Abstract
In recent years, Mycoplasma pneumoniae strains that are clinically resistant to macrolide antibiotics have occasionally been encountered in Japan. Of 76 strains of M. pneumoniae isolated in three different areas in Japan during 2000 to 2003, 13 strains were erythromycin (ERY) resistant. Of these 13 strains, 12 were highly ERY resistant (MIC, ≥256 μg/ml) and 1 was weakly resistant (MIC, 8 μg/ml). Nucleotide sequencing of domains II and V of 23S rRNA and ribosomal proteins L4 and L22, which are associated with ERY resistance, showed that 10 strains had an A-to-G transition at position 2063 (corresponding to 2058 in Escherichia coli numbering), 1 strain showed A-to-C transversion at position 2063, 1 strain showed an A-to-G transition at position 2064, and the weakly ERY-resistant strain showed C-to-G transversion at position 2617 (corresponding to 2611 in E. coli numbering) of domain V. Domain II and ribosomal proteins L4 and L22 were not involved in the ERY resistance of these clinical M. pneumoniae strains. In addition, by using our established restriction fragment length polymorphism technique to detect point mutations of PCR products for domain V of the 23S rRNA gene of M. pneumoniae, we found that 23 (24%) of 94 PCR-positive oral samples taken from children with respiratory infections showed A2063G mutation. These results suggest that ERY-resistant M. pneumoniae infection is not unusual in Japan.
Mycoplasma pneumoniae is a pathogen causing human respiratory infections such as atypical pneumonia, mainly in children and younger adults. In the chemotherapy of M. pneumoniae infection in children, erythromycin (ERY) and clarithromycin (CLR) among 14-membered macrolides and the 15-membered macrolide azithromycin (AZM) are usually considered the first-choice agents in Japan. Although there was no report on the isolation of ERY-resistant M. pneumoniae before 2000 in Japan, we found that ca. 20% of M. pneumoniae strains isolated from patients from 2000 to 2003 were ERY resistant. These results are consistent with pediatricians' impression that antibiotics such as ERY, CLR, and clindamycin (CLI) are not effective for some patients with M. pneumoniae infection.
It is well known that the macrolide-lincosamide-streptogramin B (MLS) antibiotics inhibit protein synthesis by binding to domain II and/or domain V of 23S rRNA (3, 26). Lucier et al. (10) and Okazaki et al. (17) found that an A-to-G transition or A-to-C transversion at position 2063 (corresponding to 2058 in Escherichia coli numbering) or 2064 of the 23S rRNA gene resulted in high resistance to macrolide antibiotics. No point mutation was found in domain II of 23S rRNA of the ERY-resistant M. pneumoniae strains used in the present study.
We report here the prevalence of macrolide-resistant M. pneumoniae infection in Japan. By using 13 ERY-resistant M. pneumoniae strains, we investigated the mechanisms of resistance to MLS antibiotics. Furthermore, we established restriction fragment length polymorphism (RFLP) techniques to detect point mutations in domain V of 23S rRNA of M. pneumoniae by using throat swabs or sputum samples.
MATERIALS AND METHODS
Mycoplasmas.
Three types of M. pneumoniae strains were used in the present study, i.e., ERY-resistant strains isolated from children infected with M. pneumoniae in Japan from 2000 to 2003, ERY-resistant strains induced with ERY in vitro, and three reference strains: M129, Mac, and FH. The ERY-resistant clinical isolates are listed in Table 1, with details regarding patient age, year of isolation, symptoms, and the administration of antibiotics. Most of the isolates were obtained during the patient's first visit to the hospital, except in a few cases in which the isolates were obtained within a week after an initial treatment failure. Modified Hayflick medium (6) were used for the isolation of M. pneumoniae from patients. The broth medium was composed of 7.5 parts PPLO broth (Difco), 1.5 parts heat-inactivated horse serum, and 1 part aqueous extract (25%) of baker's yeast, penicillin G (1,000 U/ml), thallium acetate (0.025%), glucose (0.5%), and phenol red (0.002%). The composition of agar medium was the same as that of the broth medium except that glucose and phenol red were omitted and 1.2% agar was added. A throat swab was immersed several times in 0.5 ml of PPLO broth; then, 0.2 ml of the suspension was transferred to the diphasic (agar/broth) medium, and 0.1 ml of the suspension was transferred onto the agar medium. The agar medium was incubated under 5% CO2 in air with moisture, and the diphasic medium was incubated aerobically at 37°C for 5 to 14 days. When a color change was observed in the diphasic medium, 0.1 ml of the broth was subcultured onto the agar medium. When typical colonies were observed on the agar medium, a single colony was inoculated into the broth medium. After cloning of the colonies, M. pneumoniae was identified serologically or by using PCR.
TABLE 1.
