Abstract
Nonribosomal peptide synthetases (NRPSs) catalyze the formation of structurally diverse and biologically important peptides. Given their modular organization, NRPSs provide an enormous potential for biocombinatorial approaches to generate novel bioactive compounds. Crucial for the exploitation of this potential is a profound knowledge of the intermolecular communication between partner NRPSs. The overall goal of this study was to understand the basis of protein–protein communication that facilitates the selective interaction in these multienzyme complexes. On this account, we studied the relevance of short regions at the termini of the NRPSs tyrocidine (Tyc) synthetases TycA, TycB, and TycC, constituting the Tyc biosynthetic template. In vitro and in vivo investigations of C-terminal deletion mutants of the initiation module TycA provided evidence for the existence and impact of short communication-mediating (COM) domains. Their decisive role in protein–protein recognition was subsequently proven by means of COM domain-swapping experiments. Substitution of the terminal COM domains between the donor modules TycA and TycB3, as well as between the acceptor modules TycB1 and TycC1, clearly demonstrated that matching pairs of COM domains are both necessary and sufficient for the establishment of communication between partner NRPSs in trans. These results corroborated the generality of COM domains, which were subsequently exploited to induce crosstalk, even between NRPSs derived from different biosynthetic systems. In conclusion, COM domains represent interesting tools for biocombinatorial approaches, which, for example, could be used for the generation of innovative natural product derivatives.
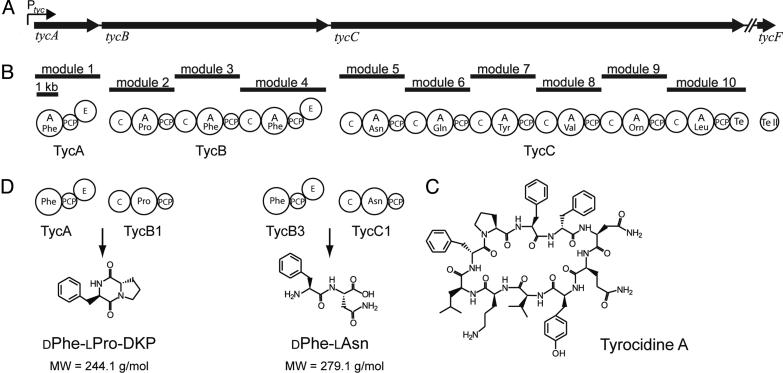
Nonribosomal peptide synthetases (NRPSs) are large multidomain enzymes responsible for the biosynthesis of many pharmacologically important bioactive compounds of great structural diversity (1–3). Prominent examples are the antibiotics penicillin, vancomycin, and actinomycin D, the immunosuppressant cyclosporine A, the siderophore enterobactin, and the antitumor drug bleomycin. As illustrated in Fig. 1 for the biosynthesis of the nonribosomal peptide (NRP) tyrocidine, NRPSs are organized into distinct modules, each of them responsible for the incorporation of one amino acid into the nascent peptide chain. A module can be further subdivided into catalytic domains, which are responsible for the coordinated recognition and activation [adenylation (A) domain] (4), covalent binding and transfer [peptidyl carrier protein (PCP) domain] (5), and incorporation [condensation (C) domain] of a certain substrate amino acid into the peptide chain (6). In addition to these so-called core domains, optional domains catalyze the modification of incorporated residues, i.e., by epimerization (E) or N-methylation (MT) domains (7). Product release is normally effected by a thioesterase (Te) domain, catalyzing the formation of linear, cyclic, or branched cyclic products, representative for the class of NRPs (8).
Fig. 1.
The Tyc biosynthetic system. The enzymatic assembly line of the cyclic decapeptide antibiotic TycA (C) consists of three NRPSs: TycA (124 kDa), TycB (405 kDa), and TycC (724 kDa), which are encoded by the polycistronic genes tycABC (A). The NRPSs are composed of one, three, and six modules, respectively, each one responsible for the incorporation of one monomeric amino acid (B). In accordance with the colinearity principle, protein–protein communication has to occur between TycA and TycB1, as well as between TycB3 and TycC1. (D) Artificial dimodular NRPS systems TycA/TycB1 and TycB3/TycC1 had been constructed, which catalyze in vitro synthesis of the dipeptide products DPhe-LPro-DKP and DPhe-LAsn, respectively.
