Abstract
Massive releases of organic substrates during marine algal blooms trigger growth of many clades of heterotrophic bacteria. Algal polysaccharides represent the most diverse and structurally complex class of these substrates, yet their role in shaping the microbial community composition is poorly understood. We investigated, whether polysaccharide utilization capabilities contribute to niche differentiation of Polaribacter spp. (class Flavobacteriia; known to include relevant polysaccharide-degraders) that were abundant during 2009–2012 spring algal blooms in the southern North Sea. We identified six distinct Polaribacter clades using phylogenetic and phylogenomic analyses, quantified their abundances via fluorescence in situ hybridization, compared metagenome-assembled genomes, and assessed in situ gene expression using metaproteomics. Four clades with distinct polysaccharide niches were dominating. Polaribacter 2-a comprised typical first responders featuring small genomes with limited polysaccharide utilization capacities. Polaribacter 3-a were abundant only in 2010 and possessed a distinct sulfated α-glucoronomannan degradation potential. Polaribacter 3-b responded late in blooms and had the capacity to utilize sulfated xylan. Polaribacter 1-a featured high numbers of glycan degradation genes and were particularly abundant following Chattonella algae blooms. These results support the hypothesis that sympatric Polaribacter clades occupy distinct glycan niches during North Sea spring algal blooms.
Subject terms: Marine microbiology, Metagenomics, Microbial ecology
Introduction
One of the fundamental questions in microbial ecology is which factors shape the composition of a given microbial community. According to the neutral theory, all species are ecologically equivalent and community structure is only shaped by stochastic processes. The niche theory, on the other hand, postulates that environmental conditions regulate the species composition by a process known as habitat filtering [1]. The niche is defined as a set of biotic and abiotic factors that an organism needs to thrive. Thus, a given habitat selects a group of organisms sharing similar niches that compete for the same resources. Stable conditions provided the best-adapted species ultimately prevails and supplants less adapted species [1]. However, many habitats are subjected to constant changes, which preclude establishment of stable communities.
Marine algal blooms are an essential part of the global carbon cycle [2] and are characterized by a dynamically changing environment with a large supply of diverse and structurally complex substrates [3]. In particular, the terminal phases of these blooms result in massive releases of algal polysaccharides [4], triggering significant and often successive shifts in microbial community composition [5–7]. Algal polysaccharides are too manifold for that a single bacterial species could carry the genes to decompose them all. Therefore, this decomposition is carried out by various co-occurring specialized bacteria with similar ecological niches—a process known as resource partitioning [8]. Carbohydrate-active enzymes (CAZymes) catalyze the actual polysaccharide decomposition [9]. Initial substrate binding is usually mediated by a dedicated carbohydrate-binding module (CBM). Besides, CAZymes can be classified into glycoside hydrolase (GH), polysaccharide lyase (PL), carbohydrate esterase (CE), and glycosyltransferase (GT) categories. Polysaccharide degradation in Bacteroidetes is usually encoded in specialized genomic islands that are referred to as polysaccharide utilization loci (PULs). To orchestrate binding, uptake, and degradation of a particular polysaccharide, PULs contain a gene tandem coding for a SusD-like glycan-binding protein and a SusC-like TonB-dependent transporter as well as dedicated CAZyme and accessory proteins such as sulfatases [10]. Therefore, the genetic composition of a PUL provides hints about the targeted polysaccharide, and thus, enables predictions about the polysaccharide niche of a bacterium [11].
During previous studies on spring algal blooms in the southern North Sea from 2009 to 2012, we observed the succession and annual recurrence of distinct clades of Flavobacteriia, Gammaproteobacteria, and alphaproteobacterial Roseobacter clade members [12, 13]. The flavobacterial genus Polaribacter constituted the most abundant and diverse recurrent bacterial clade [13]. Catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH) analysis using the Polaribacter-specific oligonucleotide probe POL740 showed that Polaribacter relative abundances reached up to ~27% (2009), ~26% (2010), ~14% (2011), and ~25% (2012), respectively [13]. Four different Polaribacter oligotypes with relative read abundances above 1% were detected within the planktonic fraction (0.2–3 µm) using minimum entropy decomposition (MED) of 16S rRNA tag sequences [14]. These data demonstrated a high intragenus diversity of the recurrent Polaribacter. The detected oligotypes were confined to spring and summer algal blooms with rapid shifts in abundance, suggesting that algae-derived substrates triggered growth of the corresponding Polaribacter clades [14]. This was corroborated by analyses of CAZyme gene repertoires of distinct bacterioplankton clades during bloom seasons [13]. These analyses revealed that members of the genus Polaribacter were enriched in genes of the GH16, 17, and 30 families (constituting PULs targeting the diatom storage glycan laminarin), as well as genes of families GH13 (e.g., α-amylases) and GH92 (exo-α-mannosidases) [13]. Based on analysis of deeply sequenced metagenomes, Krüger et al. recently suggested that during spring algae blooms in the southern North Sea the bulk of algal glycan remineralization is mediated by only about a dozen of dominant Bacteroidetes clades, including prominently Polaribacter [15]. For this reason, we performed a fine-scale analysis of polysaccharide-based niche differentiation of the respective Polaribacter clades.
Ecological theory states that sympatric species within one genus co-occur due to niche differentiation [16–18]. We previously compared Polaribacter strains Hel1_33_49 and Hel1_85, both of which were isolated during a phytoplankton bloom off the coast of Helgoland island. Strain Hel1_33_49 is a planktonic isolate supposed to mainly feed on proteins and a small subset of algal polysaccharides, whereas strain Hel1_85 is likely associated with algae and has the genetic potential to decompose a larger spectrum of polysaccharides [18]. However, two isolates are insufficient to describe the large niche space of spring bloom-associated Polaribacter clades.
In this study, we present polysaccharide niches of all Polaribacter clades that were recurrently abundant during spring algal blooms from 2009 to 2012 in the southern North Sea. We (i) identified six distinct Polaribacter clades using phylogenetic analyses, (ii) quantified their abundances using fluorescence in situ hybridization with novel oligonucleotide probes and metagenome read recruitment, (iii) compared metagenome-assembled genomes (MAGs) for all six clades, and (iv) assessed the in situ gene expression by reanalyzing published metaproteome data. We tested whether distinct Polaribacter clades exhibit differences in their PUL repertoires that might explain their occurrences during different bloom periods.
Materials and methods
16S rRNA clone libraries and phylogenetic analysis
Almost full-length Polaribacter-related 16S rRNA gene sequences were retrieved from a clone library constructed from Helgoland spring bloom bacterioplankton (0.2–3 µm) sampled on April 8th, 2010 as described in [12] (ENA project: PRJEB32146). In addition, 88 Polaribacter-related 16S rRNA sequences (>1450 bp, pintail quality >70%), retrieved from the same sampling site on April 14th, 2009 were also analyzed [12]. Using the Silva Incremental Aligner [19], these sequences were aligned to the Silva SSURef v.132 dataset [20] and manually curated in Arb v.6.1 [21]. Together with all Polaribacter-related sequences in the Silva SSURef NR 99 v.132 dataset, phylogenetic tree reconstruction was done with the RAxML v.7 [22] maximum likelihood method (GTR-GAMMA rate distribution model, rapid bootstrap algorithm, 100 repetitions) and the neighbor-joining method (Jukes–Cantor substitution model, 1000 bootstrap repetitions). Both methods were run with and without 10, 30, and 50% positional conversation filters for all Flavobacteriia. A consensus tree was generated following the recommendations described in [23].
Phytoplankton identification, oligonucleotide probe design, and CARD-FISH analysis
Phytoplankton was classified and quantified microscopically by experts as described in previous publications [12, 13]. In order to detect and quantify the most abundant clades of Polaribacter bacterioplankton, we designed four oligonucleotide probes (POL405, POL1270, POL183a, and POL180) using the probe design tool implemented in Arb with Silva SSURef NR 99 v128 (Supplementary Table S1). Hybridization conditions for these probes were optimized using environmental samples with varying formamide concentrations between 0 and 50% (5% increments) at 46 °C. The highest possible formamide concentration providing sufficient brightness for signal detection was used for further hybridizations (Supplementary Table S1). The combined total numbers detected by the four probes were compared with the genus-specific probe POL740 to evaluate probe specificities. Samples for CARD-FISH analyses were taken as described in [12] and were processed according to [24]. Cell counting was performed with an automatic microbial cell enumeration system [25] and ACMETOOL2.0 image analysis software (http://www.technobiology.ch/index.php?id=acmetool). Cell counting results and additional abundance data are summarized in Supplementary Table S2.
