosynthetic operons in enteric bacteria, normally repressed by H-NS, use RfaH-mediated transcription–translation coupling as a mechanism to transcribe efficiently through potential H-NS roadblocks [212].
The DNA repair protein called Mutation frequency decline (Mfd) can either positively or negatively affect roadblock strength by interacting with backtracked ECs (Fig. 5), in addition to its function in DNA repair and genome maintenance [210, 213–215]. Mfd can increase roadblock strength by targeting stalled ECs and stimulating dissociation of the EC [17], as observed at the LacI [216], B. subtilis CodY [178], and B. subtilis CcpA [180] roadblocks. Mfd thus aids gene silencing, but the removal of the stalled RNAP is also necessary to ensure genomic integrity during replication [210]. In contrast, recent single-molecule results show that Mfd can associate with and aid backtracked ECs [217, 218], suggesting that Mfd may aid in transcription through some roadblocks and thereby decrease roadblock strength in these cases. In either case, Mfd appears tuned to ensure a roadblocked RNAP does not block other DNA-dependent processes either by assisting RNAP or removing it from DNA.
This same set of dynamics and assisting factors likely operate when RNAP encounters chromatin protein roadblocks on DNA. Despite knowledge of these basic features of roadblocking, many important questions await further study: (i) which bacterial chromatin proteins are capable of roadblocking RNAP; (ii) what are the precise binding characteristics that determine whether DNA-binding proteins create transcriptional roadblocks in vivo; (iii) which roadblocks are passively displaced due to relatively fast off-rates and which might be affected by positive supercoiling in front of RNAP (see next section, “DNA topology mediates transcription–bacterial chromatin interplay”); and (iv) if RNAP physically engages chromatin roadblocks, what is the structure of the RNAP-roadblock complex (i.e., the analog to RNAPII-nucleosomes structures recently determined by cryo-electron microscopy (cryo-EM) [219, 220])? These promising areas for future research will benefit from synergistic applications of new genome-scale methods, improved cell imaging, advanced single-molecule strategies, and new structural approaches (e.g., cryo-EM) now becoming available.
III. DNA topology mediates transcription–bacterial chromatin interplay
Bacterial chromatin can also affect transcription through effects on DNA topology. These effects arise because transcription-generated (+) and (−) supercoiling preceding and following ECs, respectively, can be either ameliorated or exacerbated by chromatin proteins, which thus either aid or inhibit transcription initiation, elongation, and termination (Figs. 2a & 3iii). To illustrate these connections among topology, chromatin proteins, and transcription, we will discuss (i) how Fis and HU affect supercoiling to aid transcription; (ii) how bridged H-NS filaments may create a topological domain that promotes transcriptional pausing and ρ-dependent termination; and (iii) how topology can coordinate transcription within domains (i.e., CIDs and SDs).
Topological effects on initiation of transcription
Changes in supercoiling at a promoter can affect initiation by either altering alignment of the − 10 and −35 elements or making DNA melting easier or harder ((−) or (+) supercoiling, respectively). These effects have been documented in E. coli, where expression of ~300 genes changes in response to supercoiling introduced by transcription, gyrase, or topoisomerase I [42, 108] and are regulated by proteins, such as Fis or HU, that modulate or respond to supercoiling. Levels of supercoiling throughout the genome are set by a negative feedback loop between gyrase, which increases (−) supercoiling, and topoisomerase I, which relaxes (−) supercoiling because their respective promoters are conversely activated or inhibited by (−) supercoiling due to effects of twist on −35 and −10 element alignment [66, 221, 222].
Fis can directly or indirectly regulate gene expression via supercoiling effects. Fis directly represses the gyrase promoter by occlusion [223], which decreases (−) supercoiling throughout the genome. Fis thereby indirectly affects expression of many supercoiling-sensitive genes by altering supercoiling, which is one mechanism to coordinate gene regulation with cell growth (Fis levels correlate directly with growth rate) [66]. At other promoters sensitive to supercoiling, Fis may directly alter supercoiling at promoters, but the mechanism has not been established [224].
The role of DNA-binding proteins in coordinating DNA topology and gene regulation is also exemplified by dramatic changes induced by a HU mutant that constrains (+) supercoiling. Wild-type HU constrains (−) supercoils generated by transcription throughout the genome [80]; however, when HU is altered by mutation, it can preferentially constrain (+) supercoils [225, 226]. When expressed in E. coli K-12, such an HU mutant induces compaction of the nucleoid, a decrease in supercoiling throughout the genome (i.e., the genome becomes less negatively supercoiled), and extensive changes in the transcriptome, all of which result in radical changes in cell morphology, physiology, and metabolism [225, 226]. Assays using differentially supercoiled plasmids in vitro showed that a decrease in supercoiling was responsible for activating promoters, such as the hlyE promoter, which are normally repressed by (−) supercoiling (hlyE encodes hemolysin E) [226]. Therefore, the overall decrease in supercoiling caused by the alteration of HU to preferentially constrain (+) supercoils influences which promoters are most active, presumably by changes in both the thermodynamics of melting and the relative alignments of −35 and −10 promoter elements. The change in supercoiling also may displace other chromatin proteins, like H-NS, bound at the promoter [54, 227]. These results support a model in which the constraint of supercoiling by HU, and possibly other chromatin proteins, is a fundamental mechanism of gene regulation.
H-NS creates closed topological domains that decrease transcription elongation
In addition to occluding promoters, H-NS inhibits elongation by stimulating backtrack pausing and ρ-dependent termination [98]. The mechanism by which H-NS affects elongation appears to depend on topological inhibition rather than on direct roadblocking of RNAP, providing an excellent illustration of how chromatin proteins can affect transcription through topological constraint. Although H-NS can form either linear or bridged filaments (Fig. 4a), somewhat surprisingly, H-NS switches from inhibition of RNAP elongation when present at ~66 H-NS dimers/kb DNA to having little effect when present at ≥200 H-NS dimers/kb DNA (i.e., a high H-NS concentration is less inhibitory than a lower H-NS concentration; [98, 100]). This effect can be explained because lower H-NS-to-DNA ratios favor bridging and bridged but not the linear filaments, which form at higher H-NS-to-DNA ratios, inhibit RNAP elongation. Further, H-NS modifiers that stimulate bridging greatly increase inhibition of RNAP elongation even at high H-NS to DNA ratios [100]. The pattern with which bridged filaments slow RNAP also is remarkable; they enhance pausing by RNAP at a subset of pause sites, which turn out to be sites at which RNAP backtracks; in contrast, bridged H-NS has little effect on nonbacktracked pauses [98, 100]. These observations can be explained if bridged filaments trap RNAP in a topologically closed domain (Fig. 4c). If H-NS acted via direct roadblocking of RNAP, then the linear filament would be expected to slow elongation and, as described above, H-NS dwell times would be expected to be longer than measured values (koff = 1.5 s−1 [228]). However, linear filaments do not pose the same barrier to DNA rotation as bridged filaments, which are expected to prevent DNA rotation upstream and downstream of RNAP. As a result, the (+) supercoils generated in front of RNAP and (−) supercoils generated behind RNAP may not easily be relieved by writhe within a bridged filament; both supercoiling effects energetically favor backtracking by RNAP. H-NS filament-stimulated pausing was relieved by addition of GreB [98, 100], which suppresses backtrack pausing [229] and is known to increase transcription opposed by topologically generated torque [34]. This pause stimulation also appears to explain the ability of H-NS to stimulate ρ-dependent termination (because ρ dissociates ECs at pause sites). The topological mechanism of inhibition of transcription remains to be verified in vivo, but it can explain the high correlation between sites of ρ-dependent termination and H-NS binding in E. coli K-12.
Transmission of signals via DNA topology within domains
Transcription-generated supercoiling also can impact how genes transcribed near each other within a chromosomal domain (e.g., a SD) affect each other’s expression [230] (Fig. 2a). In these domains, transcription of one gene can change the supercoiling level at nearby promoters because the diffusion of supercoils is restricted to within the domain by the SD boundaries (Fig. 2). The magnitude of effects will depend on the level of transcription, its distance from the affected promoter, the supercoiling-sensitivity of the promoter, and its orientation relative the promoter (which determines whether (+) or (−) supercoiling propagates toward the promoter; Fig. 2a). Specifically, transcription in an SD will stimulate expression of upstream promoters via diffusion of (−) supercoils and will decrease expression of downstream promoters via diffusion of (+) supercoils. Analysis of gene expression data in E. coli and Streptococcus pneumoniae by Sobetzko [230] showed a correlation in expression of operons within ~10 kb regions, consistent with the average size of a SD [42]. Given the abundance of promoters that are sensitive to supercoiling levels, coordinating transcription through supercoiling diffusion is likely to be an important way to coordinate gene regulation within a topological domain that is independent of the action of TFs. Additionally, the similarity of orientation of genes among bacteria suggests that the organization of genes within domains is conserved, and thus that SDs may play key roles in gene regulation (reviewed in [30, 31]). A prediction from this model is that arbitrarily shifting genes among topological domains may have large effects on expression. Studies have confirmed this prediction [231–233]; moving genes around the chromosome alters their expression levels, suggesting the local environment, which includes the level of supercoiling at a particular location, affects expression.
Overall, the dynamic nature of supercoiling and constraints on supercoiling provide a key mechanism by which chromatin topology affects transcription. Although local levels of supercoiling vary significantly throughout a bacterial genome, the average supercoiling among different bacteria also differs, suggesting a connection between supercoiling and evolved patterns of gene expression [36].
Many mechanistic questions remain about how HU and H-NS can act through topological effects on transcription including: (i) how dynamic is HU binding throughout the genome, especially near active transcription; (ii) can factors, such as other proteins or post-translational modifications, influence the ability of HU to constrain (−) supercoils (or (+) supercoils); (iii) how do changes to HU binding affect gene expression; (iv) do bridged H-NS filaments trap RNAP in a topological domains in vivo; (v) do H-NS modulators bind preferentially to certain genes to target gene silencing by H-NS; and (vi) do HU or H-NS influence the size of transcriptionally-isolated domains? New single-molecule and genome-scale approaches will be needed to shed light on the mechanisms by which DNA topology, chromatin structure, and transcription affect each other in bacteria.