Because of the modular organization of NRPSs and the colinearity between biosynthetic template and product, the NRP assembly line mechanism accommodates an enormous potential for biocombinatorial approaches. Crucial for such approaches is a profound knowledge about the substrate selectivity of catalytic domains and the determinants of selective communication between modules. In this context, little is known about the intermolecular communication between NRPSs within the same biosynthetic complex. In fact, the tyrocidine biosynthetic systems (Fig. 1) consists of three distinct NRPSs [tyrocidine (Tyc) synthetases TycA, TycB, and TycC], responsible for the incorporation of one, three, and six amino acid residues into the growing peptide, respectively (9). Formation of the full-length product requires the productive interaction between TycA and the first module of TycB (TycB1), as well as between the third module of TycB (TycB3) and the first module of TycC (TycC1). To ensure a coordinated channeling of all reaction intermediates along the biosynthetic assembly line, undesired interactions such as between TycA and TycC, which would lead to the formation of a shortened heptapeptide product, must be prevented. The goal of this study was to enlighten the basis of protein–protein communication, which enables the selective interaction between partner NRPSs and prevents the false contact between nonpartner enzymes. For this purpose, we studied the importance of short terminal sequence motifs in mediating the specific channeling of reaction intermediates between partner enzymes. This function had been postulated by analogy to organizationally and functionally related enzymes [i.e., polyketide synthases (PKSs)] as well as sequence comparisons (10, 11).
First, C-terminal deletions were introduced into the initiation module TycA and investigated in vitro and in vivo for their impact on protein–protein communication with the partner elongation module TycB1. These studies led to the identification of short communication-mediating (COM) domains, whose importance and generality were further substantiated on the basis of COM domain-swapping experiments. Here, hybrid initiation and elongation modules were constructed and investigated, in comparison with the corresponding wild-type enzymes, for their ability to facilitate a productive interaction between natural partner and nonpartner NRPSs. In addition, the NRPS communication model established was corroborated and expanded to enforce crosstalk between NRPSs derived from different NRP biosynthetic systems.
Materials and Methods
Strains, Culture Media, and General Methods. The Escherichia coli strains were grown in LB medium, supplemented, if applicable, with 50 μg/ml ampicillin, 25 μg/ml kanamycin, and/or 20 μg/ml chloramphenicol (final concentrations). Standard procedures were applied for all DNA manipulations (12). Oligonucleotides were purchased from MWG Biotech (Ebersberg, Germany). Sequences of all oligonucleotides used in this study are listed in Table 2, which is published as supporting information on the PNAS web site. DNA sequencing confirmed the identity of all plasmids constructed.
Construction of Expression Plasmids. All recombinant gene fragments were PCR-amplified from chromosomal DNA of Bacillus brevis (ATCC 8185) by using the Expand long template PCR system (Roche Diagnostics, Mannheim, Germany) or Pfu-Turbo polymerase (Stratagene) following the manufacturer's protocols. Gene fragments encoding TycA or C-terminal mutants thereof were amplified by using the primer combinations of 5′ tycA and different 3′ primers (see Table 2) and cloned into pTrcHis-TOPO expression vector (Invitrogen), providing an N-terminal hexahistidine tag. Similarly, the gene fragment encoding for TycB3 (domain organization A-PCP-E) was amplified by using oligonucleotides 5′ tycB3 and 3′ tycB3 and cloned into pTrcHis-TOPO.
Substitutions of proposed COM domains were constructed by using gene splicing by overlapping extension (13). The genes of the donor modules TycA(B3) and TycB3(A) were cloned into pTrcHis-TOPO. The genes of the acceptor modules (C1)TycB1 and (B1)TycC1 were cloned into pTrcHis2-TOPO, providing a C-terminal hexahistidine tag. The construction of the plasmids pQE60-tycB1 and pQE60-tycC1-Te was described (14, 15).
Construction of Two-Plasmid Systems. In vivo production of DPhe-LPro-diketopiperazine (DKP) in the heterologous host E. coli was performed by the expression of module pairs from two compatible plasmids as described elsewhere (16). Accordingly, genes of the TycA deletion mutants were subcloned from pTrcHis TOPO expression plasmids in the medium-copy vector pSU18 by using the restriction enzyme HincII.
Production of Recombinant Enzymes. pTrcHis- and pTrcHis2-based expression plasmids were transformed into E. coli M15(pREP4). Expression and purification of the corresponding gene products were carried out as described (6). Fractions containing the recombinant proteins were identified by SDS/7.5% PAGE analysis, pooled, and dialyzed against assay buffer [50 mM Hepes (pH 8.0)/100 mM NaCl] supplemented with 2 mM DTT (final concentration). Protein concentrations were determined by using the calculated extinction coefficients for the absorbance at 280 nm.
General Enzyme Assays and Radiolabeled Substrates. Standard assays were applied for in vitro apo-to-holo conversion of NRPS PCP domains (17), amino acid-dependent ATP-pyrophosphate exchange reactions, and radioactive thioester formation assays (6). Alkaline thioester cleavage reactions were performed as described elsewhere (18). The activity of NRPS E domains was tested by TLC as described (19).
Radiolabeled [32P]pyrophosphate for the ATP-pyrophosphate reactions was purchased from PerkinElmer Life Science (Rodgau-Juegesheim, Germany). Radiolabeled amino acids L[14C]Phe [specific activity, 453 mCi/mmol (1 Ci = 37 GBq)], L[14C]Pro (253 mCi/mmol), and L[3H]Leu (141 Ci/mmol) were purchased from Amersham Pharmacia Biosciences (Braunschweig, Germany).