Macrolide-resistant M. pneumoniae strains isolated from patients, along with patient information
| Strain no. | Patient
|
Antimicrobial agent(s)a
|
||
|---|---|---|---|---|
| Age (yr) | Symptoms and/ or disease | First choice/effect | Second choice/ effect | |
| 350 | 9 | Pneumonia | CLI/− | CLR/+ |
| 374 | 3 | Pneumonia | Unknown | Unknown |
| 375 | 4.5 | Pneumonia | Unknown | Unknown |
| 376 | 12 | Pneumonia | CLR/− | AZM/+ |
| 377 | 7 | Fever and cough | AZM/+ | |
| 378 | 2 | Fever and cough | Cefditoren pivoxil/− | AZM/+ |
| 379 | 9 | Pneumonia | CLR/− | AZM/− |
| 380 | 11 | Pneumonia | CLR/− | Minocycline/+ |
| 381 | 11 | Pneumonia | AZM/+ | |
| 382 | 7 | Pneumonia | RKM/− | AZM/− |
| 383 | 5 | Bronchitis | Cefaclor/− | ERY/+ |
| 384 | 7 | Pneumonia | Cefdinir, Fosfomycin/− | ERY/+ |
| 385 | NIb | Pneumonia, pleurisy | CLR/+ | |
−, No effect from antimicrobial agent; +, improvement of symptoms.
NI, no information.
MIC determination.
MICs of MLS antibiotics were determined by a broth microdilution method based on the method of the National Committee for Clinical Laboratory Standards. Serial twofold dilutions of MLS antibiotics prepared in PPLO broth containing 104 to 105 CFU/ml of M. pneumoniae were put in 96-well microplates (17). The microplates were sealed with adhesive sheets and incubated at 37°C. The MIC was determined as the lowest concentration of antimicrobial agent at which the color of the control medium was changed. A number of antibiotics were tested. ERY, oleandomycin (OL), josamycin (JM), spiramycin (SPM), midekamycin (MDM), leucomycin (LM), and lincomycin (LCM) were purchased from Wako Pure Chemical Industries, Ltd., Japan; roxithromycin (RXM) and quinupristin-dalfopristin were provided by Aventis Pharm Japan, Ltd.; CLR was provided by Abbott Co., Ltd. (Japan); rokitamycin (RKM) was provided by Asahi Kasei Co. Japan; CLI was provided by Upjohn Co. (Japan); and AZM was provided by Pfizer Japan, Inc.
PCR amplification and sequencing of domains II and V of the 23S rRNA gene and L4 and L22 ribosomal protein genes.
The ERY-resistant M. pneumoniae strains were screened on the basis of MIC of ERY. A 0.5-ml aliquot of growth culture of M. pneumoniae was centrifuged at 17,500 × g for 20 min at 4°C. After removal of the supernatant, the sediment was suspended in 20 μl of TE (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) buffer containing 1.0% (vol/vol) Triton X-100 and boiled for 5 min. Specific primers were designed for the detection of the point mutations of domain II of 23S rRNA and of L4 (rplD) and L22 (rplV) ribosomal proteins (Table 2). Primers for domain V of 23S rRNA were as reported by Lucier et al. (10). To identify the mutation in domain II containing nucleotide A752 interacting with the macrolide 3-cladinose moiety, 23SDIIF-23SDIIR primer pairs were used. For domain V (peptidyltransferase region), MH23SDVF-MH23SDVR primer pairs were used. Amplification of ribosomal protein L4 and L22 fragments was performed with the MNL4F-MNL4R and MNL22F-MNL22R primer pairs, respectively. The composition of the PCR mixture was as follows: 2 μl of template, 30 pmol of forward and reverse primers, and 25 μl of premix Taq (TaKaRa Ex Taq Version; Takara Bio, Inc.) and water in a final reaction volume of 50 μl. PCR conditions were 2 min at 94°C first, followed by 45 s at 94°C for denaturation, 1 min at 55°C for annealing, and 80 s at 72°C for elongation for 30 cycles, and followed finally by 5 min at 72°C. The products were purified with a MiniElute PCR purification kit (Qiagen, Hilden, Germany), labeled with a BigDye Terminator V3.1 cycle sequencing kit (Applied Biosystems), and applied to an ABI Prism 3100 genetic analyzer (Applied Biosystems) according to the manufacturer's instructions. The primers used for sequencing were the same as those used for PCR (Table 2). DNA sequences of PCR products were compared to the sequence of M. pneumoniae M129 (accession no. X68422) by using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
TABLE 2.