IV. RNA interactions also may contribute to effects of bacterial chromatin on transcription
Although many new roles of RNA have been defined over the past two decades, the role of RNA in the interplay between bacterial chromatin and transcription remains unclear, underexplored, and probably underappreciated (Fig. 3iv). RNAs play key roles in this interplay in eukaryotes [234], and there are good reasons to suspect that much remains to be learned about their roles in bacterial chromatin. To illustrate this point, we will describe examples in which either nascent RNA or small RNAs (sRNAs) appear to play roles in the bacterial chromatin–transcription interface.
Interactions between the nascent RNA and RNA-binding proteins contribute to gene regulation in many ways. RNA-binding TFs, such as NusA [16] and Nun [235], bind to both RNAP and to the nascent RNA to affect pausing, translocation, or termination. Interestingly, abundant chromatin proteins, including H-NS [236], StpA [237], and HU [74], also possess RNA-binding activity. Indeed, StpA was discovered as an RNA folding chaperone [238]. Hfq is the most conserved and abundant RNA chaperone in bacteria, and Hfq both binds [239] and bridges DNA [240]. Additionally, Hfq associates with ECs via nascent transcripts in vitro [241] and crosslinks to DNA in promoter-proximal regions in vivo [242]. Hfq is also proposed to interact with ρ and RNA to modulate termination [243]. Thus, the potential for bridging contacts between the nascent RNA and DNA mediated by these proteins seems obvious, although it remains to be documented or even tested carefully (Fig. 3iv). In addition to the documented roles of StpA in RNA folding [238], H-NS also is capable of binding RNA near translation initiation sites and promoting efficient ribosome loading [244, 245]. These observations, while tentative, are intriguing because H-NS is primarily located in the nucleoid not the surrounding cytoplasm where most translation occurs. Thus, effects of H-NS on ribosome recruitment could principally affect the pioneering ribosome involved in transcription–translation coupling. Although much more work will be required to understand such a role, this could be a highly productive area for future research.
Another example of the role of RNA in bacterial chromatin comes from the recent identification of an sRNA, ncRNA4, that mediates HU–DNA interactions and aids chromatin compaction [74]. Deletion of this RNA changes the shape of the nucleoid dramatically and alters the patterns of CIDs detected by 3C assay [246], suggesting it plays a central role in chromatin organization. Many questions remain, however, including ncRNA4’s possible effect on gene expression patterns and its relationship to DNA topology. Continued investigation of ncRNA4 and possibly other sRNAs that affect bacterial chromatin in E. coli or other species is a compelling avenue of research.
Finally, nascent RNA plays an already documented, albeit uncharacterized, role in the interplay of bacterial chromatin and transcription by virtue of its ability to form R-loops upstream of RNAP. R-loops are increasingly appreciated as central mediators of many DNA-anchored processes [209, 247] and the ability for the nascent RNA strand to invade and pair with template DNA should be directly correlated with the strength of template DNA–nontemplate DNA reannealing upstream from RNAP [209]. Thus, transcription-generated topological stresses, which may be enhanced by H-NS or relieved by HU (see above), should favor R-loop formation. R-loops are proposed to act as loading sites for the bacterial SMC condensin complex [248] and could also create targets for other bacterial chromatin proteins. Despite the recent increase in interest in R-loop biology, much remains to be learned about whether R-loops help mediate communication between transcription and bacterial chromatin structure.
V. Phase separation of the nucleoid could regulate global gene expression
The role of biological condensates, whose formation is driven by macromolecular demixing or heterogeneity, has become increasingly clear in eukaryotic cellular processes over the past five years [127]. Recent evidence suggests that these condensates play organizing roles in eukaryotic transcriptional activation [249–251], raising the question of whether similar processes mediate gene regulation in bacteria. In eukaryotes, formation of super-enhancer condensates appears to be driven by association of intrinsically disordered regions (IDRs) of co-activators and transcription factors. Although IDRs are less well-documented in bacterial transcriptional regulators, it is currently unclear if this reflects a general property of bacterial proteins or simply the limited number of models that have been studied in detail. Recently, the IDR of a histone-like protein MDP-1 in Mycobacterium smegmatis was shown to be required for both formation of a compacted chromosome and expression of some genes, suggesting that IDR-driven condensates may also play important roles in bacterial gene regulation [252].
In addition to IDRs, either the transcription machinery or chromatin proteins could drive condensate formation in bacteria. As described above, evidence exists that regions of high transcriptional activity may segregate into distinct subregions of the nucleoid (Fig. 1). Early super-resolution microscopy experiments as well as 3C assays suggested that condensates of H-NS-silenced genes formed in E. coli [58], but subsequent studies revealed that dimerization of the fluorescent-tag proteins may have confounded the microscopy results [58]; there is an urgent need to repeat these experiments using monomeric photoactivatable tags now available and to show the tags do not alter H-NS properties. Additionally, a recent Hi-C study suggested that H-NS only creates boundaries between neighboring domains [110] and not connections between distal DNA segments [58]; however, this experiment needs to be repeated in a ΔstpA background to avoid confounding effects of StpA. Other label-free experiments, such as ChIA-PET [253], might be useful to detect bridging interactions by H-NS and StpA in vivo.
Although speculative at present, there appears to be good reason to consider how substructuring of the bacterial nucleoid by demixing forces that include contributions of bacterial chromatin proteins might influence gene regulation. For example, segregation of the nucleoid into highly active and inactive regions could influence partition of key mediators of transcription like Gre proteins, topoisomerase, HU, NusA, NusG, and ρ into regions of active transcription (Figs. 1&3v). Conversely, if repressive areas of the nucleoid exist, they could attract H-NS and its paralogs and tend to exclude factors associated with active transcription. Continued improvements in super-resolution microscopy may bring such phenomena into the observable realm in the not-too-distant future.
Dps condenses DNA without inhibiting transcription
Whatever the role of macromolecular demixing and phase separation proves to be in actively growing bacteria, recent results suggest it is likely to operate in stationary phase (non-growing) bacteria where the conserved, chromatin protein Dps is induced to high levels and compacts DNA into a compact structure strongly resembling a phase-separated droplet [254] (Fig. 1). Dps is thought to improve survival in slow growth by both sequestering iron and protecting the DNA [255, 256]. Dps induces strong DNA compaction through the oligomerization of Dps dodecamers [62]. Like other DNA compacting proteins, it had long been thought that Dps would block gene expression [257, 258]. However, compelling recent studies from Meyers and co-workers show that Dps has little effect on transcription either in vivo and in vitro [254]. The results are consistent with Dps forming a phase-separated DNA condensate that allows access by RNAP even though it excludes some other proteins, like restriction endonucleases, in vitro.
These findings raise two intriguing possibilities for the interplay between bacterial chromatin and transcription. First, DNA compaction does not always silence transcription. Many proteins, like H-NS or the oxidative response protein WhiB4 from Mycobacterium [259], that bridge or compact DNA will repress gene expression, whereas Dps can compact DNA dramatically without affecting gene expression. Thus, DNA compaction and gene silencing cannot be equated in general. Second, DNA-protein condensates in bacteria may indeed attract some proteins (e.g., RNAP) and exclude others (e.g., restriction endonucleases), consistent with the properties of eukaryotic condensates known to exclude some molecules but not others [127].
Interestingly, a recently described Pseudomonas transcription factor that activates gene expression in stationary phase cells binds RNAP and contains essential IDRs of unknown function [260, 261]. It seems possible such a factor might work in part by aiding partition of RNAP into Dps-generated condensates. Although further evidence for formation of a phase-separated liquid condensate by Dps is needed [262], its discovery is a major step forward in understanding the possible roles of macromolecular demixing in bacterial gene regulation. Dps or Dps-like proteins in other species, such as Mycobacterium [263], also can condense DNA, suggesting that transcription within a separated phase could be a widespread phenomenon in bacterial chromatin.
Nucleoid substructuring could co-localize active genes to aid expression
An interesting implication for a role of phase separation in regulating gene expression is its ability to coordinate co-expressed genes in a 3D cluster. This strategy might be beneficial for the cell because it keeps RNAP nearby all the promoters that need to be co-expressed to increase initiation events. In other words, the 3D search for promoters used by RNAP could be facilitated by segregation of RNAP and co-regulated genes into a condensate [264, 265]. The close 3D proximity of co-expressed genes could also facilitate the putative phase separation of those loci from the rest of the nucleoid during times of high transcription [117]. Additionally, effects on gene expression when genes are moved around the chromosome [231, 232] could be explained by a model in which genes shift among phase-separated domains in addition to shifting among SDs. Although data does not currently exist for such phenomena in bacteria, they are plausible given the growing evidence that eukaryotic cells use condensates to coordinate expression of genes regulated by super-enhancers [249]. Experiments to identify characteristics of condensates along with technologies to map gene loci in 3D space, like PAINT [266] and a CRISPR-based technique [267], can help determine if phase separation aids in gene regulation in bacteria.
Conclusions & Perspectives
The current literature paints an incompletely understood but compelling picture of the layers of interactions between bacterial chromatin and transcription in which transcription both organizes the nucleoid and is regulated by chromatin. Since the discovery of DNA and elucidation of the central dogma, mechanistic studies of transcriptional regulation have been dominated by analyses using purified DNAs devoid of their natural chromatin protein and RNA complements. With the advent of powerful new methods for super-resolution imaging, genome-scale methods exploiting HT-sequencing, cryo-EM, and more complete in vitro reconstitution, the tide has begun to turn toward understanding the mechanisms that govern transcription in its natural chromatin context.