Product Formation Assays. Product formation assays were performed by using radiolabeled substrates and subsequent liquid scintillation counting analysis and/or nonradiolabeled substrates and subsequent HPLC or HPLC/MS analysis. Assays were carried out mainly as described (14). For time-dependent radioactive assays, a total of 1 ml contained each enzyme at 500 nM and 10 mM MgCl2 in assay buffer. At various time points, 100-μl aliquots were removed and treated congruously.
Nonradioactive product formation assays were performed in a 2- to 5-fold scale-up and incubated for 90 min or overnight at 37°C, respectively. The mixture was quenched by adding half the reaction volume of 1-butanol/chloroform (4:1 ratio; vol/vol) and evaporated, and the pellet was dissolved in 200 μl of 90% buffer A (0.05% formic acid/water, vol/vol) and 10% buffer B (0.045% formic acid/methanol, vol/vol), 90 μl of which was applied to HPLC/MS (Hewlett Packard 1100 series; 250/3-Nucleodur-C18 reversed-phase column from Macherey and Nagel, Dueren, Germany) and analyzed as described (17).
In case of turnover experiments, 300-μl aliquots were removed at various time points, directly quenched with 150 μl of 1-butanol/chloroform (4:1 ratio; vol/vol), and evaporated, and the pellet was resolved in 100 μl of methanol (25%, vol/vol), 30 μl of which was applied to HPLC (Beckmann Coulter System Gold; 120/3-Nucleosil-C18 reversed-phase column, Macherey and Nagel) with a flow rate of 0.6 ml/min-1. Samples were separated under isocratic conditions (25% methanol).
Preparation and detection of DKP produced by two-plasmid systems were carried out as described (16).
Results
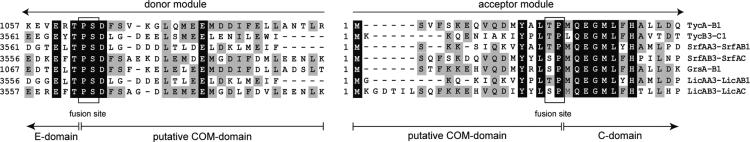
Construction and Characterization of C-Terminal TycA Deletion Mutants. Previous studies revealed that TycA and TycB1 catalyze the formation of the dipeptide DPhe-LPro-S-Ppant. In the absence of additional modules or substrates, the enzyme-bound dipeptide is readily released under autocatalytic formation of cyclic product DPhe-LPro-DKP. Strikingly, a TycA deletion mutant (domain organization: A-PCP) lacking the C-terminal E domain was completely inactive in the product formation assay with TycB1, even when loaded with DPhe in thioester linkage to the Ppant cofactor, whereas TycA mutants with nonfunctional E domains were still able to trigger dipeptide and DKP formation (6, 19). Based on sequence comparison and mutational analysis, only the first 450 amino acid residues of the 480 amino acid residues comprising the E domain appear to inherit the actual epimerization activity, whereas the remaining most C-terminal 20–30 amino acid residues possess no obvious function and may be important for the selective protein–protein communication (see Fig. 2).
Fig. 2.
Sequence comparison of the junction among seven partner NRPSs derived from four different biosynthetic systems [Tyc, Srf, gramicidin S (Grs), and lichenysin(Lic)]. Only the most C-terminal residues of the donor module and the most N-terminal residues of corresponding acceptor module are shown. Invariant amino acid residues are shaded in black, and residues that are conserved in the majority of the COM domains are shaded in gray. Localization of the proposed C- and N-terminal COM domains is indicated.
To challenge this theory, a set of six TycA deletion mutants was constructed, lacking the most C-terminal 3, 6, 9, 12, 15, and 23 amino acid residues, respectively. The corresponding gene fragments were PCR-amplified and cloned into the expression plasmid pTrcHis-TOPO. The TycA deletion mutants were produced in the heterologous host E. coli M15 and purified to apparent homogeneity by Ni2+-affinity chromatography (data not shown). All constructs were subjected to LPhe-dependent ATP-pyrophosphate exchange reaction and thioester formation assay, which assess the activity of unmutated catalytic domains (A and PCP). These tests revealed that the six mutant proteins maintained full substrate activation activity when compared with the wild-type enzyme TycA (data not shown).