Metagenome sequencing and binning
Surface seawater samples were taken at the long-term ecological research station “Kabeltonne” (54° 11.3ʹ N, 7° 54.0′ E) at the North Sea island Helgoland as described previously [12]. For DNA extraction, the biomass of free-living bacteria (0.2 µm) was harvested after prefiltration with 10 µm and 3 µm filters. Sequencing of 38 surface seawater metagenomes was performed at the Department of Energy Joint Genome Institute (Walnut Creek, CA, USA) (Supplementary Table S3) [13]. Quality filtering and trimming of raw reads, metagenome assembly, and binning were performed as described previously [15, 26]. In brief, adapters and low-quality reads were removed using BBDuk v35.14 (http://bbtools.jgi.doe.gov). SPAdes v3.10.0 was used for the individual assembly of metagenome datasets [27]. Contigs from each assembly were separately binned in CONCOCT [28] integrated into anvi’o v3 [29]. BBMap v35.14 (http://bbtools.jgi.doe.gov) was used for read mapping. Phylogenomic placement and quality estimates of MAGs were investigated in CheckM v1.0.7 [30] and a subset of automatically binned CONCOCT MAGs was selected for manual bin refinement in anvi’o. Refined bins were analyzed a second time with further Bacteroidetes reference genomes, chosen based on 16S rRNA sequences. Mash v1.1.1 [31] was used to reduce redundancy and to cluster MAGs into approximate species clusters and the ones placed within the genus Polaribacter by CheckM were kept. The MAGs having (i) a relative abundance >0.4% in at least one of 38 metagenomes based on read recruitment, (ii) contamination <5%, and (iii) completeness >69% were considered for further analyses of environmentally relevant Polaribacter (Supplementary Table S4). The corresponding MAG sequences are available at the European Nucleotide Archive (PRJEB28156).
MAG analyses
Average nucleotide identity (ANI) values between Polaribacter MAGs and genomes were calculated using the ani Ruby script from the enveomics toolbox [32]. For phylogenomic analysis, the protein sequences of 43 conserved marker genes (Supplementary Table S5) were extracted and aligned in CheckM v1.0.8. Using the concatenation of these gene sequences, a protein tree was computed with RAxML v7 (GAMMA-WAG substitution model, rapid bootstrap algorithm, 100 repetitions). Metagenome reads from 2009 to 2012 North Sea spring phytoplankton blooms were recruited by Polaribacter MAGs and genomes with BBMap v35.14 using fast mode. For 38 metagenomes sequenced on the Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA, USA) (Supplementary Table S3), read recruitments were done with minimum mapping identity of one (minid = 1) and identity filter for reporting mappings of one (idfilter = 1). Reads were recruited with the most complete and least contaminated MAGs out of those having ANI >99% as well as the complete genome sequence of strain Hel1_33_96 of Polaribacter clade 3-a (Supplementary Table S6). For clades with low micro-diversity (Polaribacter 2-a and 2-b), minid and idfilter were both set to 0.99. For four metagenomes sequenced on the now defunct 454 FLX Ti platform (454 Life Sciences, Branford, CT, USA) (Supplementary Table S3), all read recruitments were carried out with minid = 0.97 and idfilter = 0.99 using the most complete MAG or genome from each clade (see Supplementary Table S6). To calculate the clade-specific final abundance values, number of the reads mapped on MAGs or genomes in each clade were summed and reported as reads per kilobase million. For each Polaribacter clade, the correlation between the relative abundances obtained from (i) metagenome read recruitment, (ii) CARD-FISH analysis, and (iii) MED was calculated using the Spearman rank correlation test implemented in R v3.2.3 [33] (Supplementary Table S7).
Gene prediction and annotation
Gene prediction was performed using Prodigal [34] as implemented in Prokka v1.12 [35] including prediction of partial genes (omission of –c and –m options). Genes were annotated in RAST v2 [36] (Supplementary Table S8). CAZymes were annotated as described in [37] using the dbCAN v6 [38], Pfam v31 [39], and CAZy (as of March 15th, 2017) [9] databases. In addition, we included CAZyme-coding genes in our analysis that were only predicted by RAST when they were located in the vicinity of susCD genes. Peptidases were annotated based on best-hits of BLASTp v2.6.0+ [40] searches against the MEROPS database (merops_scan database v.12) [41] with default settings. For annotations of SusC-like proteins, SusD-like proteins, and sulfatases, HMMer v3.1 [42] searches with TIGRFAM (profile TIGR04056), Pfam (profiles PF07980, PF12741, PF12771, PF14322, and PF00884) were used (e-value: E−10). Putative substrate-specificities of PULs were predicted based on their CAZyme patterns as well as SusCD gene sequence similarity to PULs annotated in genomes of 53 North Sea Flavobacteriia [43]. These predictions were also crossed-checked with PULDB [44]. Amino acid sequences of SusC-like proteins were aligned in MAFFT v7 (G-INS-i algorithm, BLOSUM62 scoring matrix) [45] and subjected to phylogenetic analysis with the neighbor-joining method (JTT substitution model).
Core and pan genome analysis
Core and pan genomes of the Polaribacter clades were determined with EDGAR v2.2 [46, 47], which implements both reciprocal best BLAST hits and BLAST score ratio values [47] to calculate gene orthology. Pan genomes were determined for each Polaribacter clade (MAGs + genomes) and all of these pan genomes were subsequently used to determine common genes in-between clades as well as clade-specific genes (Supplementary Table S9). Genes other than CAZymes, peptidases, and hypothetical proteins were clustered according to COG categories [48] using eggNOG-mapper v4.5.1 [49] (mapping mode: DIAMOND, taxonomic scope: Bacteroidetes).
Metaproteomics
A total of 14 metaproteomes from 2009 [12] and 2010–2012 [43] Helgoland spring blooms were reanalyzed to investigate the in situ gene expression of distinct Polaribacter clades (23,917 proteins). Using standalone BLASTp v2.6.0+, the metaproteome sequences were queried for protein gene sequences from representative Polaribacter MAGs and genomes using the following cut-offs: percent identity >99%, e-value = 0 (Supplementary Table S10).
Results
Phylogenetic analysis of North Sea Polaribacter spp
Phylogenetic analysis of Polaribacter 16S rRNA clone sequences from 2009 to 2010 spring blooms revealed affiliation with four distinct clusters (Supplementary Fig. S1). Three clusters comprised clone sequences from April 14th, 2009, indicating a large diversity of Polaribacter species during the 2009 bloom. In contrast, clone sequences obtained on April 8th, 2010 fell into a single cluster, demonstrating presence of a distinct Polaribacter clade during the early phase of the 2010 bloom. The cultivated North Sea Polaribacter strains KT15, KT25b, and Hel1_88 branched in a separate cluster together with many other validly described species. Mean sequence similarities between these North Sea Polaribacter clades ranged from 96.7 to 98.4% (Supplementary Table S11), corroborating that they represent distinct species [50].