The years ahead promise a golden age for understanding the interplay of transcription and bacterial chromatin. The poorly understood roles of RNA interactions, phase separation, and putative post-translational modifications [268] may be particularly fertile areas for study. Although many advances will come from focused efforts to study the proteins discussed here, studies of the diverse pool of chromatin proteins found in other bacterial species, especially those that have uncharacterized gene regulatory mechanisms, may be especially exciting. Although the near-bewildering heterogeneity among these proteins has been a daunting challenge in studies of bacterial chromatin and transcription to date, it may ultimately provide an unusually rich source of information about the evolution of the genetic machinery as unifying mechanisms emerge.
Highlights.
Bacterial transcription and chromatin affect each other topologically and sterically
Bacterial chromatin can both silence (H-NS) and facilitate (HU, Fis) transcription
The complex structure and dynamic nature of bacterial chromatin complicate its study
Occlusion, roadblocking, topology, RNA & phase separation mediate chromatin effects
New methods allow a paradigm shift to study gene transcription in its native state
ACKNOWLEDGEMENTS
Research in the authors’ lab is supported by NIH Grant GM38660 to R.L. We also thank members of the Landick lab, Peter Freddolino, Michael Wolfe, and the anonymous reviewers for many helpful comments that improved the manuscript substantially.
Abbreviations:
- (+)
positive
- (−)
negative
- 3D
three dimensional
- bp
base pair
- CID
chromosomal interaction domain
- dsDNA
double-stranded DNA
- EC
elongation complex
- EM
electron microscopy
- HT
high throughput
- IDR
intrinsic disordered region
- TF
transcription factor
- RNAP
RNA polymerase
- SD
supercoil domain
- sRNA
small RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Finch JT, Lutter LC, Rhodes D, Brown RS, Rushton B, Levitt M, et al. , Structure of nucleosome core particles of chromatin, Nature, 269 (1977) 29–36. [DOI] [PubMed] [Google Scholar]
- [2].Luger K, Mader AW, Richmond RK, Sargent DF, Richmon TJ, Crystal structure of the nucleosome core particle at 2.8 Å resolution, Nature, 389 (1997) 251–260. [DOI] [PubMed] [Google Scholar]
- [3].Mattiroli F, Bhattacharyya S, Dyer PN, White AE, Sandman K, Burkhart BW, et al. , Structure of histone-based chromatin in Archaea, Science, 357 (2017) 609–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sanders TJ, Marshall CJ, Santangelo TJ, The role of archaeal chromatin in transcription, J. Mol. Biol, 10.1016/j.jmb.2019.05.006 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dillon SC, Dorman CJ, Bacterial nucleoid-associated proteins, nucleoid structure and gene expression, Nat. Rev. Microbiol, 8 (2010) 185–195. [DOI] [PubMed] [Google Scholar]
- [6].Bakshi S, Choi H, Weisshaar JC, The spatial biology of transcription and translation in rapidly growing Escherichia coli, Front. Microbiol, 6 (2015) 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jin DJ, Cagliero C, Martin CM, Izard J, Zhou YN, The dynamic nature and territory of transcriptional machinery in the bacterial chromosome, Front Microbiol., 6 (2015) 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Joyeux M, Compaction of bacterial genomic DNA: clarifying the concepts, J. Phys.: Condens. Matter, 27 (2015) 383001. [DOI] [PubMed] [Google Scholar]
- [9].Lagomarsino MC, Espeli O, Junier I, From structure to function of bacterial chromosomes: Evolutionary perspectives and ideas for new experiments, FEBS Lett, 589 (2015) 2996–3004. [DOI] [PubMed] [Google Scholar]
- [10].Dame RT, Tark-Dame M, Bacterial chromatin: converging views at different scales, Curr. Opin. Cell Biol, 40 (2016) 60–65. [DOI] [PubMed] [Google Scholar]
- [11].Badrinarayanan A, Le TB, Laub MT, Bacterial chromosome organization and segregation, Annu. Rev. Cell. Dev. Biol, 31 (2015) 171–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Krogh TJ, Moller-Jensen J, Kaleta C, Impact of Chromosomal Architecture on the Function and Evolution of Bacterial Genomes, Front. Microbiol, 9 (2018) 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Surovtsev IV, Jacobs-Wagner C, Subcellular Organization: A Critical Feature of Bacterial Cell Replication, Cell, 172 (2018) 1271–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Browning DF, Grainger DC, Busby SJ, Effects of nucleoid-associated proteins on bacterial chromosome structure and gene expression, Curr. Opin. Microbiol, 13 (2010) 773–780. [DOI] [PubMed] [Google Scholar]
- [15].Landick R, The regulatory roles and mechanisms of transcriptional pausing, Biochemical Society Transactions, 34 (2006) 1062–1067. [DOI] [PubMed] [Google Scholar]
- [16].Zhang J, Landick R, A Two-Way Street: Regulatory Interplay between RNA Polymerase and Nascent RNA Structure, Trends Biochem. Sci, 41 (2016) 293–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ray-Soni A, Bellecourt MJ, Landick R, Mechanisms of Bacterial Transcription Termination: All Good Things Must End, Annu. Rev. Biochem, 85 (2016) 319–347. [DOI] [PubMed] [Google Scholar]
- [18].Mazumder A, Kapanidis AN, Recent Advances in Understanding sigma70-Dependent Transcription Initiation Mechanisms, J. Mol. Biol, 10.1016/j.jmb.2019.04.046 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Roberts JW, Mechanisms of Bacterial Transcription Termination, J. Mol. Biol, 10.1016/j.jmb.2019.04.003 (2019) [DOI] [PubMed] [Google Scholar]
- [20].Feklistov A, Darst SA, Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit, Cell, 147 (2011) 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, et al. , A pause sequence enriched at translation start sites drives transcription dynamics in vivo, Science, 344 (2014) 1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Imashimizu M, Takahashi H, Oshima T, McIntosh C, Bubunenko M, Court DL, et al. , Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo, Genome Biol, 16 (2015) 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, et al. , Interactions between RNA polymerase and the “core recognition element” counteract pausing, Science, 344 (2014) 1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Komissarova N, Kashlev M, RNA Polymerase Switches between Inactivated and Activated States By Translocating Back and Forth along the DNA and the RNA, J. Biol. Chem, 272 (1997) 15329–15338. [DOI] [PubMed] [Google Scholar]
- [25].Nudler E, RNA polymerase backtracking in gene regulation and genome instability, Cell, 149 (2012) 1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Borukhov S, Sagitov V, Goldfarb A, Transcript Cleavage Factors from E. coli, Cell, 72 (1993) 459–466. [DOI] [PubMed] [Google Scholar]
- [27].Liu LF, Wang JC, Supercoiling of the DNA template during transcription, Proc. Natl. Acad. Sci. U. S. A, 84 (1987) 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ma J, Bai L, Wang MD, Transcription under torsion, Science, 340 (2013) 1580–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ma J, Killian JL, Wang MD. RNA Polymerase as a Torsional Motor RNAP as a Molecular Motor. 2nd ed: Royal Society of Chemistry; 2019. p. in press. [Google Scholar]
- [30].Dorman CJ, Dorman MJ, DNA supercoiling is a fundamental regulatory principle in the control of bacterial gene expression, Biophys. Rev, 8 (2016) 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Meyer S, Reverchon S, Nasser W, Muskhelishvili G, Chromosomal organization of transcription: in a nutshell, Curr. Genet, 64 (2018) 555–565. [DOI] [PubMed] [Google Scholar]
- [32].Higgins NP, RNA polymerase: chromosome domain boundary maker and regulator of supercoil density, Curr. Opin. Microbiol, 22 (2014) 138–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Muskhelishvili G, Travers A. Intrinsic In vivo Modulators: Negative Supercoiling and the Constituents of the Bacterial Nucleoid In: Buc H, Strick T, editors. RNA Polymerases as Molecular Motors. Cambridge, UK: The Royal Society of Chemistry; 2009. p. 69–95. [Google Scholar]
- [34].Ma J, Tan C, Gao X, Fulbright RM Jr., Roberts JW, Wang MD, Transcription factor regulation of RNA polymerase’s torque generation capacity, Proc. Natl. Acad. Sci. U. S. A, 116 (2019) 2583–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Guo MS, Haakonsen DL, Zeng W, Schumacher MA, Laub MT, A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication, Cell, 175 (2018) 583–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Higgins NP, Species-specific supercoil dynamics of the bacterial nucleoid, Biophys. Rev, 8 (2016) 113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Booker BM, Deng S, Higgins NP, DNA topology of highly transcribed operons in Salmonella enterica serovar Typhimurium, Mol. Microbiol, 78 (2010) 1348–1364. [DOI] [PubMed] [Google Scholar]
- [38].Land M, Hauser L, Jun SR, Nookaew I, Leuze MR, Ahn TH, et al. , Insights from 20 years of bacterial genome sequencing, Funct. Integr. Genomics, 15 (2015) 141–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].diCenzo GC, Finan TM, The Divided Bacterial Genome: Structure, Function, and Evolution, Microbiol. Mol. Biol. Rev, 81 (2017) e00019–00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Levin PA, Angert ER, Small but Mighty: Cell Size and Bacteria, Cold Spring Harbor Perspect. Biol, 7 (2015) a019216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cunha S, Woldringh CL, Odijk T, Polymer-mediated compaction and internal dynamics of isolated Escherichia coli nucleoids, J. Struct. Biol, 136 (2001) 53–66. [DOI] [PubMed] [Google Scholar]
- [42].Postow L, Hardy CD, Arsuaga J, Cozzarelli NR, Topological domain structure of the Escherichia coli chromosome, Genes & Development, 18 (2004) 1766–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Le TB, Imakaev MV, Mirny LA, Laub MT, High-resolution mapping of the spatial organization of a bacterial chromosome, Science, 342 (2013) 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].de Vries R, DNA condensation in bacteria: Interplay between macromolecular crowding and nucleoid proteins, Biochimie, 92 (2010) 1715–1721. [DOI] [PubMed] [Google Scholar]