Primers used for PCR amplification and sequencing of domains II and V of 23S rRNA and ribosomal proteins of L4 and L22 in M. pneumoniae
| PCR and primer designation | Sequence (5′ to 3′) | Positiona | Amplicon size (bp) |
|---|---|---|---|
| Domain II of 23S rRNA | |||
| MN23SDIIF | AGTACCGTGAGGGAAAGGTG | 491-510 | 816 |
| MN23SDIIR | TCCCAAGCGTTACTCATGCC | 1287-1306 | |
| Domain V of 23S rRNA | |||
| MN23SDVF | GCAGTGAAGAACGAGGGG | 1758-1775 | 927 |
| MN23SDVR | GTCCTCGCTTCGGTCCTCTCG | 2664-2684 | |
| Ribosomal protein L4 | |||
| MNL4F | AAAAGCAGCACCAGTTGTAG | 1231-1250 | 722 |
| MNL4R | GGTTAGAACTGGTTTTAGCA | 1933-1952 | |
| Ribosomal protein L22 | |||
| MNL22F | GTACATAACGGCAAGACCTT | 3640-3659 | 627 |
| MNL22R | GCAAGCCGTTGGAGTTTACT | 4247-4266 | |
| Nested PCR for 23S rRNA of 2063, 2064 region | |||
| MN23SF1937 | ACTATAACGGTCCTAAGGTA | 1918-1937 | 210 |
| MN23SR2128 | ACCTATTCTCTACATGATAA | 2108-2177 | |
| Nested PCR for 23S rRNA of 2617 region | |||
| MN23SF2577 | TACGTGAGTTGGGTTCAAA | 2577-2595 | 108 |
| MN23SR2664 | GTCCTCGCTTCGGTCCTCTCG | 2664-2684 |
RFLP analysis of point mutation in domain V of 23S rRNA.
To detect the point mutations A2063G, A2063C, A2064G, and A2617G in domain V of 23S rRNA, BbsI, BceAI, BsaI, and BsmFI (New England BioLabs) were used. Second PCR products from domain V for tested M. pneumoniae strains were used for digestion with the four restriction enzymes. After the first PCR product (927 bp) was obtained with the MH23SDVF-MH23SDVR primer pair, a second PCR product (210 bp) was obtained with the MN23SF1937-MN23SR2128 primer pair to detect the point mutation at 2063 or 2064 in domain V of 23S rRNA. For the detection of point mutation at 2617 in domain V, the primer set of MN23SF2577 and MN23SF2664 was used, and a 108-bp PCR product was obtained. A portion of the second PCR product was digested with BbsI (5 U for 1 μl of PCR product) for the A2063G mutation, BceAI (1 U for 1 μl of PCR product) was used for the A2063C mutation, BsaI (10 U for 1 μl of PCR product) was used for the A2064G mutation, and BsmFI (2 U for 1 μl of PCR product) was used for the C2617G mutation. Digested products were electrophoresed on a 10 to 15% gradient polyacrylamide gel (Nikkyo Technos Co., Ltd.) or on a 4% Nusieve 3:1 agarose gel (BioWhittaker Molecular Applications, Rockland, Maine).
RESULTS
Antimicrobial susceptibility.
In all, 13 (17%) of the 76 clinical isolates obtained in Japan during the period from 2000 to 2003 showed various degrees of elevation of MICs against macrolides, including the ERY MIC. The in vitro activities of the MLS antibiotics against ERY-resistant clinical isolates and reference strains of M. pneumoniae are summarized in Table 3. M. pneumoniae reference strains, including M129, showed low ERY, OL, RXM, CLR, AZM, JM, MDM, LM, RKM, and SPM (0.0156 to 0.25 μg/ml) MICs. Of the ERY-resistant strains, strain 377 (C2617G) showed low resistance to macrolide antibiotics except for OL. The 15-membered macrolide AZM and most of the 16-membered macrolides were more effective than the 14-membered macrolides for strain 377. Although ERY-resistant clinical strains, except for strain 377, tended to show resistance to all of the macrolides, some of them showed different responses to RKM. That is, for strains with an A-to-G mutation at position 2063 the RKM MICs were not so high (<1 μg/ml). LCM and CLI, lincosamide antibiotics, and streptogramin antibiotics showed no marked activity toward the reference strains or some of the clinical isolates.
TABLE 3.