Classification of Polaribacter MAGs
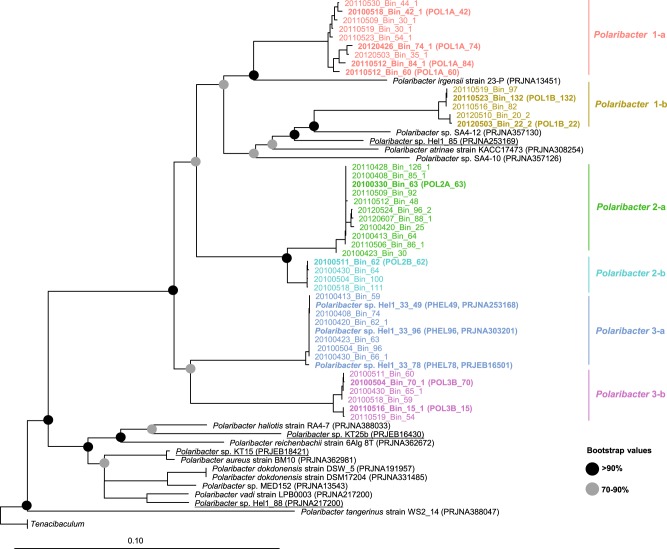
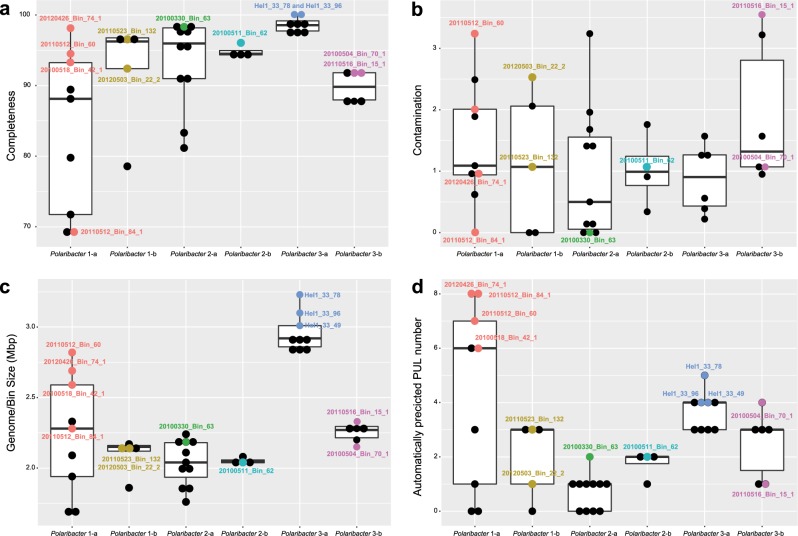
We obtained 41 Polaribacter-related MAGs (Supplementary Table S4) via binning of 38 metagenomes sampled from 2010 to 2012 bloom events (Supplementary Table S3) [15]. Phylogenetic survey of 43 conserved marker genes and ANI comparisons of the Polaribacter MAGs together with seven genomes of cultivated North Sea Polaribacter strains revealed affiliation with six distinct clades (henceforth termed Polaribacter 1-a, 1-b, 2-a, 2-b, 3-a, and 3-b) (Fig. 1). The Polaribacter 3-a clade also included the three sequenced North Sea strains Hel1_33_49, Hel1_33_78, and Hel1_33_96 [18, 43]. The overall tree topology was similar to the one inferred by 16S rRNA gene analysis (Supplementary Fig. S1). ANI values between these six clades were <95% (Supplementary Fig. S2), suggesting that they represent distinct species [51]. ANI values within the clades ranged from ~95 to 100% indicating some intraclade heterogeneity, apart from the clades Polaribacter 2-a and 2-b, which were seemingly homogeneous with ANI values around 99% (Supplementary Fig. S2). For further analysis, we selected the most complete and least contaminated MAGs with ANI >99% from clades 1-a, 1-b, 2-a, 2-b, and 3-b whereas for clade 3-a we used the genomes of strains Hel1_33_49, He1_33_78, and Hel1_33_96 (Fig. 2).
Fig. 1. Maximum likelihood phylogenetic tree of 43 conserved marker gene protein sequences from spring bloom-associated Polaribacter genomes and MAGs.
The names and abbreviations of Polaribacter MAGs that were used in further analyses are depicted in bold. For Polaribacter genomes, the corresponding NCBI, BioProject numbers are shown in brackets. The range of bootstrap values is indicated with black and gray circles. Bar: 0.1 substitutions per nucleotide position. Tenacibaculum spp. were used as outgroup.
Fig. 2. Completeness, contamination, sizes, and PUL numbers of spring-bloom-associated Polaribacter MAGs and genomes.
PULs were automatically predicted as it has been described in [15]. The MAGs that were considered for further analyses are labeled. The values for other MAGs are shown in Supplementary Table 4.
Polaribacter in situ abundances
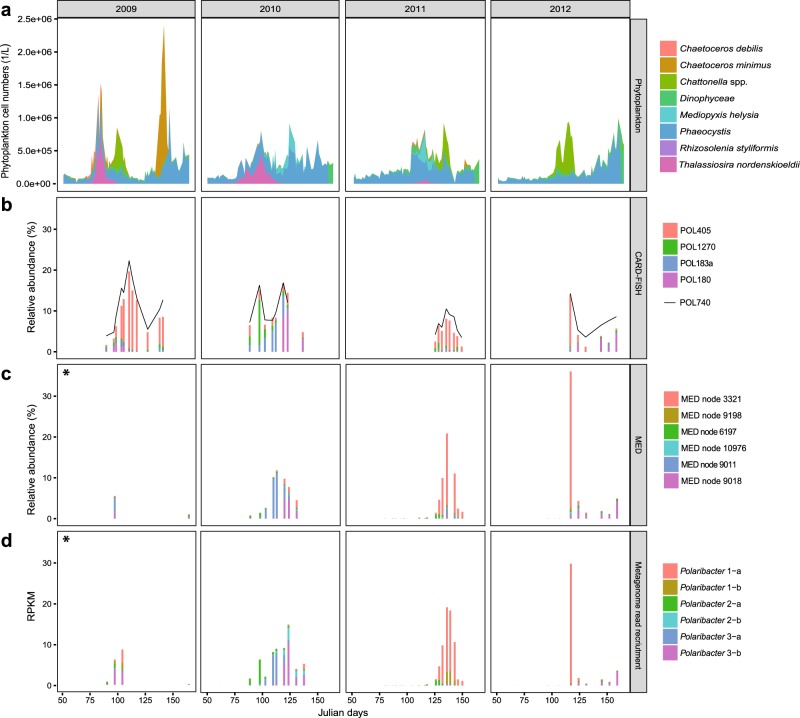
In situ abundances of the six Polaribacter clades during 2009–2012 North Sea spring blooms were assessed together with the phytoplankton community composition [13]. We (i) used CARD-FISH with newly designed oligonucleotide probes (Supplementary Table S1, Fig. 3b), (ii) reassessed previously published MED analyses of 16S rRNA amplicon data (Fig. 3c), and (iii) performed metagenome read recruitments on Polaribacter MAGs and genomes (Supplementary Table S6, Fig. 3d). CARD-FISH using specific oligonucleotide probes (coverage >82%, outgroup hits <15, Supplementary Table S1) provided cell numbers for individual Polaribacter clades (Supplementary Table S2). Comparison of these cell abundances with read frequencies detected by tag and metagenome sequencing enabled us to assess the temporal dynamics of Polaribacter clades with different methods.
Fig. 3. Temporal succession of six distinct Polaribacter clades during North Sea spring algal blooms.
a Cell counts of eight dominant phytoplankton clades as reported in [13]. b CARD-FISH analysis using newly designed oligonucleotide probes targeting four major Polaribacter clades and the genus-specific probe POL740. c Relative abundances of the six most abundant minimum entropy decomposition (MED) nodes as retrieved from [14]. d Metagenome read recruitments on Polaribacter MAGs and genomes. Recruited reads are normalized to the bin/genome sizes and reported as reads per kilobase million (RPKM). Taxonomic units with high levels of correlation across methods (Supplementary Table S7) are depicted in identical colors. An asterisk indicates data from the 2009 spring bloom are sparse, consisting of only two and four sampling dates for MED and metagenome analyses, respectively.