- [45].Joyeux M, A segregative phase separation scenario of the formation of the bacterial nucleoid, Soft Matter, 14 (2018) 7368–7381. [DOI] [PubMed] [Google Scholar]
- [46].Record MTJ, Courtenay ES, Cayley S, Guttman HJ, Biophysical compensation mechanisms buffering E. coli protein-nucleic acid interactions against changing environments, Trends Biochem. Sci, 23 (1998) 190–194. [DOI] [PubMed] [Google Scholar]
- [47].Record MTJ, Courtenay ES, Cayley S, Guttman HJ, Responses of E. coli to osmotic stress: large changes in amounts of cytoplasmic solutes and water, Trends Biochem. Sci, 23 (1998) 143–148. [DOI] [PubMed] [Google Scholar]
- [48].Chan CL, Landick R, Effects of Neutral Salts on RNA Chain Elongation and Pausing by Escherichia coli RNA Polymerase, J. Mol. Biol, 268 (1997) 37–53. [DOI] [PubMed] [Google Scholar]
- [49].Sun Z, Cagliero C, Izard J, Chen Y, Zhou YN, Heinz WF, et al. , Density of sigma70 promoter-like sites in the intergenic regions dictates the redistribution of RNA polymerase during osmotic stress in Escherichia coli, Nucleic Acids Res, 47 (2019) 3970–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hurme R, Rhen M, Temperature sensing in bacterial gene regulation - what it all boils down to, Mol. Microbiol, 30 (1998) 1–6. [DOI] [PubMed] [Google Scholar]
- [51].Luijsterburg MS, White MF, van Driel R, Dame RT, The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes, Crit. Rev. Biochem. Mol. Biol, 43 (2008) 393–418. [DOI] [PubMed] [Google Scholar]
- [52].Ross ED, Hardwidge PR, Maher LJ, HMG Proteins and DNA Flexibility in Transcription Activation, Molecular and Cellular Biology, 21 (2001) 6598–6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Podesta A, Indrieri M, Brogioli D, Manning GS, Milani P, Guerra R, et al. , Positively charged surfaces increase the flexibility of DNA, Biophys. J, 89 (2005) 2558–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, et al. , Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli, Nucleic Acids Res, 39 (2011) 2073–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Fukui N, Oshima T, Ueda T, Ogasawara N, Tobe T, Gene Activation through the Modulation of Nucleoid Structures by a Horizontally Transferred Regulator, Pch, in Enterohemorrhagic Escherichia coli, PLoS One, 11 (2016) e0149718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Smits WK, Grossman AD, The transcriptional regulator Rok binds A+T-rich DNA and is involved in repression of a mobile genetic element in Bacillus subtilis, PLoS Genet., 6 (2010) e1001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Grainger DC, Hurd D, Goldberg MD, Busby SJ, Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome, Nucleic Acids Res, 34 (2006) 4642–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wang W, Li GW, Chen C, Xie XS, Zhuang X, Chromosome Organization by a Nucleoid-Associated Protein in Live Bacteria, Science, 333 (2011) 1445–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Prieto AI, Kahramanoglou C, Ali RM, Fraser GM, Seshasayee AS, Luscombe NM, Genomic analysis of DNA binding and gene regulation by homologous nucleoid-associated proteins IHF and HU in Escherichia coli K12, Nucleic Acids Res, 40 (2012) 3524–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Dame RT, Wyman C, Goosen N, H-NS mediated compaction of DNA visualised by atomic force microscopy, Nucleic Acids Res, 28 (2000) 3504–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Song D, Loparo JJ, Building bridges within the bacterial chromosome, Trends Genet, 31 (2015) 164–173. [DOI] [PubMed] [Google Scholar]
- [62].Kim J, Yoshimura SH, Hizume K, Ohniwa RL, Ishihama A, Takeyasu K, Fundamental structural units of the Escherichia coli nucleoid revealed by atomic force microscopy, Nucleic Acids Res, 32 (2004) 1982–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lim CJ, Whang YR, Kenney LJ, Yan J, Gene silencing H-NS paralogue StpA forms a rigid protein filament along DNA that blocks DNA accessibility, Nucleic Acids Res, 40 (2012) 3316–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Japaridze A, Muskhelishvili G, Benedetti F, Gavriilidou AF, Zenobi R, De Los Rios P, et al. , Hyperplectonemes: A Higher Order Compact and Dynamic DNA Self-Organization, Nano Lett, 17 (2017) 1938–1948. [DOI] [PubMed] [Google Scholar]
- [65].Kroner GM, Wolfe MB, Freddolino PL, Escherichia coli Lrp regulates one-third of the genome via direct, cooperative, and indirect routes, J. Bacteriol, 201 (2018) e00411–00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Dorman CJ, Function of nucleoid-associated proteins in chromosome structuring and transcriptional regulation, J. Mol. Microbiol. Biotechnol, 24 (2014) 316–331. [DOI] [PubMed] [Google Scholar]
- [67].Grainger DC, Overton TW, Reppas N, Wade JT, Tamai E, Hobman JL, et al. , Genomic studies with Escherichia coli MelR protein: applications of chromatin immunoprecipitation and microarrays, J. Bacteriol, 186 (2004) 6938–6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Macvanin M, Adhya S, Architectural organization in E. coli nucleoid, Biochim. Biophys. Acta, 1819 (2012) 830–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hammel M, Amlanjyoti D, Reyes FE, Chen JH, Parpana R, Tang HY, et al. , HU multimerization shift controls nucleoid compaction, Sci. Adv, 2 (2016) e1600650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bonnefoy E, Rouviere-Yaniv J, HU and IHF, two homologous histone-like proteins of Escherichia coli, form different protein-DNA complexes with short DNA fragments, EMBO J, 10 (1991) 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kamashev D, Rouviere-Yaniv J, The histone-like protein HU binds specifically to DNA recombination and repair intermediates, EMBO J, 19 (2000) 6527–6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kamashev D, Agapova Y, Rastorguev S, Talyzina AA, Boyko KM, Korzhenevskiy DA, et al. , Comparison of histone-like HU protein DNA-binding properties and HU/IHF protein sequence alignment, PLoS One, 12 (2017) e0188037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Balandina A, Claret L, Hengge-Aronis R, Rouviere-Yaniv J, The Escherichia coli histone-like protein HU regulates rpoS translation, Mol. Microbiol, 39 (2001) 1069–1079. [DOI] [PubMed] [Google Scholar]
- [74].Macvanin M, Edgar R, Cui F, Trostel A, Zhurkin V, Adhya S, Noncoding RNAs binding to the nucleoid protein HU in Escherichia coli, J. Bacteriol, 194 (2012) 6046–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT, Dual architectural roles of HU: formation of flexible hinges and rigid filaments, Proc. Natl. Acad. Sci. U. S. A, 101 (2004) 6969–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Skoko D, Wong B, Johnson RC, Marko JF, Micromechanical Analysis of the Binding of DNA-Bending Proteins HMGB1, NHP6A, and HU Reveals Their Ability To Form Highly Stable DNA-Protein Complexes, Biochemistry, 43 (2004) 13867–13874. [DOI] [PubMed] [Google Scholar]
- [77].Broyles SS, Pettijohn DE, Interaction of the Escherichia coli HU protein with DNA, J. Mol. Biol, 187 (1986) 47–60. [DOI] [PubMed] [Google Scholar]
- [78].Kobryn K, Lavoie BD, Chaconas G, Supercoiling-dependent site-specific binding of HU to naked Mu DNA, J. Mol. Biol, 289 (1999) 777–784. [DOI] [PubMed] [Google Scholar]
- [79].Kar S, Choi EJ, Guo F, Dimitriadis EK, Kotova SL, Adhya S, Right-handed DNA supercoiling by an octameric form of histone-like protein HU: modulation of cellular transcription, J. Biol. Chem, 281 (2006) 40144–40153. [DOI] [PubMed] [Google Scholar]
- [80].Lal A, Dhar A, Trostel A, Kouzine F, Seshasayee AS, Adhya S, Genome scale patterns of supercoiling in a bacterial chromosome, Nat. Commun, 7 (2016) 11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Qian Z, Zhurkin VB, Adhya S, DNA-RNA interactions are critical for chromosome condensation in Escherichia coli, Proc. Natl. Acad. Sci. U. S. A, 114 (2017) 12225–12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ghosh S, Mallick B, Nagaraja V, Direct regulation of topoisomerase activity by a nucleoid-associated protein, Nucleic Acids Res, 42 (2014) 11156–11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Dame RT, Hall MA, Wang MD, Single-molecule unzipping force analysis of HU-DNA complexes, ChemBioChem, 14 (2013) 1954–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Teves SS, Henikoff S, Transcription-generated torsional stress destabilizes nucleosomes, Nat. Struct. Mol. Biol, 21 (2014) 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Muskhelishvili G, Travers A, The regulatory role of DNA supercoiling in nucleoprotein complex assembly and genetic activity, Biophys. Rev, 8 (2016) 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Rouviere-Yaniv J, Yaniv M, coli E DNA Binding Protein HU Forms Nucleosome-like Structure with Circular Double-Stranded DNA, Cell, 17 (1979) 265–274. [DOI] [PubMed] [Google Scholar]
- [87].Claret L, Rouviere-Yaniv J, Vairation in HU Composition During Growth of Escherichai coli: the Heterodimer is Required for Long Term Survival, J. Mol. Biol, 273 (1997) 93–104. [DOI] [PubMed] [Google Scholar]
- [88].Tendeng C, Bertin PN, H-NS in Gram-negative bacteria: a family of multifaceted proteins, Trends in Microbiology, 11 (2003) 511–518. [DOI] [PubMed] [Google Scholar]
- [89].Tendeng C, Soutourina OA, Danchian A, Bertin PN, MvaT proteins in Pseudomonas spp.: a novel class of H-NS-like proteins, Microbiology, 149 (2003) 3047–3050. [DOI] [PubMed] [Google Scholar]
- [90].Chen JM, Ren H, Shaw JE, Wang YJ, Li M, Leung AS, et al. , Lsr2 of Mycobacterium tuberculosis is a DNA-bridging protein, Nucleic Acids Res, 36 (2008) 2123–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Leonard PG, Ono S, Gor J, Perkins SJ, Ladbury JE, Investigation of the self-association and hetero-association interactions of H-NS and StpA from Enterobacteria, Mol. Microbiol, 73 (2009) 165–179. [DOI] [PubMed] [Google Scholar]