MICs of MLS antibiotics for M. pneumoniae isolated from patients and reference strains
| Strain no. | 23S rRNA mutationa | MIC (μg/ml)
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERY | OL | RXM | CLR | AZM | JM | MDM | LM | RKM | SPM | LCM | CLI | Q-Db | ||
| 350 | A2063G | >256 | >256 | >256 | 256 | 32 | 8 | 16 | 4 | 0.5 | 8 | >256 | >256 | 1 |
| 374 | A2063G | >256 | >256 | >256 | >256 | 64 | 8 | 16 | 4 | 0.5 | 16 | >256 | 256 | 0.5 |
| 375 | A2063G | >256 | >256 | >256 | >256 | 32 | 16 | 16 | 8 | 0.5 | 16 | >256 | 256 | 0.5 |
| 376 | A2063C | >256 | >256 | >256 | >256 | 16 | 64 | 64 | 64 | 4 | 256 | 64 | 32 | 1 |
| 377 | C2617G | 8 | 64 | 8 | 1 | 0.031 | 0.25 | 0.25 | 0.25 | 0.0625 | 1 | 16 | 2 | 0.25 |
| 378 | A2063G | >256 | >256 | >256 | >256 | 64 | 8 | 16 | 4 | 0.5 | 16 | 256 | 256 | 1 |
| 379 | A2063G | >256 | >256 | >256 | >256 | 64 | 8 | 16 | 4 | 0.5 | 16 | 256 | 256 | 0.5 |
| 380 | A2063G | >256 | >256 | >256 | >256 | 64 | 8 | 16 | 8 | 0.5 | 16 | 256 | 256 | 0.5 |
| 381 | A2063G | >256 | >256 | >256 | >256 | 64 | 8 | 16 | 8 | 0.5 | 16 | 256 | 256 | 0.5 |
| 382 | A2063G | >256 | >256 | >256 | >256 | 64 | 8 | 16 | 8 | 0.5 | 16 | 256 | 256 | 1 |
| 383 | A2064G | 256 | >256 | 128 | 32 | 16 | 256 | >256 | >256 | 32 | >256 | 64 | 32 | 0.25 |
| 384 | A2063G | >256 | >256 | >256 | >256 | 64 | 8 | 16 | 8 | 0.5 | 16 | >256 | 256 | 1 |
| 385 | A2063G | >256 | >256 | >256 | >256 | 64 | 16 | 16 | 16 | 1 | 16 | >256 | 256 | 1 |
| FH | 0.0625 | 0.25 | 0.0625 | 0.0156 | 0.00098 | 0.0156 | 0.25 | 0.0625 | 0.0625 | 0.25 | 16 | 4 | 0.0625 | |
| M129 | 0.0156 | 0.125 | 0.0156 | 0.0156 | 0.00195 | 0.125 | 0.0625 | 0.0625 | 0.0625 | 0.125 | 8 | 4 | 0.25 | |
| Mac | 0.0156 | 0.25 | 0.0156 | 0.0156 | 0.00098 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 4 | 4 | 0.25 | |
According to M. pneumoniae numbering.
Q-D, quinupristin-dalfopristin.
Sequencing analysis of ribosomal protein and 23S rRNA genes.
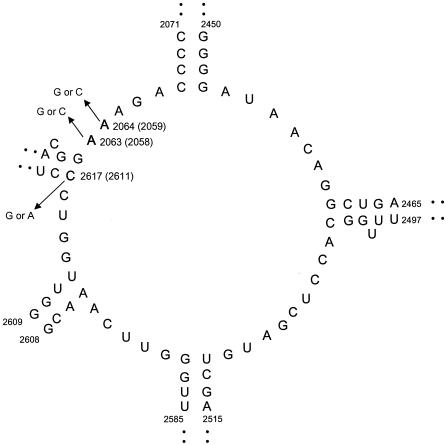
PCR amplification and sequence analysis of ribosomal proteins and 23S rRNA were performed for all M. pneumoniae strains used in the present study. The results are summarized in Table 4. In domain II of the 23S rRNA containing position 752, there was no difference in sequence from that of M. pneumoniae M129. Figure 1 shows the results of the nucleotide sequence analysis of domain V, called the peptidyltransferase region, in the 23S rRNA of the M. pneumoniae strains. Five ERY-resistant strains (1020-EMR3, 1020, 1253, 1552, and 1653) were induced with ERY in vitro, as previously reported (17). Figure 2 shows the position of a point mutation on the peptidyltransferase loop in domain V of M. pneumoniae 23S rRNA. Of 13 ERY-resistant clinical isolates, 10 (77%) showed A2063G transition, and the remaining 3 showed one A2064G transition, one A2063C transversion, and one A2617G transversion. Of the ERY-resistant strains obtained in vitro, strain 1020-EMR3 had C2617G and strain 1552 had two point mutations: A2064C and C2617A. Compared to the sequence of the M129 strain, different nucleotides were found in some strains (350, 376, 377, 378, 379, FH, and Mac) at positions 162 and 430 of L4 and 279 of L22 ribosomal protein genes. These differences are related to two different types of M. pneumoniae strains (19). Mutation T508C of the L22 ribosomal protein gene was observed in all strains used in the present study except for M129. Thus, these nucleotide differences are not involved in the ERY resistance of M. pneumoniae. Although C62A and C341T mutations were found in strain 1253, it is uncertain whether these mutations are involved in ERY resistance because of the A2064G mutation, which imparts high ERY resistance.
TABLE 4.