Polaribacter 1-a dominated the bacterial community in 2009 (19%), 2011 (7%), and 2012 (13%) based on cell counts with the clade-specific probe POL405 (Fig. 3b). Since Polaribacter MAG assemblies lacked 16S rRNA genes (Supplementary Table S8), a sequence-based link between taxonomic units of different methods could not be established. We therefore used Spearman rank tests to correlate the individual methods (Supplementary Table S7) and to interrelate different types of abundance data (Supplementary Fig. S3). For example, MED node 3321 (Fig. 3c) and metagenome read recruitment on Polaribacter 1-a MAGs (Fig. 3d) yielded a similar abundance pattern as CARD-FISH with POL405 (Fig. 3b) (Spearman’s rho = 0.96 and 0.88, respectively) (Supplementary Table S7). Polaribacter 1-a MAGs exhibited a differential abundance pattern. MAG POL1A_74 was abundant in 2009 and 2012, whereas MAGs POL1A_42, POL1A_60, and POL1A_84 were detected in 2011. MAG POL1A_42 was also found in lower abundances in the late phase of the 2010 bloom (Supplementary Table S6). Interestingly, the peak abundances of Polaribacter 1-a occurred after blooms of Chattonella (Fig. 3a). In terms of taxonomy, Chattonella spp. (phylum Ochrophyta, class Raphidophyceae) are rather distinct from diatoms (phylum Ochrophyta, class Bacillariophyceae), which usually dominate Helgoland spring blooms. In 2010, the phytoplankton community was dominated by Phaeocystis spp., Thalassiosira nordenskioeldii, and Mediopyxis helysia. In this year, Polaribacter 2-a, Polaribacter 3-a, and Polaribacter 3-b successively reached high abundances (6%, 8%, and 10%) (Fig. 3d). Polaribacter 2-a and Polaribacter 3-b were also present during the early and late bloom phases in 2009, 2011, and 2012, albeit with lower abundances (1%, 1%, 0.4 and 4%, 0.5%, 3%), whereas Polaribacter 3-a was only abundant in 2010 (Fig. 3d). In contrast, Polaribacter 1-b and Polaribacter 2-b exhibited lower relative abundances than the other Polaribacter clades (Fig. 3d). Polaribacter 1-b was detected at relatively low abundances of 2–3% in 2009 and 2011, while Polaribacter 2-b was present only in 2010 where it reached up to 3%.
Core and pan genomes
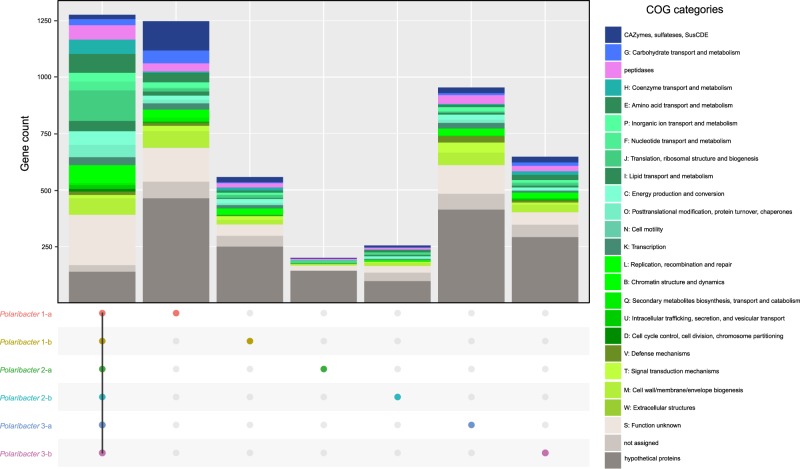
We determined common genes between the six distinct Polaribacter clades, as well as genes that were unique to each clade (Fig. 4). The combined core genome of all Polaribacter clades comprised 1275 genes and was dominated by genes involved in housekeeping and basic cellular functions (Supplementary Table S9).
Fig. 4. Core and pan genome analysis of North Sea Polaribacter clades.
Number of the genes in the core genome of all clades, and unique gene repertoire of each clade are shown. Metabolic classification was obtained according to COG categories [48]. The genes which are shared by at least two clades are listed in Supplementary Table S9.
All Polaribacter clades coded for the Embden–Meyerhof‐Parnas, and pentose‐5‐phosphate pathways as well as the tricarboxylic acid cycle. An aerobic redox chain comprising a NAD(H):ubiquinone oxidoreductase (complex I), succinate dehydrogenase (complex II), and cytochrome cbb3 and aa3 type (complex IV) terminal oxidases was also present. All clades furthermore contained proteorhodopsin, which might generate supplemental energy from light. In terms of nutrient metabolism, assimilatory sulfate, and nitrate reduction genes (e.g., sulfate adenylyltransferase and ferrodoxin-nitrate reductase) were found together with dedicated transporter systems. All Polaribacter clades also possessed polyphosphate production and hydrolysis genes (ppK and ppX). Genes associated with gliding motility (gldDE-H-B-KLMN-A-I-FGJ) were also present in all clades. As for vitamin metabolism, Polaribacter spp. possessed biotin and thiamin biosynthesis genes (bioC and apbE) as well as ABC transporters for vitamin B12 uptake (btuB-F-C). In contrast, many carbohydrate degradation and transport genes were distinct for each clade (Fig. 4), thus belonging to the pan genome. For example, Polaribacter 1-a contained 130 genes for glycan utilization and transport including CAZyme families GH92 (e.g., α-mannosidase) and GH2 (e.g., β-galactosidase) (Supplementary Table S9). Likewise, Polaribacter 3-a possessed GH92, while Polaribacter 3-b encoded GH10 (e.g., β-xylanase) and GH128 (β-glucanase) genes.
Metabolic comparison of Polaribacter MAGs and genomes
We compared the metabolic potential of Polaribacter MAGs and the genomes of cultivated Polaribacter strains based on RAST subsystem analyses [36]. Polaribacter MAGs contained higher gene proportions in the RAST categories “Amino acid and Derivatives,” “Cofactors, Vitamins, Prosthetic Groups, Pigments,” and “RNA metabolism” (99.4, 67.5, and 47.5 genes/Mbp in average) (Supplementary Fig. S4) (Supplementary Table S12). Furthermore, both MAGs and genomes possessed high gene abundances in the categories “Carbohydrates” (average: 53.9 genes/Mbp) and “Protein Metabolism” (average: 59.6 genes/Mbp), albeit with large variations (standard deviations: 13.6 and 15.5 genes/Mbp).
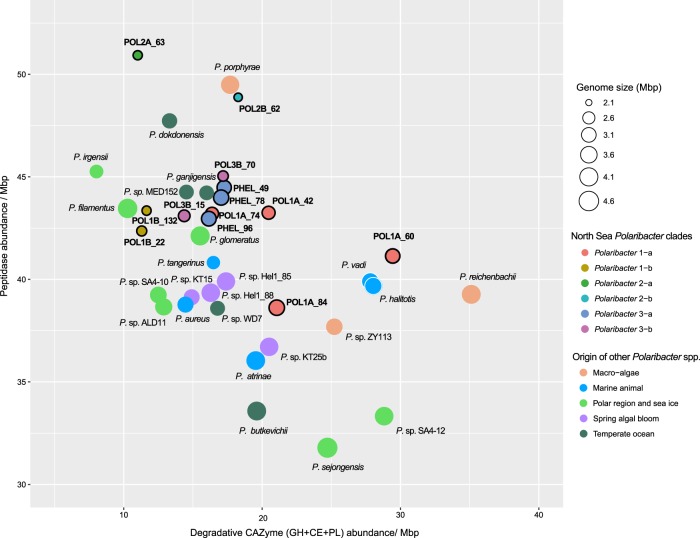
Besides polysaccharides, large amounts of proteins are also present in marine phytoplankton [52] and heterotrophic bacteria are able to utilize these proteins using peptidases [53]. Comparison of peptidase and degradative CAZyme abundances (GH + CE + PL) between Polaribacter clades and other available Polaribacter genomes showed different protein and carbohydrate utilization profiles (Supplementary Table S13). All Polaribacter clades encoded high numbers of serine (S) and metallo (M) peptidases (~80% of all peptidases), which mediate the degradation and uptake of extracellular proteins [18]. Furthermore, GHs were the most abundant CAZyme family genes. Remarkably, the range of CAZyme proportions (8–39 per Mbp) was two times higher than that of peptidases (37–51 per Mbp). With increasing genome size, Polaribacter spp. harbor proportionately fewer peptidases (Spearman’s rho: −0.65, p = 0.01) and more degradative CAZymes (Spearman’s rho: 0.45, p = 0.04), corroborating an earlier analysis of 27 Flavobacteriaceae genomes [18]. Together with Polaribacter isolates from, for example, temperate seawater and polar regions, Polaribacter 2-a and Polaribacter 2-b had the highest peptidase and the lowest degradative CAZyme abundances together with the smallest estimated MAG sizes (Fig. 5). In contrast, some MAGs in Polaribacter 1-a (POL1A_60 and POL1A_84) possessed the highest CAZyme and lowest peptidase proportions and grouped with species that were associated with macroalgae and marine animals (Fig. 5). Polaribacter 3-a and Polaribacter 3-b contained moderate CAZyme and peptidase repertoires together with North Sea spring bloom isolates, while Polaribacter 1-b harbored the lowest numbers (Fig. 5).