- [92].Ali SS, Whitney JC, Stevenson J, Robinson H, Howell PL, Navarre WW, Structural insights into the regulation of foreign genes in Salmonella by the Hha/H-NS complex, J. Biol. Chem, 288 (2013) 13356–13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Madrid C, Garcia J, Pons M, Juarez A, Molecular evolution of the H-NS protein: interaction with Hha-like proteins is restricted to enterobacteriaceae, J. Bacteriol, 189 (2007) 265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, et al. , Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella, Science, 313 (2006) 236–238. [DOI] [PubMed] [Google Scholar]
- [95].Liu Y, Chen H, Kenney LJ, Yan J, A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes, Genes & Development, 24 (2010) 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].van der Valk RA, Vreede J, Qin L, Moolenaar GF, Hofmann A, Goosen N, et al. , Mechanism of environmentally driven conformational changes that modulate H-NS DNA-bridging activity, eLife, 6 (2017) e27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Singh SS, Singh N, Bonocora RP, Fitzgerald DM, Wade JT, Grainger DC, Widespread suppression of intragenic transcription initiation by H-NS, Genes & Development, 28 (2014) 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kotlajich MV, Hron DR, Boudreau BA, Sun Z, Lyubchenko YL, Landick R, Bridged filaments of histone-like nucleoid structuring protein pause RNA polymerase and aid termination in bacteria, eLife, 4 (2015) e04970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Landick R, Wade JT, Grainger DC, H-NS and RNA polymerase: a love-hate relationship?, Curr. Opin. Microbiol, 24 (2015) 53–59. [DOI] [PubMed] [Google Scholar]
- [100].Boudreau BA, Hron DR, Qin L, van der Valk RA, Kotlajich MV, Dame RT, et al. , StpA and Hha stimulate pausing by RNA polymerase by promoting DNA-DNA bridging of H-NS filaments, Nucleic Acids Res, 46 (2018) 5525–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Rangarajan AA, Schnetz K, Interference of transcription across H-NS binding sites and repression by H-NS, Mol. Microbiol, 108 (2018) 226–239. [DOI] [PubMed] [Google Scholar]
- [102].Chandraprakash D, Seshasayee ASN, Inhibition of factor-dependent transcription termination in Escherichia coli might relieve xenogene silencing by abrogating H-NS-DNA interactions in vivo, Journal of Biosciences, 39 (2014) 53–61. [DOI] [PubMed] [Google Scholar]
- [103].Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R, Rho and NusG suppress pervasive antisense transcription in Escherichia coli, Genes & Development, 26 (2012) 2621–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Bakshi S, Choi H, Mondal J, Weisshaar JC, Time-dependent effects of transcription- and translation-halting drugs on the spatial distributions of the Escherichia coli chromosome and ribosomes, Mol. Microbiol, 94 (2014) 871–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Valens M, Penaud S, Rossignol M, Cornet F, Boccard F, Macrodomain organization of the Escherichia coli chromosome, EMBO J, 23 (2004) 4330–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Marbouty M, Le Gall A, Cattoni DI, Cournac A, Koh A, Fiche JB, et al. , Condensin- and Replication-Mediated Bacterial Chromosome Folding and Origin Condensation Revealed by Hi-C and Super-resolution Imaging, Mol. Cell, 59 (2015) 588–602. [DOI] [PubMed] [Google Scholar]
- [107].Deng S, Stein RA, Higgins NP, Transcription-induced barriers to supercoil diffusion in the Salmonella typhimurium chromosome, Proc. Natl. Acad. Sci. U. S. A, 101 (2004) 3398–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Deng S, Stein RA, Higgins NP, Organization of supercoil domains and their reorganization by transcription, Mol. Microbiol, 57 (2005) 1511–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Hacker WC, Li S, Elcock AH, Features of genomic organization in a nucleotide-resolution molecular model of the Escherichia coli chromosome, Nucleic Acids Res, 45 (2017) 7541–7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Lioy VS, Cournac A, Marbouty M, Duigou S, Mozziconacci J, Espeli O, et al. , Multiscale Structuring of the E. coli Chromosome by Nucleoid-Associated and Condensin Proteins, Cell, 172 (2018) 771–783. [DOI] [PubMed] [Google Scholar]
- [111].Trussart M, Yus E, Martinez S, Bau D, Tahara YO, Pengo T, et al. , Defined chromosome structure in the genome-reduced bacterium Mycoplasma pneumoniae, Nat. Commun, 8 (2017) 14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].van Steensel B, Furlong EEM, The role of transcription in shaping the spatial organization of the genome, Nat. Rev. Mol. Cell Biol, 10.1038/s41580-019-0114-6 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Cook PR, Marenduzzo D, Transcription-driven genome organization: a model for chromosome structure and the regulation of gene expression tested through simulations, Nucleic Acids Res, 46 (2018) 9895–9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. , Spatial partitioning of the regulatory landscape of the X-inactivation centre, Nature, 485 (2012) 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Le TB, Laub MT, New approaches to understanding the spatial organization of bacterial genomes, Curr. Opin. Microbiol, 22 (2014) 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Boccard F, Esnault E, Valens M, Spatial arrangement and macrodomain organization of bacterial chromosomes, Mol. Microbiol, 57 (2005) 9–16. [DOI] [PubMed] [Google Scholar]
- [117].Stracy M, Lesterlin C, Garza de Leon F, Uphoff S, Zawadzki P, Kapanidis AN, Live-cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid, Proc. Natl. Acad. Sci. U. S. A, 112 (2015) E4390–E4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Katayama T, Kasho K, Kawakami H, The DnaA Cycle in Escherichia coli: Activation, Function and Inactivation of the Initiator Protein, Front. Microbiol, 8 (2017) 2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Le TB, Laub MT, Transcription rate and transcript length drive formation of chromosomal interaction domain boundaries, EMBO J., 35 (2016) 1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Cagliero C, Grand RS, Jones MB, Jin DJ, O’Sullivan JM, Genome conformation capture reveals that the Escherichia coli chromosome is organized by replication and transcription, Nucleic Acids Res, 41 (2013) 6058–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Noom MC, Navarre WW, Oshima T, Wuite GJ, Dame RT, H-NS promotes looped domain formation in the bacterial chromosome, Curr. Biol, 17 (2007) R913–914. [DOI] [PubMed] [Google Scholar]
- [122].Yildirim A, Feig M, High-resolution 3D models of Caulobacter crescentus chromosome reveal genome structural variability and organization, Nucleic Acids Res, 46 (2018) 3937–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Carter SD, Sjogren C, The SMC complexes DNA and chromosome topology: right or knot?, Crit. Rev. Biochem. Mol. Biol, 47 (2012) 1–16. [DOI] [PubMed] [Google Scholar]
- [124].Hyman AA, Weber CA, Julicher F, Liquid-liquid phase separation in biology, Annu. Rev. Cell. Dev. Biol, 30 (2014) 39–58. [DOI] [PubMed] [Google Scholar]
- [125].Cayley S, Lewis BA, Guttman HJ, Record MTJ, Characterization of the Cytoplasm of Escherichia coli as a Function of External Osmolarity, J. Mol. Biol, 222 (1991) 281–300. [DOI] [PubMed] [Google Scholar]
- [126].Zimmerman SB, Trach SO, Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli, J. Mol. Biol, 222 (1991) 599–620. [DOI] [PubMed] [Google Scholar]
- [127].Banani SF, Lee HO, Hyman AA, Rosen MK, Biomolecular condensates: organizers of cellular biochemistry, Nat. Rev. Mol. Cell Biol, 18 (2017) 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Brangwynne CP, Mitchison TJ, Hyman AA, Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes, Proc. Natl. Acad. Sci. U. S. A, 108 (2011) 4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Brangwynne CP, Eckman CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, et al. , Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation, Science, 324 (2009) 1729–1732. [DOI] [PubMed] [Google Scholar]
- [130].Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH, Phase separation drives heterochromatin domain formation, Nature, 547 (2017) 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, et al. , Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome, Cell, 175 (2018) 1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Castellana M, Li SH, Wingreen NS, Spatial organization of bacterial transcription and translation, Proc. Natl. Acad. Sci. U. S. A, 113 (2016) 9286–9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Pelletier J, Halvorsen K, Ha BY, Paparcone R, Sandler SJ, Woldringh CL, et al. , Physical manipulation of the Escherichia coli chromosome reveals its soft nature, Proc. Natl. Acad. Sci. U. S. A, 109 (2012) E2649–E2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Gaal T, Bratton BP, Sanchez-Vazquez P, Sliwicki A, Sliwicki K, Vegel A, et al. , Colocalization of distant chromosomal loci in space in E. coli: a bacterial nucleolus, Genes & Development, 30 (2016) 2272–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Mata Martin C, Sun Z, Zhou YN, Jin DJ, Extrachromosomal Nucleolus-Like Compartmentalization by a Plasmid-Borne Ribosomal RNA Operon and Its Role in Nucleoid Compaction, Front. Microbiol, 9 (2018) 1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Lewis PJ, Thaker SD, Errington J, Compartmentalization of transcription and translation in Bacillus subtilis, EMBO J., 19 (2000) 710–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Endesfelder U, Finan K, Holden SJ, Cook PR, Kapanidis AN, Heilemann M, Multiscale spatial organization of RNA polymerase in Escherichia coli, Biophys. J, 105 (2013) 172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Yin Y, Zhang H, Olman V, Xu Y, Genomic arrangement of bacterial operons is constrained by biological pathways encoded in the genome, Proc. Natl. Acad. Sci. U. S. A, 107 (2010) 6310–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Wright MA, Kharchenko P, Church GM, Segre D, Chromosomal periodicity of evolutionarily conserved gene pairs, Proc. Natl. Acad. Sci. U. S. A, 104 (2007) 10559–10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Xie T, Fu LY, Yang QY, Xiong H, Xu H, Ma BG, et al. , Spatial features for Escherichia coli genome organization, BMC Genomics, 16 (2015) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Monterroso B, Zorrilla S, Sobrinos-Sanguino M, Robles-Ramos MA, Lopez-Alvarez M, Keating CD, et al. , Bacterial division FtsZ forms liquid condensates with nucleoid-associated Z-ring inhibitor SlmA, bioRxiv, 10.1101/264192 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Al-Husini N, Tomares DT, Childers WS, Schrader JM, α-proteobacterial RNA degradosomes assemble liquid-liquid phase separated RNP bodies, bioRxiv, 10.1101/272286 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Lee DJ, Minchin SD, Busby SJ, Activating transcription in bacteria, Annu. Rev. Microbiol, 66 (2012) 125–152. [DOI] [PubMed] [Google Scholar]