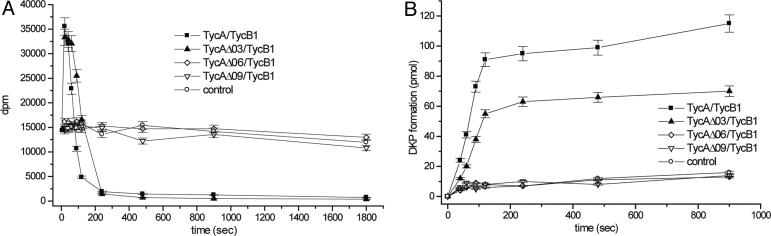
Elongation Activity of C-Terminal TycA Deletion Mutants. To examine the consequences of C-terminal deletions on the initiation module's ability to interact with the partner elongation module TycB1, we carried out product assays. Different initiation modules were preincubated with ATP and L[14C]Phe to allow for the formation of the corresponding D[14C]Phe-S-Ppant thioester intermediates and then combined with preloaded acceptor module prolyl-S-Ppant TycB1. Under these conditions, the DPhe moiety is transferred from the TycA donor to the TycB1 acceptor module to give the D[14C]Phe-LPro-S-Ppant dipeptidyl intermediate. This reaction is accompanied by a fast reloading of TycA with [14C]Phe and the slow autocatalytic release of D[14C]Phe-LPro-DKP product. Consequently, elongation can be followed by an increase of trichloroacetic acid-precipitable label as expected for a simultaneous labeling of donor (D[14C]Phe-S-Ppant TycA) and acceptor (D[14C]Phe-LPro-S-Ppant TycB1). Likewise, DKP product formation can be monitored after organic extraction by the accumulation of radiolabeled D[14C]Phe-LPro-DKP product in the organic layer.
As shown in Fig. 3A, only elongation reactions in the presence of wild-type TycA or TycAΔ03 mutant gave rise to the expected increase in acid-stable label. The subsequently observed rapid decrease of acid-stable label is explained by the use of only a low stoichiometric excess of [14C]Phe substrate and the autocatalytic release of D[14C]Phe-LPro-DKP product (see Fig. 3B). Thorough analysis revealed that the mutant system TycAΔ03/TycB1 is already impeded in DKP formation, showing a slightly reduced rate when compared with the wild-type system TycA/TycB1 (wild-type, kobs = 0.94 min-1; mutant, kobs = 0.54 min-1). In the case of mutant systems, featuring the remaining TycA deletion mutants (TycAΔ06–23), neither transfer of D[14C]Phe from the initiation to the elongation module (Fig. 3A) nor DKP formation (Fig. 3B) could be observed. All these mutant systems essentially gave the same pattern as a control reaction lacking the acceptor amino acid LPro.
Fig. 3.
Elongation activity of C-terminal TycA deletion mutants. (A) Elongation activity was determined with the partner elongation module TycB1 by measuring the amount of enzyme-bound, acid-stable label. (B) DKP product formation was monitored after organic extraction by the accumulation of radiolabeled D[14C]Phe-LPro-DKP in the organic layer. For the sake of clarity, mutant systems TycAΔ12–23/TycB1 were omitted from the presentation but gave essentially the same patterns as the control reaction.
Epimerization Activity of C-Terminal TycA Deletion Mutants. The inability of mutant systems (TycAΔ06–23/TycB1) to synthesize DPhe-LPro-DKP can be explained by either the loss of protein–protein communication between both partner NRPSs or the inactivity of the initiation module's E domain, caused by the deletion of the most C-terminal amino acid residues. Notably, the C domain of the acceptor module TycB1 is able to process only D-configured donor amino acids, but TycA mutants with nonfunctional E domains are still able to trigger dipeptide and DKP formation, once provided with DPhe as substrate (19).
To challenge both possible explanations, we carried out two sets of experiments. First, the epimerization activity of wild-type and deletion mutants was investigated by covalent loading of the substrate amino acid L[14C]Phe followed by thioester cleavage and TLC analysis. Second, the elongation and DKP formation assay was repeated by using either LPhe or DPhe and L[14C]Pro as substrate amino acids. Both tests unequivocally revealed that the E domain activity was not impaired for any of the TycA deletion mutants and that all initiation modules were equally able to convert LPhe-S-Ppant into DPhe-S-Ppant. Accordingly, the observed elongation activity was independent of the configuration of the donor substrate provided. Only TycA and TycAΔ03 were able to form the expected dipeptide DKP (data not shown). This result is clear evidence that the inactivity of the mutant systems in the DKP formation assay is caused by the interruption of protein–protein communication between donor and acceptor module and that presumably short terminal COM domains facilitate the productive interaction between partner NRPSs.
In Vivo Activity of C-Terminal TycA Deletion Mutants. To verify that the failure of protein–protein communication between TycA deletion mutants and TycB1 was not due to an artifact caused by the reaction conditions used in vitro, the influence of C-terminal TycA deletions on DKP formation was investigated in vivo. To this end, we took advantage of a system recently developed in our laboratory, which uses a set of two compatible plasmids that coexist in the same heterologous host, E. coli HM0079 (16).