Fig. 5. Comparison of peptidase and degradative CAZyme (GH + PL + CE) repertoires of North Sea Polaribacter clades (black-rimmed) and the described species or isolates with sequenced genomes in the same genus.
Genomes belonging to Polaribacter 3-a are indicated by the prefix “PHEL”. Gene abundances are normalized with the MAG or genome sizes. Complete MAG sizes are estimated based on the completeness values calculated via CheckM [30].
PUL repertoires
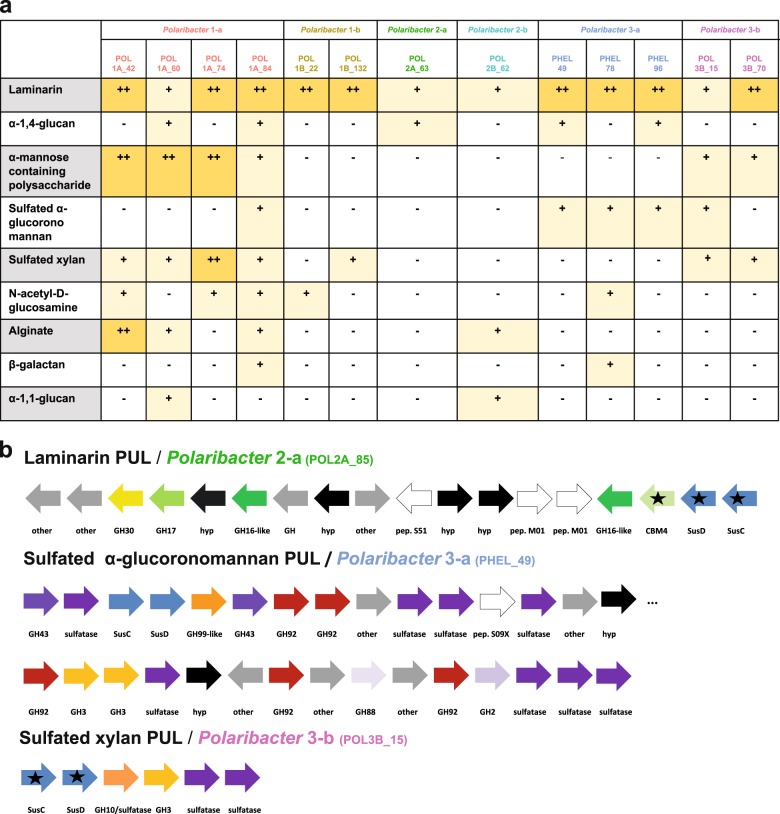
We investigated the PUL repertoires of Polaribacter MAGs and genomes to predict the glycan niches of North Sea Polaribacter spp. (Fig. 6a). Phylogenetic analysis of the translated susC gene sequences encoded in these PULs indicated substrate-specific clustering [43] and enabled to identify variants of PULs that putatively target identical or at least similar substrates (Supplementary Figs. S5, S6).
Fig. 6. PUL repertoires of North Sea Polaribacter clades.
a Distribution of PULs with predicted substrates across six distinct Polaribacter clades. Plus sign (+) correspond to the PUL variants for individual substrates as they are suggested by phylogenetic analysis of SusC protein sequences in Supplementary Fig. S5. b Gene composition of some distinctive PULs encoded by North Sea Polaribacter clades. Black stars indicate expressed genes based on metaproteome analyses (Fig. 7). CAZymes, peptidases and Sus transport genes are depicted in different colors. Hypothetical proteins and genes involved in other metabolic functions are abbreviated with “hyp” and “other,” respectively. CAZymes annotated only by RAST are highlighted by the suffix “-like.” Compositions of all annotated PULs in the Polaribacter clades are summarized in Supplementary Fig. S6.
All Polaribacter clades possessed PULs predicted to degrade diatom storage glycan laminarin, which has a β-1,3-linked glucose backbone that sometimes includes β-1,2 or β-1,6-glucose side chains (Fig. 6a) [54]. Seven variants of laminarin PULs with different combinations of GH3, GH5, GH16, GH17, and GH30 family GH were detected (Supplementary Figs. S5, S6). The respective PUL (laminarin B1) detected in Polaribacter 3-a (strain Hel1_33_49) was also shown to be up-regulated with laminarin in a previous study [18]. Among these PULs, Polaribacter 2-a, and Polaribacter 2-b had the most complex one, which included peptidases M01 (e.g. aminopeptidase activity) and S51 (dipeptidase activity) and CBM4 (e.g., binding to β-1,3-glucans) (Fig. 6b).
Three Polaribacter clades (1-a, 2-a, and 3-a) harbored PULs predicted to target α-1,4-glucans, which are common storage compounds in marine algae, bacteria, and animals (Fig. 6a) [55]. Having two variants, these PULs contained GH13 (α-amylase), GH65 (maltose phosphorylase), GH31 (e.g., α-glucosidase) as core GH genes (Supplementary Fig. S6). A glucose-induced PUL with high synteny was shown in the North Sea isolate Gramella forsetii KT0803T [56]. Alpha-1,4-glucan PULs also encoded two susE genes, which direct the uptake of maltooligosaccharides of specific lengths and likely facilitate the selection of particular glycans from the environment [57].
Moreover, Polaribacter clades 1-a and 3-b had putative PULs to utilize α-mannose-rich polysaccharides (Fig. 6a). These PULs featured two variants and encoded proteins of CAZymes families, for example, GH92 (e.g., α-mannosidase) and GH130 (e.g., mannooligosaccharide phosphorylase) (Supplementary Fig. S6). Specificity for α-mannan of a PUL containing GH92, GH130, and GH76 has been shown in a bacterium from the human gut [58]. Polysaccharides rich in α-mannose have been identified as constituents in the frustules of some diatoms [59].
Polaribacter 1-a, 3-a, and 3-b harbored PULs presumably targeting sulfated α-glucoronomannans (Fig. 6a). Among these, Polaribacter 3-a carried the most complex PUL of all clades. This PUL contained twelve CAZymes including five GH92 (endo-α-mannosidases), two GH3 and a GH99 (putative endo-α-mannanase) together with nine sulfatases (Fig. 6b). Sulfated α-glucoronomannans are found in diatom cell walls [60] and sulfatase- and GH92-rich PULs have been previously detected in Polaribacter–affiliated fosmids from the North Atlantic [61], suggesting a potential high prevalence in the world’s oceans.
Polaribacter clades 1-a, 1-b, and 3-b possessed PULs predicted to target sulfated xylans (Fig. 6a). Polaribacter 3-b harbored all variants and the most complex form of putative sulfated xylan PUL (Fig. 6b, Supplementary Fig. S6). This PUL comprised a GH3 (e.g., xylosidase), a GH10 (e.g., β-xylanase) and two sulfatase genes, together with an adjacent putative PUL containing a GH128 (β-glucanase) and four sulfatases genes. A similar PUL with GH3 and GH10 was shown to be upregulated with xylan in the human gut bacterium Bacteroides xylanisolvens [62]. Xylans are also components of marine phytoplankton [63], and high xylanase activities have been reported for many ocean provinces [64]. In addition to these substrates, the glycan niches of the Polaribacter clades were distinct with respect to the utilization of N-acetyl-D-glucosamine, alginate, β-galactans, and α-1,1-glucans (Fig. 6a, Supplementary Fig. S6, Supplementary Information).