- [144].Browning DF, Butala M, Busby SJW, Bacterial Transcription Factors: Regulation by Pick “N” Mix, J. Mol. Biol, 10.1016/j.jmb.2019.04.011 (2019) [DOI] [PubMed] [Google Scholar]
- [145].Duprey A, Muskhelishvili G, Reverchon S, Nasser W, Temporal control of Dickeya dadantii main virulence gene expression by growth phase-dependent alteration of regulatory nucleoprotein complexes, Biochim. Biophys. Acta, 1859 (2016) 1470–1480. [DOI] [PubMed] [Google Scholar]
- [146].Zghidi-Abouzid O, Herault E, Rimsky S, Reverchon S, Nasser W, Buckle M, Regulation of pel genes, major virulence factors in the plant pathogen bacterium Dickeya dadantii, is mediated by cooperative binding of the nucleoid-associated protein H-NS, Res. Microbiol, 167 (2016) 247–253. [DOI] [PubMed] [Google Scholar]
- [147].Ouafa ZA, Reverchon S, Lautier T, Muskhelishvili G, Nasser W, The nucleoid-associated proteins H-NS and FIS modulate the DNA supercoiling response of the pel genes, the major virulence factors in the plant pathogen bacterium Dickeya dadantii, Nucleic Acids Res, 40 (2012) 4306–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Leonard S, Hommais F, Nasser W, Reverchon S, Plant-phytopathogen interactions: bacterial responses to environmental and plant stimuli, Environ. Microbiol, 19 (2017) 1689–1716. [DOI] [PubMed] [Google Scholar]
- [149].Ayala JC, Silva AJ, Benitez JA, H-NS: an overarching regulator of the Vibrio cholerae life cycle, Res. Microbiol, 168 (2017) 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC, H-NS mediates the silencing of laterally acquired genes in bacteria, PLoS Path, 2 (2006) e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Ono S, Goldberg MD, Olsson T, Esposito D, Hinton JC, Ladbury JE, H-NS is a part of a thermally controlled mechanism for bacterial gene regulation, Biochem. J, 391 (2005) 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Nagarajavel V, Madhusudan S, Dole S, Rahmouni AR, Schnetz K, Repression by binding of H-NS within the transcription unit, J. Biol. Chem, 282 (2007) 23622–23630. [DOI] [PubMed] [Google Scholar]
- [153].Falconi M, Colonna B, Prosseda G, Micheli G, Gualerzi CO, Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS, EMBO J, 17 (1998) 7033–7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Prosseda G, Falconi M, Giangrossi M, Gualerzi CO, Micheli G, Colonna B, The virF promoter in Shigella: more than just a curved DNA stretch, Mol. Microbiol, 51 (2004) 523–537. [DOI] [PubMed] [Google Scholar]
- [155].Shahul Hameed UF, Liao C, Radhakrishnan AK, Huser F, Aljedani SS, Zhao X, et al. , H-NS uses an autoinhibitory conformational switch for environment-controlled gene silencing, Nucleic Acids Res, 47 (2019) 2666–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Lim CJ, Lee SY, Kenney LJ, Yan J, Nucleoprotein filament formation is the structural basis for bacterial protein H-NS gene silencing, Sci. Rep, 2 (2012) 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Nieto JM, Madrid C, Prenafeta A, Miquelay E, Balsalobre C, Carrascal M, et al. , Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS Mol. Gen. Genet, 263 (2000) 349–358. [DOI] [PubMed] [Google Scholar]
- [158].Lucchini S, McDermott P, Thompson A, Hinton JC, The H-NS-like protein StpA represses the RpoS (sigma 38) regulon during exponential growth of Salmonella Typhimurium, Mol. Microbiol, 74 (2009) 1169–1186. [DOI] [PubMed] [Google Scholar]
- [159].Singh SS, Grainger DC, H-NS can facilitate specific DNA-binding by RNA polymerase in AT-rich gene regulatory regions, PLoS Genet., 9 (2013) e1003589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Hengge-Aronis R, Stationary Phase Gene Regulation: What Makes an Escherichia coli promoter sigmaS-selective?, Curr. Opin. Microbiol, 5 (2002) 591–595. [DOI] [PubMed] [Google Scholar]
- [161].van Ulsen P, Hillebrand M, Kainz M, Collard R, Zulianello L, van de Putte P, et al. , Function of the C-Terminal Domain of the Alpha Subunit of Escherichia coli RNA Polymerase in Basal Expression and Integration Host Factor-Mediated Activation of the Early Promoter of Bacteriophage Mu, J. Bacteriol, 179 (1997) 530–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Dorman MJ, Dorman CJ, Regulatory Hierarchies Controlling Virulence Gene Expression in Shigella flexneri and Vibrio cholerae, Front. Microbiol, 9 (2018) 2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].van Ulsen P, Hillebrand M, Zulianello L, van de Putte P, Goosen N, Integration host factor alleviates the H-NS-mediated repression of the early promoter of bacteriophage Mu, Mol. Microbiol, 21 (1996) 567–578. [DOI] [PubMed] [Google Scholar]
- [164].Stoebel DM, Free A, Dorman CJ, Anti-silencing: overcoming H-NS mediated repression of transcription in Gram-negative enteric bacteria, Microbiology, 154 (2008) 2533–2545. [DOI] [PubMed] [Google Scholar]
- [165].Will WR, Navarre WW, Fang FC, Integrated circuits: how transcriptional silencing and counter-silencing facilitate bacterial evolution, Curr. Opin. Microbiol, 23 (2015) 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Newman SL, Will WR, Libby SJ, Fang FC, The curli regulator CsgD mediates stationary phase counter-silencing of csgBA in Salmonella Typhimurium, Mol. Microbiol, 108 (2018) 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Cordeiro TN, Schmidt H, Madrid C, Juarez A, Bernado P, Griesinger C, et al. , Indirect DNA readout by an H-NS related protein: structure of the DNA complex of the C-terminal domain of Ler, PLoS Path., 7 (2011) e1002380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Stonehouse EA, Hulbert RR, Nye MB, Skorupski K, Taylor RK, H-NS binding and repression of the ctx promoter in Vibrio cholerae, J. Bacteriol, 193 (2011) 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Ali SS, Beckett E, Bae SJ, Navarre WW, The 5.5 protein of phage T7 inhibits H-NS through interactions with the central oligomerization domain, J. Bacteriol, 193 (2011) 4881–4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Santiago AE, Yan MB, Hazen TH, Sauder B, Meza-Segura M, Rasko DA, et al. , The AraC Negative Regulator family modulates the activity of histone-like proteins in pathogenic bacteria, PLoS Path., 13 (2017) e1006545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Williamson HS, Free A, A truncated H-NS-like protein from enteropathogenic Escherichia coli acts as an H-NS antagonist, Mol. Microbiol, 55 (2005) 808–827. [DOI] [PubMed] [Google Scholar]
- [172].Levine JA, Hansen AM, Michalski JM, Hazen TH, Rasko DA, Kaper JB, H-NST induces LEE expression and the formation of attaching and effacing lesions in enterohemorrhagic Escherichia coli, PLoS One, 9 (2014) e86618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Walthers D, Li Y, Liu Y, Anand G, Yan J, Kenney LJ, Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription, J. Biol. Chem, 286 (2011) 1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. , Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression, Cell, 152 (2013) 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Hao N, Krishna S, Ahlgren-Berg A, Cutts EE, Shearwin KE, Dodd IB, Road rules for traffic on DNA-systematic analysis of transcriptional roadblocking in vivo, Nucleic Acids Res, 42 (2014) 8861–8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Voros Z, Yan Y, Kovari DT, Finzi L, Dunlap D, Proteins mediating DNA loops effectively block transcription, Protein Sci, 26 (2017) 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Lewis DE, Komissarova N, Le P, Kashlev M, Adhya S, DNA sequences in gal operon override transcription elongation blocks, J. Mol. Biol, 382 (2008) 843–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Belitsky BR, Sonenshein AL, Roadblock repression of transcription by Bacillus subtilis CodY, J. Mol. Biol, 411 (2011) 729–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Choi SK, Saier MH Jr., Regulation of sigL expression by the catabolite control protein CcpA involves a roadblock mechanism in Bacillus subtilis: potential connection between carbon and nitrogen metabolism, J. Bacteriol, 187 (2005) 6856–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Zalieckas JM, Wray LV Jr., Ferson AE, Fisher SH, Transcription-repair coupling factor is involved in carbon catabolite repression of the Bacillus subtilis hut and gnt operons, Mol. Microbiol, 27 (1998) 1031–1038. [DOI] [PubMed] [Google Scholar]
- [181].Teves SS, Weber CM, Henikoff S, Transcribing through the nucleosome, Trends Biochem. Sci, 39 (2014) 577–586. [DOI] [PubMed] [Google Scholar]