Nucleotide substitution by point mutation of genes of ribosomal protein and 23S rRNA for macrolide-resistant M. pneumoniae strains and M. pneumoniae FH and Mac compared to M. pneumoniae M129a
| Strain no. | Substitution(s) in ribosomal protein
|
Mutation in 23S rRNA
|
Type of P1 gene | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Position of L4
|
Position of L22
|
||||||||
| 162 | 430 | 62 | 279 | 341 | 508 | Domain II | Domain V | ||
| M129 | C | A | C | T | C | T | - | - | I |
| 350 | C→A | A→G | - | T→C | - | T→C | - | A2063G | II |
| 374 | - | - | - | - | - | T→C | - | A2063G | I |
| 375 | - | - | - | - | - | T→C | - | A2063G | I |
| 376 | C→A | A→G | - | T→C | - | T→C | - | A2063C | II |
| 377 | C→A | A→G | - | T→C | - | T→C | - | C2617G | II |
| 378 | C→A | A→G | - | T→C | - | T→C | - | A2063G | II |
| 379 | C→A | A→G | - | T→C | - | T→C | - | A2063G | II |
| 380 | - | - | - | - | - | T→C | - | A2063G | I |
| 381 | - | - | - | - | - | T→C | - | A2063G | I |
| 382 | - | - | - | - | - | T→C | - | A2063G | I |
| 383 | - | - | - | - | - | T→C | - | A2064G | I |
| 384 | - | - | - | - | - | T→C | - | A2063G | I |
| 385 | - | - | - | - | - | T→C | - | A2063G | I |
| 1020-EMR3 | - | - | - | - | - | T→C | - | C2617G | I |
| 1020 | - | - | - | - | - | T→C | - | A2064G | I |
| 1253 | - | - | C→A | - | C→T | T→C | - | A2064G | I |
| 1552 | - | - | - | - | - | T→C | - | A2064C/C2617A | I |
| 1653 | - | - | - | - | - | T→C | - | A2064G | I |
| FH | C→A | A→G | - | T→C | - | T→C | - | - | II |
| Mac | C→A | A→G | - | T→C | - | T→C | - | - | II |
-, No mutation compared to the sequence of M. pneumoniae M129.
FIG. 1.
Multiple alignment of 23S rRNA gene of ERY-resistant M. pneumoniae strains and M. pneumoniae M129, FH, and Mac. Partial sequences of the peptidyltransferase (domain V) from positions 2051 to 2081 and 2601 to 2630 are presented. The nucleotides are numbered on the basis of M. pneumoniae. The nucleotide sequence of M. pneumoniae M129 was according to GenBank accession no. X68422. Identical nucleotides are indicated by dashes. The positions of 2063, 2064, and 2617 are underlined.
FIG. 2.
Secondary structure of the peptidyltransferase loop in domain V of M. pneumoniae 23S rRNA. Positions of the newly found mutations (A2063C and C2617G), as well as previously reported in vitro mutations (A2063G, A2064G, and A2064C), in clinical isolates are indicated by using the numbering for M. pneumoniae 23S rRNA (accession no. X68422). The numbers in parentheses indicate E. coli numbering.
RFLP analysis of ERY-resistant M. pneumoniae strains.
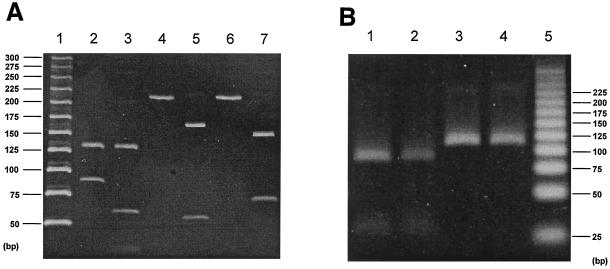
To detect a point mutation at position 2063 or 2064 of the 23S rRNA gene, a second PCR product (210 bp) was digested from the first PCR product (927 bp) with suitable restriction enzymes. Digestion with BsaI generated two fragments of 124 and 86 bp for ERY-susceptible strain M129, whereas three fragments of 124, 57, and 29 bp were obtained in the case of the A2063G mutation (lanes 2 and 3 in Fig. 4A). Two fragments of 158 and 52 bp were generated with BceAI in the case of the A2063G mutation (lane 5 in Fig. 4A), and two fragments were generated with BsaI in the case of the A2064G mutation (lane 7 in Fig. 4A). Strain M129 has no cut site for the second PCR product with BceAI and BsaI (lanes 4 and 6 in Fig. 4A). To detect a point mutation at position 2617, the PCR primer pair MN23SF2577 and MN23SDVR was used, generating a 108-bp product (Fig. 3). Although there was no restriction enzyme to digest C2617A or C2617G mutation, the M129 strain had a restriction site with BsmFI and generated two fragments of 81 and 27 bp (Fig. 4B).
FIG. 4.