Metaproteomics
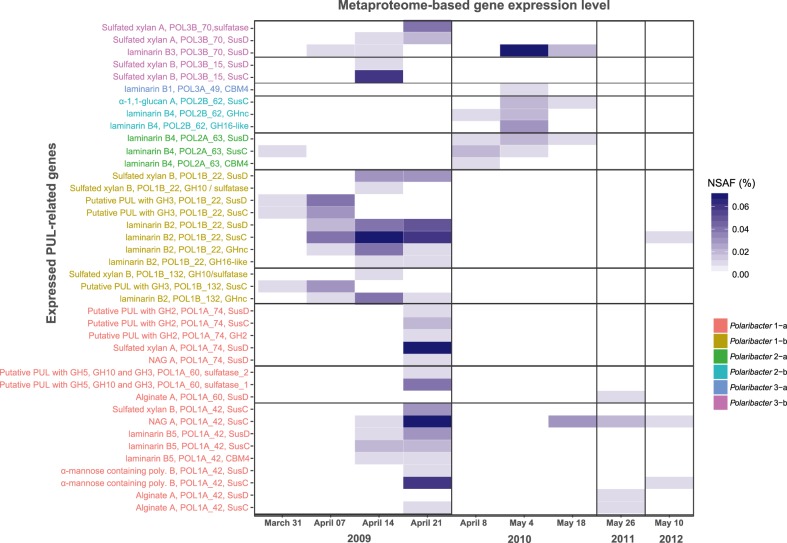
We analyzed 14 metaproteomes from 2009 to 2012 spring algal blooms [12, 43] to investigate in situ expression profiles of representative MAGs and genomes in each Polaribacter clade (Fig. 7) (Supplementary Table S10). In total, 845 Polaribacter proteins were found to be expressed (3.5% of the total proteome). Polaribacter clades 1-b and 3-b exhibited high expression of genes related to the degradation of laminarin and sulfated xylan during the late phase of the 2009 bloom (Fig. 7). Furthermore, Polaribacter 1-a showed expression of proteins associated with utilization of N-acetyl-D-glucosamine, α-mannose-rich polysaccharides and alginate during this time. In 2010, we could detect mainly laminarin degradation by the clades 2-a, 2-b, 3-a, and 3-b. In 2011 and 2012, the metaproteome sampling dates did not coincide with Polaribacter peak abundances, which is why no relevant Polaribacter protein expression profiles could be detected (Fig. 7). Analysis of expressed SusC genes from all abundant Bacteroidetes species during North Sea algal blooms revealed that Polaribacter spp. dominated alginate and sulfated xylan degradation, while other clades were also involved in the degradation of laminarin, α-mannose-rich polysaccharide and α-glucan-containing polysaccharides [15].
Fig. 7. Metaproteome-based expression analysis of PUL-related genes in Polaribacter clades.
All expressed genes in Polaribacter MAGs/genomes are summarized in Supplementary Table S10. Metaproteomes without any detected PUL-related protein are not shown. NSAF normalized spectral abundance factor.
Discussion
Temporal dynamics and distinct polysaccharide niches of abundant Polaribacter clades during spring phytoplankton blooms in the southern North Sea suggest that changes in the composition and quantity of available algal polysaccharides play a prominent role in their differential occurrences.
Co-occurring bacteria during algal blooms that feed on algal high-molecular weight compounds (HMW) are expected to specialize on different substrates to avoid direct competition. The six Polaribacter clades that we analyzed occupied different niches with respect to utilization of HMW compounds as they differ with respect to genome sizes, peptidase numbers, and PUL spectra (Fig. 8). Polaribacter strains isolated from seawater outside of bloom events such as P. dokdonensis feature only few degradative CAZymes and high relative peptidase numbers [65]. On the other hand, Polaribacter strains that were isolated from the North Sea on agar plates feature notably higher proportions of CAZymes and lower proportions of peptidases [18, 43], e.g., strains KT25b, Hel1_85, and Hel1_88 (Fig. 5). However, these strains are not representative for any of the abundant Polaribacter clades during North Sea spring blooms, which occupy more of a middle position in terms of peptidases and CAZymes (Fig. 5), indicating elevated relevance of both, proteins and polysacharides.
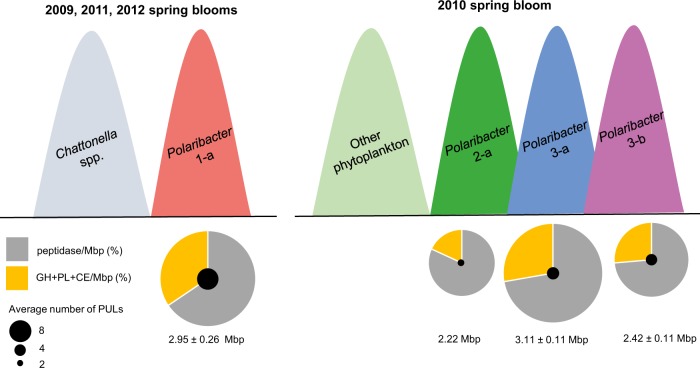
Fig. 8. Conceptual model describing the proposed temporal and metabolic basis for polysaccharide niche partitioning between four major Polaribacter clades.
A simplified scheme of temporal occurrences in sampled bloom events is shown. For the representative MAGs or genomes in each clade, the average number of PULs and average peptidase and degradative CAZyme (GH + PL + CE) proportions are depicted. Pie charts are normalized with the average genome or complete MAG sizes.
Polaribacter 2-a was abundant in the early phases of sampled bloom events (Fig. 3). Owing to its high peptidase proportion, small genome size (2.18 Mbp) and limited polysaccharide utilization capacity (Fig. 5), it could be characterized as a typical first responder (Fig. 8). Polaribacter 2-a encoded a unique laminarin PUL variant with a combination of CAZymes and peptidases (Fig. 6b). Respective SusC-like, SusD-like and CBM44 proteins were expressed during the 2010 bloom (Fig. 7). Coupling of carbohydrate and protein degradation could enable rapid growth. Such a mechanism has been recently described for North Sea spring-bloom associated Formosa strain Hel1_33_131 that also features a comparably small genome (2.7 Mbp) [66].
Polaribacter 3-a and Polaribacter 3-b responded later in the sampled spring blooms and were predicted to target more complex sulfated polysaccharides. Polaribacter 3-a was highly abundant only during the mid-phase of the 2010 bloom (Fig. 3) and had a distinctive PUL predicted to utilize sulfated α-glucoronomannan (Fig. 6b). However, expression of proteins related to this PUL was not detected, likely since the metaproteome sampling dates in 2010 (April 4th and May 4th) did not coincide with Polaribacter 3-a peak abundances (April 23rd) (Fig. 7). Polaribacter 3-b was detected in the late phase of bloom events (Fig. 3). This clade had a distinctive PUL predicted to target sulfated xylan together with a putative GH128-containing PUL (Fig. 6b). Expression of susCD genes of this PUL was detected during the 2009 spring bloom (Fig. 7). Sulfated polysaccharides are more recalcitrant, since the sulfate groups reduce the accessibility for degrading CAZymes (steric hinderance). Removal of these sulfate groups requires dedicated sulfatases. The flavobacterium Zobellia galactanivorans DsijT, for instance, harbors no less than 71 sulfatase genes, which constitutes a considerable genetic investment [67, 68]. Maximum sulfatase expression has been also linked to the final stage of algal blooms [12]. Therefore, the capacity to degrade sulfated polysaccharides could provide an ecological advantage for Polaribacter 3-a and 3-b to dominate the late phases of bloom events.
Of all investigated clades, the analyzed Polaribacter 1-a MAGs featured among the highest CAZyme gene frequencies and PUL numbers together with the lowest relative peptidases proportions (Fig. 5). Four representative MAGs within Polaribacter 1-a possessed PULs targeting nine different substrate classes, including laminarin, α-mannose-rich polysaccharides, sulfated xylans, N-acetyl-D-glucosamine, and alginate (Fig. 6a, Supplementary Fig. S6). A recent study analyzing the PUL repertoire of 53 sequenced North Sea flavobacterial isolates suggested an average of 3.8 targeted classes of glycans per genome [43]. Hence, nine substrates are indicative for a pronounced specialization of this clade on glycans. Moreover, alginate has so far only been described in brown macroalgae [69], indicating that Polaribacter 1-a might not only associate with planktonic microalgae during transient bloom events. This observation was also corroborated by peptidase and CAZyme abundance patterns of Polaribacter 1-a, which were similar to those of Polaribacter species that are known to associate with macroalgae and marine animals. Remarkably, peaks in Polaribacter 1-a abundances were detected after blooms of Chattonella microalgae in 2009, 2011, and 2012 (Fig. 3). However, co-occurrence in just three years (n = 3) is insufficient for meaningful statistical analyses. Thus, targeted studies under controlled conditions with representative Polaribacter and Chattonella strains are required to elucidate whether a specific interaction indeed exists as suggested by our in situ data.