First, the shortened genes of the TycA mutants were subcloned into the medium-copy plasmid pSU18. Subsequently, a set of seven two-plasmid systems was constructed by simultaneously transforming E. coli HM0079 with pTrc99a-tycB1 and the respective pSU18-tycA derivative. The different strains were grown in M9 minimal medium supplemented with 0.1% of casamino acids. At certain time points, samples were taken and analyzed by HPLC/MS. The results confirmed the outcome of the in vitro experiments (data not shown), substantiating the observation that TycA and TycAΔ03 are capable to form DKP, but an initiation module, lacking as few as six C-terminal amino acid residues, is completely incapable of triggering product formation when incubated with its partner, NRPS.
Strategy for the Exchange of COM Domains. Given the results of the TycA deletion studies, there is clear evidence for the presence of a COM domain at the C terminus of the initiation module TycA. Unfortunately, the proposed partner COM domain at the N terminus of the elongation module of TycB1 could not be investigated by the introduction of N-terminal deletion, because the activity of the C domain can be determined only by the elongation (and DKP formation) assay. Consequently, it was not possible to distinguish whether a possible lack of elongation activity were due to the interruption of the COM domain or inactivation of the C domain of TycB1. Thus, we decided to take an alternative route, exchanging the proposed COM domains of partner and nonpartner NRPSs.
As a model system for the investigation of N-terminal COM domains, the Tyc biosynthetic assembly line was chosen, featuring three NRPSs with two compatible pairs of COM domains at the junctions between TycA/TycB1 and TycB3/TycC1 (see Figs. 1 and 2). From previous studies, it has been known that there is no crosstalk between the nonpartner NRPSs TycA and TycC1 or between TycB3 and TycB1 (14, 15). To test the postulated communication model, a set of four hybrid NRPSs was constructed, containing substitutions of the proposed COM domains. Suitable fusion sites were determined by means of sequence comparisons of seven junctions between partner NRPSs derived from different biosynthetic systems (Fig. 2). The sequence alignment revealed highly conserved sequence motifs in close proximity to the termini of both donor and acceptor modules, whereas the regions of the proposed COM domains show only low similarity. Approximately 20 amino acid residues in front of the C terminus of donor modules, the highly conserved sequence motif TPSD was detected within the E domain (Fig. 2) and used as fusion site (precise fusion site is in italics) between E domain and proposed C-terminal COM domain. Likewise, ≈15 amino acid residues behind the N terminus of acceptor modules, the conserved motif L(T/S)P(M/L)QEG, was determined and used as a fusion site between proposed N-terminal COM and C domains (Fig. 2).
Based on the proposed location of COM domains, the following hybrid NRPSs were constructed: (i) the donor modules TycA(B3) and TycB3(A) and (ii) the acceptor modules (C1)TycB1 and (B1)TycC1 (origin of the proposed COM domains are in parentheses). Gene fragments encoding donor modules were cloned into the expression plasmid pTrcHis-TOPO, whereas gene fragments encoding for acceptor modules were cloned in pTrcHis2-TOPO. The recombinant proteins were heterologously produced in E. coli M15 and purified to apparent homogeneity by single-step Ni2+-nitrilotriacetic acid chromatography. To test for correct folding, all constructs were subjected to amino acid-dependent ATP-pyrophosphate exchange assays, which assess the activity of the unmutated A domain of the corresponding enzymes. These tests revealed that the four hybrid NRPSs maintained full substrate activation activity when compared with the corresponding wild-type enzymes (data not shown).
Investigation of Product Formation Between Partner and Nonpartner NRPSs. To investigate the importance for COM domains for the establishment of protein–protein communication between partner NRPSs, as well as the suppression of interaction between nonpartner NRPSs, we carried out product formation assays. The donor modules TycA and TycB3 [and the hybrid NRPSs derived thereof: TycA(B3) and TycB3(A)] activate DPhe, whereas the acceptor modules TycB1 and TycC1 [and the hybrid NRPSs derived thereof: (C1)TycB1 and (B1)TycC1] provide the amino acids LPro and LAsn, respectively. Consequently, depending on the acceptor module used, formation of two different dipeptide products was expected: (i) DPhe-LPro-DKP ([M+H]+ = 245.1 m/z) and (ii) DPhe-LAsn ([M+H]+ = 280.1 m/z). All 16 possible combinations of acceptor and donor modules were incubated with the respective substrate amino acids and subsequently analyzed by HPLC/MS (Table 1). Most strikingly, we observed that a matching pair of COM domains, rather than a matching pair of partner NRPSs, was the only criterion required for the establishment of protein–protein communication (Table 1). Product formation between partner NRPSs completely fails when donor and acceptor modules are carrying nonmatching COM domains, i.e., TycA/(C1)TycB1 and TycB3(A)/TycC1. This deficiency can be overcome by providing also the second partner NRPS with the unnatural, but matching, COM domain, i.e., TycA(B3)/(C1)TycB1 and TycB3(A)/(B1)TycC1. Likewise, productive interaction between nonpartner NRPSs can be enforced by the presence of a matching pair of COM domains, i.e., TycA(B3)/TycC1 and TycB3(A)/TycB1.