Restriction analysis of 210-bp (A) and 108-bp (B) amplicons from the peptidyltransferase region (domain V) in 23S rRNA of M. pneumoniae. (A) Restriction profile for detection of the A2063G, A2063C, and A2064G mutations. Lanes: 1, DNA size marker (25-bp DNA step ladder; Promega); 2, 4, and 6, M. pneumoniae M129 (susceptible strain) treated with BbsI (lane 2, 124-, and 86-bp products) and BceAI and BsaI (lanes 4 and 6, respectively; uncut 210-bp product); 3, strain 375 (A2063G) treated with BbsI (124-, 57-, and 52-bp products); 5, strain 376 (A2063C) treated with BceAI (158- and 52-bp products); 7, strain 1020 (A2064G) treated with BsaI (141- and 69-bp products). (B) Restriction profile for detection of C2617 mutation with BsmFI digestion. Although M. pneumoniae M129 and strain 375 (A2063G) produced two fragments of 81 and 27 bp (lanes 1 and 2), the 108-bp fragment remained uncut in strains 377 and 1020-EMR3 (C2617G) as a result of loss of the restriction site for BsmF1 (lanes 3 and 4). Lane 5, DNA size marker (25-bp DNA step ladder; Promega).
FIG. 3.
Nucleotide sequence of the 927-bp amplicon from positions 1758 to 2684 of the 23S rRNA gene from M. pneumoniae M129. A long arrow indicates a primer sequence with direction. A short arrow indicates a site of mutation with a substituted base, i.e., A2063G, A2063C, A2064G, or C2617A. A newly constructed restriction site and the responsible base change with underline is shown in parentheses with the corresponding restriction enzyme.
DISCUSSION
In general, macrolides such as ERY, CLR, and AZM are used as the first-choice therapeutic agent for treating M. pneumoniae infections in children, as well as in adults. We isolated 76 M. pneumoniae strains from three geographically distant regions in Japan (Hokkaido in the northern island, Kanagawa in the central region, and Kochi in south) and found that 13 strains (17%) were ERY resistant. Although resistance to ERY was observed many years ago in a few M. pneumoniae strains (16, 20), when we investigated the ERY MICs for 296 M. pneumoniae strains isolated in Japan from 1983 to 1998, no ERY-resistant strain was found among them (data not shown). Thus, we concluded that ERY-resistant M. pneumoniae had appeared in 2000 and spread rapidly in Japan. We applied our established RFLP analysis to ca. 1,000 sputum samples taken from patients with respiratory infections from 2000 to 2002 and found that 23 (24%) of 94 PCR-positive samples for M. pneumoniae DNA had the ERY resistance-inducing point mutation A2063G (unpublished data). Whether or not the prevalence of ERY-resistant M. pneumoniae and the predominance of A2063G among the isolates are peculiar to Japan needs to be clarified by future studies outside Japan.
The mechanisms of resistance to MLS antibiotics in various microorganisms have been reviewed and include modification of the target site, active efflux, or inactivation (13, 24-26). The MLS antibiotics inhibit protein synthesis by binding to domains II and V of 23S rRNA (3, 26). In particular, it has been clearly shown that ribosomal mutations in domains II and V of 23S rRNA and mutations in ribosomal protein L4 (rplD) and L22 (rplV) are related to resistance to MLS antibiotics (2, 4). In L4 and L22 ribosomal proteins, no mutation that clearly contributed to resistance to macrolide antibiotics was found, although one strain (strain 1253) exhibited mutations of the L22 protein, such as C62A and C341T, in vitro. We found several point mutations in domain V of 23S rRNA in ERY-resistant M. pneumoniae but none in domain II of 23S rRNA. Among them, the point mutations at position 2063 or 2064 in domain V have been reported in several pathogens such as E. coli, H. pylori, Mycobacterium spp., and S. pneumoniae (24) and generated strong resistance to macrolide antibiotics. Transversions of C to G and C to A at position 2617 of domain V were observed in a clinical isolate (strain 377) and ERY-induced strains (1020-EMR3 and 1552), respectively. On the other hand, it has been reported that C-to-U transition at position 2611 (corresponding to 2617 in M. pneumoniae numbering) in clinical pathogens such as Neisseria gonorrhoeae (15), Streptococcus pyogenes (11), Mycoplasma hominis (18), Chlamydia trachomatis (12), and E. coli (23) was associated with macrolide resistance. M. pneumoniae strain 1552, derived by incubation with ERY in vitro, showed A2064C transversion and C2617A transversion. The mutation at position 2617 produced less resistance to macrolide antibiotics than did the mutation at position 2063 or 2064 of domain V. Based on our results, it is considered that transition is the predominant type of mutation in M. pneumoniae. This may be due to the structural difference between purine and pyrimidine. These results support the observation in E. coli that the apparent dissociation constant (Kd) for ERY of C2611U (corresponding to 2617 in M. pneumoniae) [Kd = (4.4 ± 0.9) × 10−7] is ca. 480 times higher than that of the A2058G (2063 in M. pneumoniae) E. coli strain [Kd = (1.9 ± 0.3) × 10−4] (3). As mentioned above, macrolide resistance of M. pneumoniae has been explained thus far in terms of mutation of 23S rRNA. However, M. hominis was associated with an absence of intracellular accumulation and ribosomal binding of macrolide antibiotics (18). These results suggest that several different mechanisms of macrolide resistance exist in Mycoplasma species.