Polaribacter 1-b and 2-b represented lower abundant rare clades with distinctive polysaccharide utilization capacities. Polaribacter 1-b was detected during 2009 and 2011 blooms (Fig. 3) and expressed genes within predicted sulfated xylan and laminarin targeting PULs and a putative PUL containing a GH3 family gene (Fig. 7). Polaribacter 2-b was furthermore detected in the late phase of 2010 bloom where it expressed the SusC-like protein of its predicted α-1,1-glucan PUL (Figs. 6a, 7, Supplementary Information).
North Sea Polaribacter MAGs had smaller estimated genome sizes (average: 2.51 Mbp) than cultivated strains (average: 3.47 Mbp) (Fig. 5). They also possessed substantially higher gene abundances for cofactor, vitamin, and amino acid synthesis and transport metabolisms than North Sea spring bloom isolates (Supplementary Fig. S4). These differences could result from imperfect binning of recently transferred genes [70]. This might also apply to PULs, which have been shown to be horizontally transferred [71]. Unfortunately, extensive cultivation efforts during the bloom events did not result in the isolation of representatives of most of the environmentally relevant Polaribacter clades [43]. We hence suggest that future cultivation efforts should consider the metabolic differences indicated by comparison of MAGs and strain genomes.
The intragenus phylogenetic diversity and corresponding metabolic diversification that we show for the genus Polaribacter highlights how different clades within a genus can forgo direct competition in a nutrient-rich environment with rapidly changing conditions. These clades are likely yet-uncultivated Polaribacter species, which should be future targets of isolation efforts and taxonomic characterization. Such studies will allow for a more complete picture of the distinct niches of closely related bacteria that co-occur during algal blooms and ultimately will lead to a better understanding of the ecological and evolutionary processes that determine the microbial community composition in such dynamic environments.
Supplementary information
Acknowledgements
We thank Jörg Wulf and Dolma Michellod for technical support, Ivaylo Kostadinov of GFBio (https://www.gfbio.org) for support with sequence data deposition, T. Ben Francis for help in bioinformatics, and Carol Arnosti for critical reading. Jochen Blom’s support in core and pan genome analysis is acknowledged. Metagenome sequencing was conducted in the framework of the Community Sequencing Project COGITO (CSP 998) by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, and is supported by the Office of Science of the US Department of Energy under Contract No. DE-AC02-05CH11231. This study was funded by the Max Planck Society and supported by the Deutsche Forschungsgemeinschaft (DFG) in the framework of the research unit FOR2406 ‘Proteogenomics of Marine Polysaccharide Utilization (POMPU)’ by grants of RA (AM 73/9-1), HT (TE 813/2-1), and BF (FU 627/2-1). Burak Avcı and Karen Krüger are members of the International Max Planck Research School of Marine Microbiology (MarMic). Open access funding provided by Projekt DEAL.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hanno Teeling, Email: hteeling@mpi-bremen.de.
Rudolf I. Amann, Email: ramann@mpi-bremen.de
Supplementary information
The online version of this article (10.1038/s41396-020-0601-y) contains supplementary material, which is available to authorized users.
References
- 1.Nemergut DR, Schmidt SK, Fukami T, O’Neill SP, Bilinski TM, Stanish LF, et al. Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev. 2013;77:342–56. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998;281:237–40. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 3.Mühlenbruch M, Grossart H-P, Eigemann F, Voss M. Phytoplankton-derived polysaccharides in the marine environment and their interactions with heterotrophic bacteria. Environ Microbiol. 2018;20:2671–85.. doi: 10.1111/1462-2920.14302. [DOI] [PubMed] [Google Scholar]
- 4.Becker S, Scheffel A, Polz MF, Hehemann J-H. Accurate quantification of laminarin in marine organic matter with enzymes from marine microbes. Appl Environ Microbiol. 2017;83:e03389–16.. doi: 10.1128/AEM.03389-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camarena-Gomez MT, Lipsewers T, Piiparinen J, Eronen-Rasimus E, Perez-Quemalinos D, Hoikkala L, et al. Shifts in phytoplankton community structure modify bacterial production, abundance and community composition. Aquat Microb Ecol. 2018;81:149–70.. [Google Scholar]
- 6.Niu Y, Shen H, Chen J, Xie P, Yang X, Tao M, et al. Phytoplankton community succession shaping bacterioplankton community composition in Lake Taihu, China. Water Res. 2011;45:4169–82. doi: 10.1016/j.watres.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 7.Tada Y, Taniguchi A, Nagao I, Miki T, Uematsu M, Tsuda A, et al. Differing growth responses of major phylogenetic groups of marine bacteria to natural phytoplankton blooms in the Western North Pacific Ocean. Appl Environ Microbiol. 2011;77:4055–65. doi: 10.1128/AEM.02952-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hehemann J-H, Arevalo P, Datta MS, Yu X, Corzett CH, Henschel A, et al. Adaptive radiation by waves of gene transfer leads to fine-scale resource partitioning in marine microbes. Nat Commun. 2016;7:e12860. doi: 10.1038/ncomms12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–5. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemsworth GR, Déjean G, Davies Gideon J, Brumer H. Learning from microbial strategies for polysaccharide degradation. Biochem Soc Trans. 2016;44:94–108. doi: 10.1042/BST20150180. [DOI] [PubMed] [Google Scholar]
- 11.Grondin JM, Tamura K, Déjean G, Abbott DW, Brumer H. Polysaccharide utilization loci: fueling microbial communities. J Bacteriol. 2017;199:e00860–16.. doi: 10.1128/JB.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336:608–11. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 13.Teeling H, Fuchs BM, Bennke CM, Krüger K, Chafee M, Kappelmann L, et al. Recurring patterns in bacterioplankton dynamics during coastal spring algae blooms. eLIFE. 2016;5:e11888. doi: 10.7554/eLife.11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chafee M, Fernandez-Guerra A, Buttigieg PL, Gerdts G, Eren AM, Teeling H, et al. Recurrent patterns of microdiversity in a temperate coastal marine environment. ISME J. 2018;12:237–52.. doi: 10.1038/ismej.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krüger K, Chafee M, Francis TB, Glavina del Rio T, Becher D, Schweder T, et al. In marine Bacteroidetes the bulk of glycan degradation during algae blooms is mediated by few clades using a restricted set of genes. ISME J. 2019;13:2800–16.. doi: 10.1038/s41396-019-0476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prosser JI, Bohannan BJM, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, et al. The role of ecological theory in microbial ecology. Nat Rev Microbiol. 2007;5:384–92. doi: 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- 17.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–5. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- 18.Xing P, Hahnke RL, Unfried F, Markert S, Huang SX, Barbeyron T, et al. Niches of two polysaccharide-degrading Polaribacter isolates from the North Sea during a spring diatom bloom. ISME J. 2015;9:1410–22. doi: 10.1038/ismej.2014.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–9. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 23.Peplies J, Kottmann R, Ludwig W, Glöckner FO. A standard operating procedure for phylogenetic inference (SOPPI) using (rRNA) marker genes. Syst Appl Microbiol. 2008;31:251–7. doi: 10.1016/j.syapm.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Thiele S., Fuchs B.M., Amann R.I. Treatise on Water Science. 2011. Identification of Microorganisms Using the Ribosomal RNA Approach and Fluorescence In Situ Hybridization; pp. 171–189. [Google Scholar]
- 25.Bennke CM, Reintjes G, Schattenhofer M, Ellrott A, Wulf J, Zeder M, et al. Modification of a high-throughput automatic microbial cell enumeration system for shipboard analyses. Appl Environ Microbiol. 2016;82:3289–96. doi: 10.1128/AEM.03931-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francis TB, Krüger K, Fuchs BM, Teeling H, Amann RI. Candidatus Prosiliicoccus vernus, a spring bloom associated member of the Flavobacteriaceae. Syst Appl Microbiol. 2019;42:41–53. doi: 10.1016/j.syapm.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, et al. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11:1144–6. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 29.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-R LM, Konstantinidis K. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016;4:e1900v1. [Google Scholar]
- 33.R Core Team. R: a language and environment for statistical computing. Austria: R Found Stat Comput Vienna; 2017.