Table 1. Investigation of product formation between partner and nonpartner NRPSs.
| Acceptor modules
|
||||
|---|---|---|---|---|
| DPhe-LPro-DKP,*m/z
|
DPhe-LAsn,†m/z
|
|||
| Donor modules | TycB1 | (C1)TycB1 | TycC1-Te | (B1)TycC1 |
| TycA | 245.1, 267.1 | ND | ND | 280.1, 302.1 |
| TycA(B3) | ND | 245.1, 267.1 | 280.1, 302.1 | ND |
| TycB3 | ND | 245.1, 267.1 | 280.1, 302.1 | ND |
| TycB3(A) | 245.1, 267.1 | ND | ND | 280.1, 302.1 |
All 16 possible combinations of the donor modules TycA, TycA(B3), TycB3, and TycB3(A) and the acceptor modules TycB1, (C1)TycB1, TycC1-Te, and (B1)TycC1 were tested and analyzed by HPLC/MS. Formation of the expected dipeptide product DPhe-LPro-DKP or DPhe-LAsn could be observed and verified only for systems featuring matching pairs of COM domains. ND, not detected.
Expected mass: [M+H]+ = 245.1 m/z; [M+Na]+ = 267.1 m/z
Expected mass: [M+H]+ = 280.1 m/z; [M+Na]+ = 302.1 m/z
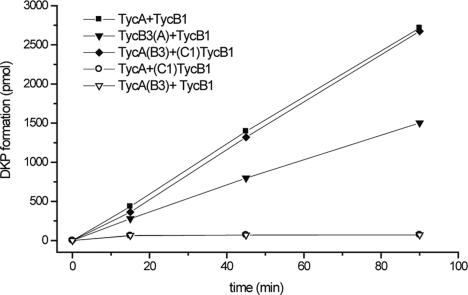
To further analyze the communication-mediating effect of COM domains, we carried out a time-dependent DKP formation assay by using the partner NRPSs TycA and TycB1 and all possible combinations of COM domains [TycA/TycB1, TycA(B3)/TycB1, TycA(B3)/(C1)TycB1, and TycA/(C1)TycB1]. Additionally, the natural nonpartner system TycB3(A)/TycB1 was tested, and all samples were analyzed by quantitative HPLC.
As shown in Fig. 4, this experiment clearly verified that productive protein–protein interaction and, as a consequence, efficient dipeptide formation, were observed only in systems harboring a matching pair of COM domains. Moreover, communication and product formation can be induced even between natural nonpartner NRPSs, although at a slightly reduced rate [TycA/TycB1, kobs = 0.62 min-1; TycA(B3)/(C1)TycB1, kobs = 0.59 min-1; and TycB3(A)/TycB1, kobs = 0.38 min-1]. This latter observation indicates that the main body of acceptor and donor module may also contribute somehow to the system's overall catalytic efficiency. Nonetheless, COM domains apparently provide the very first contact between donor and acceptor modules and thus are decisive for whether a system is productive at all.
Fig. 4.
Time-dependent formation of DPhe-LPro-DKP. Quantitative HPLC analysis was carried out by using the partner NRPS system TycA/TycB1 with all possible combinations of COM domains as well as the nonpartner system TycB3(A)/TycB1.
Crosstalk Between Different NRP Biosynthetic Systems. To demonstrate the biocombinatorial potential of COM domains (but also their generality), we investigated the crosstalk between the Tyc and surfactin (Srf) biosynthetic systems or, more precisely, between the initiation module TycA and the LLeu-activating termination module SrfAC (domain order: C-A-PCP-Te). From previous studies (6), it was known that the gramicidin S initiation module GrsA can trigger Tyc biosynthesis in vitro by the transfer of the activated DPhe moiety onto the first elongation module of TycB (TycB1). Thus, GrsA can form a productive complex with TycB1 because, according to our model, the N-terminal COM domains of the acceptor modules TycB1 and GrsB1 share a remarkable 77% similarity (71% identity in Fig. 2). Interestingly, the homology between the proposed COM domains of the Tyc elongation module TycB1 and the Srf termination module SrfAC is even higher (88–77%), indicating that TycA should likewise be capable of forming a productive biosynthetic complex with the acceptor module SrfAC. Given the substrate selectivity of the participating modules, this complex should result in the formation of the dipeptide DPhe-LLeu, which could be monitored by qualitative HPLC/MS analysis.
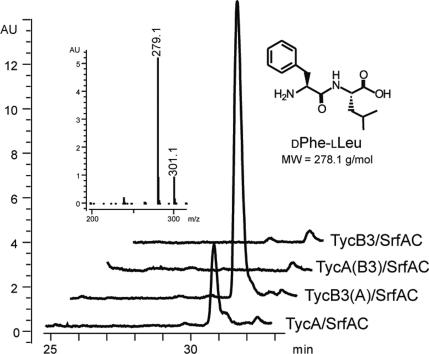
As shown in Fig. 5, formation of the expected dipeptide product was, in fact, observed for the artificial hybrid system TycA/SrfAC. Protein–protein communication and formation of a productive biosynthetic complex relied on the presence of a compatible pair of COM domains, as was verified by control reactions [TycA(B3)/SrfAC and TycB3/SrfAC]. Again, communication could be enforced by providing the complementary N-terminal COM domain [TycB3(A)/SrfAC].