Table 1 summarizes information about the patients from whom ERY-resistant M. pneumoniae strains were isolated. Although these patients were actually infected with ERY-resistant M. pneumoniae, macrolides were apparently effective after their first administration in six (ERY in cases 383 and 384, CLR in case 350, and AZM in cases 377, 378, and 381) of the ten patients for whom the clinical course was known. One possible explanation may be the anti-inflammatory effects of macrolides, which inhibit the production of cytokines such as proinflammatory tumor necrosis factor alpha, interleukin-1β (IL-1β), IL-6, IL-8, and so on rather than the antimicrobial effect (1, 7, 8, 21). Much more information is available about the immunopathological mechanisms of M. pneumoniae pneumonia, particularly with regard to a wide variety of cytokines. Among them, Th1-type cytokines (22) and IL-8 (14) might play significant roles in the pathomechanism. In this context, recent investigations have revealed that macrolides modulate the actions of these cytokines (5, 9). It is therefore a reasonable proposition that macrolides, particularly 14- and 15-membered macrolides, exert their clinical efficacy in the treatment of M. pneumoniae pneumonia through immunomodulation. Our results obtained for patients with ERY-resistant M. pneumoniae infection strongly suggest that the beneficial effects of macrolides in the treatment of M. pneumoniae pneumonia are not solely due to direct antimicrobial activity and support the idea that immunomodulatory effects of macrolides play an important role in recovery from the illness.
In conclusion, we found 13 strains of macrolide-resistant M. pneumoniae among 76 clinical isolates obtained during the period from 2000 to 2003, despite the fact that no resistant strain was found among 296 isolates from 1983 to 1998. The predominant mutation was A2063G in domain V of 23S rRNA (10 of 13 resistant strains), and mutations involving either A2063 or A2064 resulted in high MICs to macrolide antibiotics. On the other hand, mutations involving C2617 in domain V of 23S rRNA generated less resistance to ERY than mutations involving A2063 or A2064. Our results indicate that macrolide-resistant M. pneumoniae is spreading in Japan, and it will be necessary to reconsider the effectiveness of macrolides in the treatment of patients with M. pneumoniae pneumonia.
Acknowledgments
We thank Satoshi Yamada of Tonden Children's Clinic, Sapporo, Japan, and Shohei Harada of Ikeda Municipal Hospital for isolating the M. pneumoniae clinical strains.
This study was partly supported by a Grant for Studies on Emergency and Re-emergency Infectious Diseases (H15-Shinko-24) from the MHLW.
REFERENCES
- 1.Abe, S., H. Nakamura, S. Inoue, H. Takeda, H. Saito, S. Kato, N. Mukaida, K. Matsushima, and H. Tomoike. 2000. Interleukin-8 gene repression by clarithromycin is mediated by the activator protein-1 binding site in human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 22:51-60. [DOI] [PubMed] [Google Scholar]
- 2.Canu, A., B. Malbruny, M. Coquemont, T. A. Davies, P. C. Appelbaum, and R. Leclercq. 2002. Diversity of ribosomal mutations conferring resistance to macrolides, clindamycin, streptogramin, and telithromycin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Douthwaite, S., L. H. Hansen, and P. Mauvais. 2000. Macrolide-ketolide inhibition of MLS-resistant ribosomes is improved by alternative drug interaction with domain II of 23S rRNA. Mol. Microbiol. 36:183-193. [DOI] [PubMed] [Google Scholar]
- 4.Gregory, S. T., and A. E. Dahlberg. 1999. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S rRNA. J. Mol. Biol. 289:827-834. [DOI] [PubMed] [Google Scholar]
- 5.Hardy, R. D., A. M. Rios, S. Chavez-Bueno, H. S. Jafri, J. Hatfield, B. B. Rogers, G. H. McCracken, and O. Ramilo. 2003. Antimicrobial and immunologic activities of clarithromycin in a murine model of M. pneumoniae-induced pneumonia. Antimicrob. Agents Chemother. 47:1614-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayflick, L. 1965. Tissue cultures and mycoplasmas. Tex. Rep. Biol. Med. 23(Suppl. 1):285-303. [PubMed] [Google Scholar]