- 34.Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 36.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avcı B, Hahnke RL, Chafee M, Fischer T, Gruber-Vodicka H, Tegetmeyer HE, et al. Genomic and physiological analyses of ‘Reinekea forsetii’ reveal a versatile opportunistic lifestyle during spring algae blooms. Environ Microbiol. 2017;19:1209–21.. doi: 10.1111/1462-2920.13646. [DOI] [PubMed] [Google Scholar]
- 38.Huang Le, Zhang Han, Wu Peizhi, Entwistle Sarah, Li Xueqiong, Yohe Tanner, Yi Haidong, Yang Zhenglu, Yin Yanbin. dbCAN-seq: a database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Research. 2017;46(D1):D516–D521. doi: 10.1093/nar/gkx894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finn Robert D., Coggill Penelope, Eberhardt Ruth Y., Eddy Sean R., Mistry Jaina, Mitchell Alex L., Potter Simon C., Punta Marco, Qureshi Matloob, Sangrador-Vegas Amaia, Salazar Gustavo A., Tate John, Bateman Alex. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research. 2015;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 41.Rawlings ND, Alan J, Thomas PD, Huang XD, Bateman A, Finn RD. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46(D1):624–32. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mistry J, Finn RD, Eddy SR, Bateman A, Punta M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013;41:e121. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kappelmann L, Krüger K, Hehemann J-H, Harder J, Markert S, Unfried F, et al. Polysaccharide utilization loci of North Sea Flavobacteriia as basis for using SusC/D-protein expression for predicting major phytoplankton glycans. ISME J. 2019;13:76–91. doi: 10.1038/s41396-018-0242-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terrapon N, Lombard V, Drula E, Lapebie P, Al-Masaudi S, Gilbert HJ, et al. PULDB: the expanded database of Polysaccharide Utilization Loci. Nucleic Acids Res. 2018;46:677–83. doi: 10.1093/nar/gkx1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2017; 10.1093/bib/bx108. [DOI] [PMC free article] [PubMed]
- 46.Blom J, Kreis J, Spanig S, Juhre T, Bertelli C, Ernst C, et al. EDGAR 2.0: an enhanced software platform for comparative gene content analyses. Nucleic Acids Res. 2016;44:22–8. doi: 10.1093/nar/gkw255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lerat E, Daubin V, Moran NA. From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-proteobacteria. PLoS Biol. 2003;1:101–9. doi: 10.1371/journal.pbio.0000019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–6. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34:2115–22.. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12:635–45. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 51.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 52.Roy S. Distributions of phytoplankton carbohydrate, protein and lipid in the world oceans from satellite ocean colour. ISME J. 2018;12:1457–72.. doi: 10.1038/s41396-018-0054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orsi WD, Smith JM, Liu S, Liu Z, Sakamoto CM, Wilken S, et al. Diverse, uncultivated bacteria and archaea underlying the cycling of dissolved protein in the ocean. ISME J. 2016;10:2158–73. doi: 10.1038/ismej.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gügi B, Le Costaouëc T, Burel C, Lerouge P, Helbert W, Bardor M. Diatom-specific oligosaccharide and polysaccharide structures help to unravel biosynthetic capabilities in diatoms. Mar Drugs. 2015;13:5993–6018. doi: 10.3390/md13095993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viola R, Nyvall P, Pedersen M. The unique features of starch metabolism in red algae. Proc R Soc B-Biol Sci. 2001;268:1417–22. doi: 10.1098/rspb.2001.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kabisch A, Otto A, König S, Becher D, Albrecht D, Schüler M, et al. Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes ‘Gramella forsetii’ KT0803. ISME J. 2014;8:1492–502. doi: 10.1038/ismej.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foley Matthew H., Martens Eric C., Koropatkin Nicole M. SusE facilitates starch uptake independent of starch binding in B. thetaiotaomicron. Molecular Microbiology. 2018;108(5):551–566. doi: 10.1111/mmi.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cuskin F, Lowe EC, Temple MJ, Zhu YP, Cameron EA, Pudlo NA, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–86. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiovitti A, Harper RE, Willis A, Bacic A, Mulvaney P, Wetherbee R. Variations in the substituted 3-linked mannans closely associated with the silicified walls of diatoms. J Phycol. 2005;41:1154–61.. [Google Scholar]
- 60.Le Costaouëc T, Unamunzaga C, Mantecon L, Helbert W. New structural insights into the cell-wall polysaccharide of the diatom Phaeodactylum tricornutum. Algal Res. 2017;26:172–9. [Google Scholar]
- 61.Bennke CM, Krüger K, Kappelmann L, Huang S, Gobet A, Schüler M, et al. Polysaccharide utilisation loci of Bacteroidetes from two contrasting open ocean sites in the North Atlantic. Environ Microbiol. 2016;18:4456–70.. doi: 10.1111/1462-2920.13429. [DOI] [PubMed] [Google Scholar]
- 62.Despres J, Forano E, Lepercq P, Comtet-Marre S, Jubelin G, Chambon C, et al. Xylan degradation by the human gut Bacteroides xylanisolvens XB1AT involves two distinct gene clusters that are linked at the transcriptional level. BMC Genomics. 2016;17:326. doi: 10.1186/s12864-016-2680-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parsons TR, Stephens K, Strickland JDH. On the chemical composition of eleven species of marine phytoplankters. J Fish Res Board Can. 1961;18:1001–16.. [Google Scholar]
- 64.Arnosti C. Microbial extracellular enzymes and the marine carbon cycle. Annu Rev Mar Sci. 2011;3:401–25.. doi: 10.1146/annurev-marine-120709-142731. [DOI] [PubMed] [Google Scholar]
- 65.Fernández-Gómez B, Richter M, Schüler M, Pinhassi J, Acinas SG, González JM, et al. Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. 2013;7:1026–37. doi: 10.1038/ismej.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Unfried F, Becker S, Robb C, Hehemann J-H, Markert S, Heiden S, et al. Adaptive mechanisms that provide competitive advantages to marine Bacteroidetes during microalgal blooms. ISME J. 2018;12:2894–906. doi: 10.1038/s41396-018-0243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barbeyron T, Thomas F, Barbe V, Teeling H, Schenowitz C, Dossat C, et al. Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: example of the model algae-associated bacterium Zobellia galactanivorans DsijT. Environ Microbiol. 2016;18:4610–27.. doi: 10.1111/1462-2920.13584. [DOI] [PubMed] [Google Scholar]
- 68.Hettle AG, Vickers C, Robb CS, Liu F, Withers SG, Hehemann J-H, et al. The molecular basis of polysaccharide sulfatase activity and a nomenclature for catalytic subsites in this class of enzyme. Structure. 2018;26:747–58.e4. doi: 10.1016/j.str.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 69.Peteiro César. Springer Series in Biomaterials Science and Engineering. Singapore: Springer Singapore; 2017. Alginate Production from Marine Macroalgae, with Emphasis on Kelp Farming; pp. 27–66. [Google Scholar]
- 70.Ramos-Barbero MD, Martin-Cuadrado A-B, Viver T, Santos F, Martinez-Garcia M, Antón J. Recovering microbial genomes from metagenomes in hypersaline environments: the Good, the Bad and the Ugly. Syst Appl Microbiol. 2019;42:30–40. doi: 10.1016/j.syapm.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 71.Hehemann J-H, Kelly AG, Pudlo NA, Martens EC, Boraston AB. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc Natl Acad Sci USA. 2012;109:19786–91. doi: 10.1073/pnas.1211002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.