- [182].Epshtein V, Toulme F, Rachid Rahmouni A, Borukhov S, Nudler E, Transcription through the roadblocks: the role of RNA polymerase coordination, EMBO J, 22 (2003) 4719–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Proshkin S, Rahmouni AR, Mironov A, Nudler E, Cooperation between translating ribosomes and RNA polymerase in transcription elongation, Science, 328 (2010) 504–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Sellitti MA, Pavco PA, Steege DA, lac Repressor Blocks in vivo Transcription of lac Control Region DNA, Proc. Natl. Acad. Sci. U. S. A, 84 (1987) 3199–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Pavco PA, Steege DA, Elongation by Escherichia coli RNA Polymerase is Blocked in vitro by a Site-specific DNA Binding Protein, J. Biol. Chem, 265 (1999) 9960–9969. [PubMed] [Google Scholar]
- [186].Schleif RF, Modulation of DNA binding by gene-specific transcription factors, Biochemistry, 52 (2013) 6755–6765. [DOI] [PubMed] [Google Scholar]
- [187].Azam TA, Ishihama A, Twelve Species of the Nucleoid-associated Protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity, J. Biol. Chem, 274 (1999) 33105–33113. [DOI] [PubMed] [Google Scholar]
- [188].Deuschle U, Gentz R, Bujard H, lac repressor blocks transcribing RNA polymerase and terminates transcription, Proc. Natl. Acad. Sci. U. S. A, 83 (1986) 4134–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Strobel EJ, Watters KE, Nedialkov Y, Artsimovitch I, Lucks JB, Distributed biotin-streptavidin transcription roadblocks for mapping cotranscriptional RNA folding, Nucleic Acids Res, 45 (2017) e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].King K, Benkovic SJ, Modrich P, Glu-111 is required for activation of the DNA cleavage center of EcoRI endonuclease, J. Biol. Chem, 264 (1989) 11807–11815. [PubMed] [Google Scholar]
- [191].Whitson PA, Olson JS, Matthews KS, Thermodynamic analysis of the lactose repressor-operator DNA interaction, Biochemistry, 25 (1986) 3852–3858. [DOI] [PubMed] [Google Scholar]
- [192].Singh D, Sternberg SH, Fei J, Doudna JA, Ha T, Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9, Nat. Commun, 7 (2016) 12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Wilchek M, Bayer EA, The avidin-biotin complex in bioanalytical applications, Anal. Biochem, 171 (1988) 1–32. [DOI] [PubMed] [Google Scholar]
- [194].Guerin M, Leng M, Rahmouni AR, High resolution mapping of E. coli transcription elongation complex in situ reveals protein interactions with the non-transcribed strand, EMBO J, 15 (1996) 5397–5407. [PMC free article] [PubMed] [Google Scholar]
- [195].Toulme F, Guerin M, Robichon N, Leng M, Rahmouni AR, In vivo evidence for back and forth oscillations of the transcription elongation complex, EMBO J, 18 (1999) 5052–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Mosrin-Huaman C, Turnbough JR CL, Rahmouni AR, Translocation of Escherichia coli RNA polymerase against a protein roadblock in vivo highlights a passive sliding mechanism for transcript elongation, Mol. Microbiol, 51 (2004) 1471–1481. [DOI] [PubMed] [Google Scholar]
- [197].Toulme F, Mosrin-Huaman C, Artsimovitch I, Rahmouni AR, Transcriptional pausing in vivo : a nascent RNA hairpin restricts lateral movements of RNA polymerase in both forward and reverse directions, J. Mol. Biol, 351 (2005) 39–51. [DOI] [PubMed] [Google Scholar]
- [198].Mekler V, Minakhin L, Severinov K, Mechanism of duplex DNA destabilization by RNA-guided Cas9 nuclease during target interrogation, Proc. Natl. Acad. Sci. U. S. A, 114 (2017) 5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Vigouroux A, Oldewurtel E, Cui L, Bikard D, van Teeffelen S, Tuning dCas9’s ability to block transcription enables robust, noiseless knockdown of bacterial genes, Mol. Syst. Biol, 14 (2018) e7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Jensen MK, Design principles for nuclease-deficient CRISPR-based transcriptional regulators, FEMS Yeast Res, 18 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Elowitz MB, Surette MG, Wolf PE, Stock JB, Leibler S, Protein Mobility in the Cytoplasm of Escherichia coli, J. Bacteriol, 181 (1999) 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Halford SE, An end to 40 years of mistakes in DNA-protein association kinetics?, Biochemical Society Transactions, 37 (2009) 343–348. [DOI] [PubMed] [Google Scholar]
- [203].Garza de Leon F, Sellars L, Stracy M, Busby SJW, Kapanidis AN, Tracking Low-Copy Transcription Factors in Living Bacteria: The Case of the lac Repressor, Biophys. J, 112 (2017) 1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Winter RB, Berg OG, von Hippel PH, Diffusion-Driven Mechanisms of Protein Translocation on Nucleic Acids. 3. The Escherichia coli lac Repressor-Operator Interaction Kinetic Measurements and Conclusions, Biochemistry, 20 (1981) 6961–6977. [DOI] [PubMed] [Google Scholar]
- [205].Grigorova IL, Phleger NJ, Mutalik VK, Gross CA, Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA, Proc. Natl. Acad. Sci. U. S. A, 103 (2006) 5332–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Artsimovitch I, Landick R, Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals, Proc. Natl. Acad. Sci. U. S. A, 97 (2000) 7090–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Walter W, Kireeva ML, Studitsky VM, Kashlev M, Bacterial polymerase and yeast polymerase II use similar mechanisms for transcription through nucleosomes, J. Biol. Chem, 278 (2003) 36148–36156. [DOI] [PubMed] [Google Scholar]
- [208].Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M, Nature of the nucleosomal barrier to RNA polymerase II, Mol. Cell, 18 (2005) 97–108. [DOI] [PubMed] [Google Scholar]
- [209].Gowrishankar J, Leela JK, Anupama K, R-loops in bacterial transcription, Transcription, 4 (2014) 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E, Linking RNA polymerase backtracking to genome instability in E. coli, Cell, 146 (2011) 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Kohler R, Mooney RA, Mills DJ, Landick R, Cramer P, Architecture of a transcribing-translating expressome, Science, 256 (2017) 194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Burmann BM, Knauer SH, Sevostyanova A, Schweimer K, Mooney RA, Landick R, et al. , An alpha helix to beta barrel domain switch transforms the transcription factor RfaH into a translation factor, Cell, 150 (2012) 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Selby CP, Sancar A, Molecular mechanism of transcription-repair coupling, Science, 260 (1993) 53. [DOI] [PubMed] [Google Scholar]
- [214].Haines NM, Kim YI, Smith AJ, Savery NJ, Stalled transcription complexes promote DNA repair at a distance, Proc. Natl. Acad. Sci. U. S. A, 111 (2014) 4037–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Achar YJ, Foiani M, Coordinating Replication with Transcription, Adv. Exp. Med. Biol, 1042 (2017) 455–487. [DOI] [PubMed] [Google Scholar]
- [216].Chambers AL, Smith AJ, Savery NJ, A DNA translocation motif in the bacterial transcription-repair coupling factor, Mfd, Nucleic Acids Res, 31 (2003) 6409–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Le TT, Yang Y, Tan C, Suhanovsky MM, Fulbright RM Jr., Inman JT, et al. , Mfd Dynamically Regulates Transcription via a Release and Catch-Up Mechanism, Cell, 172 (2018) 344–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Ho HN, van Oijen AM, Ghodke H, The transcription-repair coupling factor Mfd associates with RNA polymerase in the absence of exogenous damage, Nat. Commun, 9 (2018) 1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Farnung L, Vos SM, Cramer P, Structure of transcribing RNA polymerase II-nucleosome complex, Nat. Commun, 9 (2018) Article number: 5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Ehara H, Kujirai T, Fujino Y, Shirouzu M, Kurumizaka H, Sekine S, Structural insight into nucleosome transcription by RNA polymerase II with elongation factors, Science, 363 (2019) 744–747. [DOI] [PubMed] [Google Scholar]
- [221].Tse-Dinh Y, Regulation of the Escherichia coli DNA topoisomerase I gene by DNA supercoiling, Nucleic Acids Res, 13 (1985) 4751–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Menzel R, Gellert M, Modulation of transcription by DNA supercoiling: A deletion analysis of the Escherichai coli gyrA and gyrB promoters, Proc. Natl. Acad. Sci. U. S. A, 84 (1987) 4185–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Schneider R, Travers A, Kutateladze T, Muskhelishvili G, A DNA architectural protein couples cellular physiology and DNA topology in Escherichia coli, Mol. Microbiol, 34 (1999) 953–964. [DOI] [PubMed] [Google Scholar]
- [224].Auner H, Buckle M, Deufel A, Kutateladze T, Lazarus L, Mavathur R, et al. , Mechanism of Transcriptional Activation by FIS: Role of Core Promoter Structure and DNA Topology, J. Mol. Biol, 331 (2003) 331–344. [DOI] [PubMed] [Google Scholar]
- [225].Kar S, Edgar R, Adhya S, Nucleoid remodeling by an altered HU protein: reorganization of the transcription program, Proc. Natl. Acad. Sci. U. S. A, 102 (2005) 16397–16402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Koli P, Sudan S, Fitzgerald D, Adhya S, Kar S, Conversion of commensal Escherichia coli K-12 to an invasive form via expression of a mutant histone-like protein, mBio, 2 (2011) e00182–0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Myers KS, Yan H, Ong IM, Chung D, Liang K, Tran F, et al. , Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding, PLoS Genet, 9 (2013) e1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Dame RT, Noom MC, Wuite GJ, Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation, Nature, 444 (2006) 387–390. [DOI] [PubMed] [Google Scholar]