Fig. 5.
Crosstalk between the Tyc and Srf biosynthetic systems. The crosstalk was investigated for the nonpartner NRPS systems TycA/SrfAC, TycB3(A)/SrfAC, TycA(B3)/SrfAC, and TycB3/SrfAC. HPLC/MS revealed, only for the first two systems, the formation of the expected DPhe-LLeu dipeptide product (retention time, 30.8 min; [M+H]+ = 279.1 m/z).
Discussion
The proper coordination of communication between partner NRPSs and the prevention of unselective interactions between nonpartner enzymes are important to assure an efficient biosynthesis of the desired peptide product. Given the experimental data presented, protein–protein communication is controlled by the interplay of matching pairs of COM domains, which are localized at the C and N termini of partner NRPSs.
The existence of C-terminal COM domains was proven for the initiation module TycA. Based on previous studies, it was known that the C-terminal half of TycA is indispensable for the formation of a heterodimeric complex with the partner elongation module TycB1. By the construction and investigation of C-terminal deletion mutants, we found that the removal of the six most C-terminal amino acids already led to complete disruption of protein–protein communication. In contrast, E domain activity was not affected even after deletion of the 23 most C-terminal amino acids.
Determination of TycA's partner COM domain, located at the N terminus of the acceptor module TycB1, was experimentally a bit more demanding because simple generation of N-terminal deletion mutants of TycB1 was not a practicable approach. Consequently, for the determination of the N-terminal COM domains of the acceptor modules, chimeric derivatives of the acceptor modules TycB1 and TycC1 and the donor modules TycA and TycB3 were constructed by substituting their putative COM domains. Studying all possible combinations of acceptor and donor modules, we found that protein–protein interaction and, as a consequence, efficient product formation were observed only in systems harboring a matching pair of COM domains. In contrast, product formation completely failed when donor and acceptor modules were carrying nonmatching COM domains. Strikingly, this outcome also concerned partner NRPSs when only one COM domain had been exchanged [i.e., TycA(B3)/TycB1]. Activity of these systems could be restored, however, when the corresponding partner NRPS was also provided with the unnatural, but matching, COM domain [i.e., TycA(B3)/(C1)TycB1]. All these experiments (i) proved the presence of N-terminal COM domain, (ii) verified the generality of COM domains at the junction between different partner NRPSs, and (iii) demonstrated that matching pairs of COM domains are essential for the establishment of productive interactions between NRPSs.
Most remarkably, matching pairs of COM domains could be used also to enforce protein–protein communication between nonpartner enzymes (i.e., TycA/(B1)TycC1 and TycB3(A)/TycB1) and even between NRPSs derived from different NRP biosynthetic systems. Consequently, COM domains are believed to accommodate a tremendous biocombinatorial potential, which could be exploited, e.g., to establish crosstalk between different biosynthetic systems to produce completely novel NRP products that combine the pharmacophores of different peptide drugs. Furthermore, COM domains could be used to do real biocombinatorial chemistry. For example, various chimeric nonpartner NRPSs with a matching pair of COM domains could be combined to randomly form an almost infinite number of unprecedented biosynthetic assembly lines.
The importance of short terminal enzyme regions for the establishment of protein–protein communication has been demonstrated recently for the functionally related class of PKSs. PKSs are the biosynthetic assembly lines of pharmacologically important polyketide antibiotics and are likewise organized in a multimodular fashion. In various studies, it had been shown that matching pairs of terminal regions, referred to as docking domains or linkers, promote the correct positioning of PKSs. The structure of N- and C-terminal linkers contain two separate four α-helix bundles, which together mediate the specific docking interactions (20). Based on the crystal structure of the free-standing NRPS C domain VibH of vibriobactin biosynthetic system (21), as well as secondary structure predictions, NRPS COM domains are believed also to possess α-helical structures that may provide the interfaces for the selective recognition of partner NRPSs.
Apart from several similarities between NRPS COM domains and PKS docking domains concerning, e.g., terminal localization, structure, and function, there are, however, also at least two striking differences. First, NMR solution structure suggests that C- and N-terminal PKS docking domains comprise 80–100 and 20–30 amino acid residues, respectively. In contrast, based on the domain-swapping experiments presented, C- and N-terminal NRPS COM domains appear to be significantly shorter, comprising only 20–30 and 15–25 amino acid residues, respectively. Second, it has been reported that docking domains, but also the donor substrate, contribute to the efficiency of protein–protein communication between PKSs. In contrast, at least in the case of the chimeric NRPS system TycA/SrfAC, no influence of the donor substrate amino acid could be determined.