- 7.Ichiyama, T., M. Nishikawa, T. Yoshitomi, S. Hasegawa, T. Matsubara, T. Hayashi, and S. Furukawa. 2001. Clarithromycin inhibits NF-κB activation in human peripheral blood mononuclear cells and pulmonary epithelial cells. Antimicrob. Agents Chemother. 45:44-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohyama, T., H. Takizawa, S. Kawasaki, N. Akiyama, M. Sato, and K. Ito. 1999. Fourteen-member macrolides inhibit interleukin-8 release by human eosinophils from atopic donors. Antimicrob. Agents Chemother. 43:907-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labro MT. 1998. Anti-inflammatory activity of macrolides: a new therapeutic potential? J. Antimicrob. Chemother. 41(Suppl. B):37-46. [DOI] [PubMed] [Google Scholar]
- 10.Lucier, T. S., K. Heitzman, S.-K. Liu, and P.-C. Hu. 1995. Transition mutations in the 23S rRNA of erythromycin-resistant isolates of Mycoplasma pneumoniae. Antimicrob. Agents Chemother. 39:2770-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malbruny, B., K. Nagai, M. Coquemont, B. Bozdogan, A. T. Andrasevic, H. Hupkova, R. Leclercq, and P. C. Appelbaum. 2002. Resistance to macrolides in clinical isolates of Streptococcus pyogenes due to ribosomal mutations. J. Antimicrob. Chem. 49:935-939. [DOI] [PubMed] [Google Scholar]
- 12.Misyurina, O. Y., E. V. Chipitsyna, Y. P. Finashutina, V. N. Lazarev, T. A. Akopian, A. M. Savicheva, and V. M. Govorun. 2004. Mutations in a 23S rRNA gene of Chlamydia trachomatis associated with resistance to macrolides. Antimicrob. Agents Chemother. 48:1347-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakajima, Y. 1999. Mechanisms of bacterial resistance to macrolide antibiotics. J. Infect. Chemother. 5:61-74. [DOI] [PubMed] [Google Scholar]
- 14.Narita, M., H. Tanak, S. Yamada, S. Abe, T. Ariga, and Y. Sakiyama. 2001. Significant role of interleukin-8 in pathogenesis of pulmonary disease due to M. pneumoniae infection. Clin. Diagn. Lab. Immunol. 8:1028-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng, L.-K., I. Martin, G. Liu, and L. Bryden. 2002. Mutation in 23S rRNA associated with macrolide resistance in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 46:3020-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niitu, Y., S. Hasegawa, T. Suetake, H. Kubota, S. Komatsu, and M. Horikawa. 1970. Resistance of Mycoplasma pneumoniae to erythromycin and other antibiotics. J. Pediatr. 76:438-443. [DOI] [PubMed] [Google Scholar]
- 17.Okazaki, N., M. Narita, S. Yamada, K. Izumikawa, M. Umetsu, K. Kenri, Y. Sasaki, Y. Arakawa, and T. Sasaki. 2001. Characteristics of macrolide-resistant Mycoplasma pneumoniae strains isolated from patients and induced with erythromycin in vitro. Microbiol. Immunol. 45:617-620. [DOI] [PubMed] [Google Scholar]
- 18.Pereyre, S., P. Gonzalez, B. de Barbeyrac, A. Darnige, H. Renaudin, A. Charron, S. Raherison, C. Bébéar, and C. M. Bébéar. 2002. Mutations in 23S rRNA account for intrinsic resistance to macrolides in Mycoplasma hominis and Mycoplasma fermentans and for acquired resistance to macrolides in M. hominis. Antimicrob. Agents Chemother. 46:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki T, T. Kenri, N. Okazaki, M. Iseki, R. Yamashita, M. Shintani, T. Sasaki, and M. Yayoshi. 1996. Epidemiological study of Mycoplasma pneumoniae infections in Japan based on PCR-restriction fragment length polymorphism of the P1 cytadhesin gene. J. Clin. Microbiol. 34:447-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stopler, T., and D. Branski. 1986. Resistance of Mycoplasma pneumoniae to macrolides, lincomycin, and streptogramin B. J. Antimicrob. Chemother. 18:359-364. [DOI] [PubMed] [Google Scholar]
- 21.Takizawa, H., M. Desaki, T. Ohitoshi, T. Kikutani, H. Okazaki, M. Sato, N. Akiyama, S. Shoji, K. Hiramatsu, and K. Ito. 1995. Erythromycin suppresses interleukin 6 expression by human bronchial epithelial cells: a potential mechanism of its anti-inflammatory action. Biochem. Biophys. Res. Commun. 210:781-786. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka, H., M. Narita, S. Teramoto, T. Saikai, K. Osahi, T. Igarashi, and S. Abe. 2002. Role of interleukin-18 and T-helper type 1 cytokines in the development of M. pneumoniae pneumonia in adults. Chest 121:1493-1497. [DOI] [PubMed] [Google Scholar]
- 23.Vannuffel, P., M. Di Giambattista, E. A. Morgan, and C. Cocito. 1992. Identification of a single base change in rRNA leading to erythromycin resistance. J. Biol. Chem. 267:8377-8382. [PubMed] [Google Scholar]
- 24.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weisblum, B. 1998. Macrolide resistance. Drug Resist. Updates 1:29-41. [DOI] [PubMed] [Google Scholar]
- 26.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]