- [229].Laptenko O, Lee J, Lomakin I, Borukhov S, Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase, EMBO J, 22 (2003) 6322–6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Sobetzko P, Transcription-coupled DNA supercoiling dictates the chromosomal arrangement of bacterial genes, Nucleic Acids Res, 44 (2016) 1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Bryant JA, Sellars LE, Busby SJ, Lee DJ, Chromosome position effects on gene expression in Escherichia coli K-12, Nucleic Acids Res, 42 (2014) 11383–11392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Brambilla E, Sclavi B, Gene regulation by H-NS as a function of growth conditions depends on chromosomal position in Escherichia coli, G3 (Bethesda), 5 (2015) 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Scholz SA, Diao R, Wolfe MB, Fivenson EM, Lin XN, Freddolino PL, High-Resolution Mapping of the Escherichia coli Chromosome Reveals Positions of High and Low Transcription, Cell Syst, 8 (2019) 212–225.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Johnson WL, Straight AF, RNA-mediated regulation of heterochromatin, Curr. Opin. Cell Biol, 46 (2017) 102–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Vitiello CL, Kireeva ML, Lubkowska L, Kashlev M, Gottesman M, Coliphage HK022 Nun protein inhibits RNA polymerase translocation, Proc. Natl. Acad. Sci. U. S. A, 111 (2014) E2368–E2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Brescia CC, Kaw MK, Sledjeski DD, The DNA binding protein H-NS binds to and alters the stability of RNA in vitro and in vivo, J. Mol. Biol, 339 (2004) 505–514. [DOI] [PubMed] [Google Scholar]
- [237].Mayer O, Rajkowitsch L, Lorenz C, Konrat R, Schroeder R, RNA chaperone activity and RNA-binding properties of the E. coli protein StpA, Nucleic Acids Res, 35 (2007) 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Doetsch M, Gstrein T, Schroeder R, Fürtig B, Mechanisms of StpA-mediated RNA remodeling, RNA Biology, 7 (2014) 735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Talukder A, Ishihama A, Growth phase dependent changes in the structure and protein composition of nucleoid in Escherichia coli, Sci. China: Life Sci, 58 (2015) 902–911. [DOI] [PubMed] [Google Scholar]
- [240].Malabirade A, Partouche D, El Hamoui O, Turbant F, Geinguenaud F, Recouvreux P, et al. , Revised role for Hfq bacterial regulator on DNA topology, Sci. Rep, 8 (2018) 16792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Sukhodolets MV, Garges S, Interaction of Escherichia coli RNA Polymerase with the Ribosomal Protein S1 and the Sm-like ATPase Hfq, Biochemistry, 42 (2003) 8022–8034. [DOI] [PubMed] [Google Scholar]
- [242].Kambara TK, Ramsey KM, Dove SL, Pervasive Targeting of Nascent Transcripts by Hfq, Cell Rep, 23 (2018) 1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Rabhi M, Espeli O, Schwartz A, Cayrol B, Rahmouni AR, Arluison V, et al. , The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators, EMBO J, 30 (2011) 2805–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Park HS, Ostberg Y, Johansson J, Wagner EG, Uhlin BE, Novel role for a bacterial nucleoid protein in translation of mRNAs with suboptimal ribosome-binding sites, Genes & Development, 24 (2010) 1345–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Silva AJ, Sultan SZ, Liang W, Benitez JA, Role of the histone-like nucleoid structuring protein in the regulation of rpoS and RpoS-dependent genes in Vibrio cholerae, J. Bacteriol, 190 (2008) 7335–7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Qian Z, Macvanin M, Dimitriadis EK, He X, Zhurkin V, Adhya S, A New Noncoding RNA Arranges Bacterial Chromosome Organization, mBio, 6 (2015) e00998–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Crossley MP, Bocek M, Cimprich KA, R-Loops as Cellular Regulators and Genomic Threats, Mol. Cell, 73 (2019) 398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Yano K, Niki H, Multiple cis-Acting rDNAs Contribute to Nucleoid Separation and Recruit the Bacterial Condensin Smc-ScpAB, Cell Rep, 21 (2017) 1347–1360. [DOI] [PubMed] [Google Scholar]
- [249].Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA, A Phase Separation Model for Transcriptional Control, Cell, 169 (2017) 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, et al. , Coactivator condensation at super-enhancers links phase separation and gene control, Science, 361 (2018) eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, et al. , Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains, Cell, 175 (2018) 1842–1855.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Savitskaya A, Nishiyama A, Yamaguchi T, Tateishi Y, Ozeki Y, Nameta M, et al. , C-terminal intrinsically disordered region-dependent organization of the mycobacterial genome by a histone-like protein, Sci. Rep, 8 (2018) 8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Li G, Cai L, Chang H, Hong P, Zhou Q, Kulakova EV, et al. , Chromatin Interaction Analysis with Paired-Eng Tag (ChIA-PET) sequencing technology and application, BMC Genomics, 15 (2014) S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Janissen R, Arens MMA, Vtyurina NN, Rivai Z, Sunday ND, Eslami-Mossallam B, et al. , Global DNA Compaction in Stationary-Phase Bacteria Does Not Affect Transcription, Cell, 174 (2018) 1188–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Haikarainen T, Papageorgiou AC, Dps-like proteins: structural and functional insights into a versatile protein family., Cell. Mol. Life Sci, 67 (2010) 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Nair S, Finkel SE, Dps protects cells against multiple stresses during stationary phase, J. Bacteriol, 186 (2004) 4192–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Almiron M, Link AJ, Furlong D, Kolter R, A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli, Genes & Development, 6 (1992) 2646–2654. [DOI] [PubMed] [Google Scholar]
- [258].Antipov SS, Tutukina MN, Preobrazhenskaya EV, Kondrashov FA, Patrushev MV, Toshchakov SV, et al. , The nucleoid protein Dps binds genomic DNA of Escherichia coli in a non-random manner, PLoS One, 12 (2017) e0182800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Chawla M, Mishra S, Anand K, Parikh P, Mehta M, Vij M, et al. , Redox-dependent condensation of the mycobacterial nucleoid by WhiB4, Redox Biol, 19 (2018) 116–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Babin BM, Bergkessel M, Sweredoski MJ, Moradian A, Hess S, Newman DK, et al. , SutA is a bacterial transcription factor expressed during slow growth in Pseudomonas aeruginosa, Proc. Natl. Acad. Sci. U. S. A, 113 (2016) E597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Bergkessel M, Babin BM, VanderVelde DG, Sweredoski MJ, Moradian A, Eggleston-Rangel R, et al. , The dormancy-specific regulator, SutA, is intrinsically disordered and modulates transcription initiation in Pseudomonas aeruginosa, bioRxiv, 10.1101/423384 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Abbondanzieri EA, Meyer AS, More than just a phase: the search for membraneless organelles in the bacterial cytoplasm, Curr. Genet, 65 (2019) 691–694. [DOI] [PubMed] [Google Scholar]
- [263].Ghatak P, Karmakar K, Kasetty S, Chatterji D, Unveiling the role of Dps in the organization of mycobacterial nucleoid, PLoS One, 6 (2011) e16019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Wang F, Redding S, Finkelstein IJ, Gorman J, Reichman DR, Greene EC, The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion, Nat. Struct. Mol. Biol, 20 (2013) 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Friedman LJ, Mumm JP, Gelles J, RNA polymerase approaches its promoter without long-range sliding along DNA, Proc. Natl. Acad. Sci. U. S. A, 110 (2013) 9740–9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Spahn CK, Glaesmann M, Grimm JB, Ayala AX, Lavis LD, Heilemann M, A toolbox for multiplexed super-resolution imaging of the E. coli nucleoid and membrane using novel PAINT labels, Sci. Rep, 8 (2018) 14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, et al. , Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system, Cell, 155 (2013) 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Dilweg IW, Dame RT, Post-translational modification of nucleoid-associated proteins: an extra layer of functional modulation in bacteria?, Biochemical Society Transactions, 46 (2018) 1381–1392. [DOI] [PubMed] [Google Scholar]
- [269].Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A, Growth Phase-Dependent Variation in Protein Composition of the Escherichia coli Nucleoid, J. Bacteriol, 181 (1999) 6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Schmidt A, Kochanowski K, Vedelaar S, Ahrne E, Volkmer B, Callipo L, et al. , The quantitative and condition-dependent Escherichia coli proteome, Nat. Biotechnol, 34 (2016) 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Grainger DC, Structure and function of bacterial H-NS protein, Biochemical Society Transactions, 44 (2016) 1561–1569. [DOI] [PubMed] [Google Scholar]
- [272].Madrid C, Balsalobre C, Garcia J, Juarez A, The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins, Mol. Microbiol, 63 (2007) 7–14. [DOI] [PubMed] [Google Scholar]
- [273].Munoz A, Valls M, de Lorenzo V. Extreme DNA Bending: Molecular Basis of the Regulatory Breadth of IHF In: Remus T Dame CJD, editor. Bacterial Chromatin: Springer Science & Business Media; 2009. p. 365–394. [Google Scholar]
- [274].Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJ, Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome, Proc. Natl. Acad. Sci. U. S. A, 102 (2005) 17693–17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Soberon-Chavez G, Alcaraz LD, Morales E, Ponce-Soto GY, Servin-Gonzalez L, The Transcriptional Regulators of the CRP Family Regulate Different Essential Bacterial Functions and Can Be Inherited Vertically and Horizontally, Front Microbiol., 8 (2017) 959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Cobbe N, Heck MM, The evolution of SMC proteins: phylogenetic analysis and structural implications, Mol. Biol. Evol, 21 (2004) 332–347. [DOI] [PubMed] [Google Scholar]
- [277].Gruber S, Errington J, Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis, Cell, 137 (2009) 685–696. [DOI] [PubMed] [Google Scholar]
- [278].Mercier R, Petit MA, Schbath S, Robin S, El Karoui M, Boccard F, et al. , The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain, Cell, 135 (2008) 475–485. [DOI] [PubMed] [Google Scholar]