In summary, here we presented the discovery of COM domains that facilitate protein–protein communication between successive partner NRPSs. These COM domains possess an enormous potential that can be exploited; i.e., to establish crosstalk between different biosynthetic systems, and to develop strategies for real biocombinatorial chemistry.
Supplementary Material
Acknowledgments
This work is dedicated to Christopher T. Walsh on the occasion of his 60th birthday. We thank Claudia Chiocchini (Phillips University of Marburg) for providing SrfAC protein, and Henning D. Mootz and Katrin Eppelmann for discussions and critical reading of the manuscript. This work was supported by the Federal Ministry of Education and Research (Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie) within the scope of its BioFuture program.
M.H. and T.S. designed research, analyzed data, and wrote the paper; and M.H. performed research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Tyc, tyrocidine; Srf, surfactin; NRP, nonribosomal peptide; NRPS, NRP synthetase; PKS, polyketide synthase; COM, communication-mediating; A, adenylation; PCP, peptidyl carrier protein; C, condensation; E, epimerization; MT, N-methylation; Te, thioesterase; DKP, diketopiperazine.
References
- 1.Marahiel, M. A., Stachelhaus, T. & Mootz, H. D. (1997) Chem. Rev. (Washington, D.C.) 97, 2651-2673. [DOI] [PubMed] [Google Scholar]
- 2.Schwarzer, D., Finking, R. & Marahiel, M. A. (2003) Nat. Prod. Rep. 20, 275-287. [DOI] [PubMed] [Google Scholar]
- 3.Cane, D. E., Walsh, C. T. & Khosla, C. (1998) Science 282, 63-68. [DOI] [PubMed] [Google Scholar]
- 4.Stachelhaus, T., Mootz, H. D. & Marahiel, M. A. (1999) Chem. Biol. 6, 493-505. [DOI] [PubMed] [Google Scholar]
- 5.Stachelhaus, T., Hüser, A. & Marahiel, M. A. (1996) Chem. Biol. 3, 913-921. [DOI] [PubMed] [Google Scholar]
- 6.Stachelhaus, T., Mootz, H. D., Bergendahl, V. & Marahiel, M. A. (1998) J. Biol. Chem. 273, 22773-22781. [DOI] [PubMed] [Google Scholar]
- 7.Walsh, C. T., Chen, H., Keating, T. A., Hubbard, B. K., Losey, H. C., Luo, L., Marshall, C. G., Miller, D. A. & Patel, H. M. (2001) Curr. Opin. Chem. Biol. 5, 525-534. [DOI] [PubMed] [Google Scholar]
- 8.Trauger, J. W., Kohli, R. M., Mootz, H. D., Marahiel, M. A. & Walsh, C. T. (2000) Nature 407, 215-218. [DOI] [PubMed] [Google Scholar]
- 9.Mootz, H. D. & Marahiel, M. A. (1997) J. Bacteriol. 179, 6843-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gokhale, R. S., Tsuji, S. Y., Cane, D. E. & Khosla, C. (1999) Science 284, 482-485. [DOI] [PubMed] [Google Scholar]
- 11.Wu, N., Tsuji, S. Y., Cane, D. E. & Khosla, C. (2001) J. Am. Chem. Soc. 123, 6465-6474. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 13.Horton, R. M., Hunt, H. D., Ho, S. N., Pullen, J. K. & Pease, L. R. (1989) Gene 77, 61-68. [DOI] [PubMed] [Google Scholar]
- 14.Mootz, H. D., Schwarzer, D. & Marahiel, M. A. (2000) Proc. Natl. Acad. Sci. USA 97, 5848-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linne, U., Stein, D. B., Mootz, H. D. & Marahiel, M. A. (2003) Biochemistry 42, 5114-5124. [DOI] [PubMed] [Google Scholar]
- 16.Gruenewald, S., Mootz, H. D., Stehmeier, P. & Stachelhaus, T. (2004) Appl. Environ. Microbiol. 70, 3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linne, U. & Marahiel, M. A. (2000) Biochemistry 39, 10439-10447. [DOI] [PubMed] [Google Scholar]
- 18.Walzel, B., Riederer, B. & Keller, U. (1997) Chem. Biol. 4, 223-230. [DOI] [PubMed] [Google Scholar]
- 19.Stachelhaus, T. & Walsh, C. T. (2000) Biochemistry 39, 5775-5787. [DOI] [PubMed] [Google Scholar]
- 20.Broadhurst, R. W., Nietlispach, D., Wheatcroft, M. P., Leadlay, P. F. & Weissman, K. J. (2003) Chem. Biol. 10, 723-731. [DOI] [PubMed] [Google Scholar]
- 21.Keating, T. A., Marshall, C. G., Walsh, C. T. & Keating, A. E. (2002) Nat. Struct. Biol. 7, 522-526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.