Abstract
Pyrosequencing of the 16S rRNA targeting RNA, community-level physiological profiles made with Biolog EcoPlates, proteolysis, and volatile component (VOC) analyses were mainly used to characterize the manufacture and ripening of the pasta filata cheese Caciocavallo Pugliese. Plate counts revealed that cheese manufacture affected the microbial ecology. The results agreed with those from culture-independent approaches. As shown by urea-PAGE, reverse-phase high pressure liquid chromatography (RP-HPLC), and free-amino-acid (FAA) analyses, the extent of secondary proteolysis mainly increased after 30 to 45 days of ripening. VOCs and volatile free fatty acids (VFFA) were identified by a purge-and-trap method (PT) and solid-phase microextraction (SPME) coupled with gas chromatography-mass spectrometry (GC-MS), respectively. Except for aldehydes, the levels of most of VOCs and VFFA mainly increased from 30 to 45 days onwards. As shown through pyrosequencing analysis, raw cows' milk was contaminated by Firmicutes (53%), Proteobacteria (39%), Bacteroidetes (7.8%), Actinobacteria (0.06%), and Fusobacteria (0.03%), with heterogeneity at the genus level. The primary starter Streptococcus thermophilus dominated the curd population. Other genera occurred at low incidence or sporadically. The microbial dynamics reflected on the overall physiological diversity. At 30 days, a microbial succession was clearly highlighted. The relative abundance of Streptococcus sp. and especially St. thermophilus decreased, while that of Lactobacillus casei, Lactobacillus sp., and especially Lactobacillus paracasei increased consistently. Despite the lower relative abundance compared to St. thermophilus, mesophilic lactobacilli were the only organisms positively correlated with the concentration of FAAs, area of hydrophilic peptide peaks, and several VOCs (e.g., alcohols, ketones, esters and all furans). This study showed that a core microbiota was naturally selected during middle ripening, which seemed to be the main factor responsible for cheese ripening.
INTRODUCTION
Pasta filata cheeses are a group of cheese varieties that originated primarily from the northern Mediterranean area, encompassing Italy, Greece, the Balkans, Turkey, and Eastern Europe. Pasta filata cheeses include soft or semisoft varieties, typically consumed fresh or after a short ripening (e.g., high- and low-moisture mozzarella and pizza cheese). Others are hard or semihard varieties that undergo considerable ageing before being consumed (e.g., provolone, ragusano, and caciocavallo) (1). Literally, the Italian term “pasta filata” means “spun paste” or “stretched curd” and refers to a unique processing step of curd plasticization and stretching. In particular, pasta filata cheeses undergo a texturization process that involves soaking of the acidified curd in hot water or salt brine until the plastic consistency is achieved. The hot plastic curd is manually or mechanically kneaded and stretched to produce a homogeneous cheese with a fiber-like structure, which is molded into a variety of shapes (2).
Caciocavallo is one of the most typical Italian pasta filata cheeses and has very wide market popularity. Several types of caciocavallo are manufactured in the Mediterranean area, which differ in their names (e.g., kashkaval, cascaval, kashar, and kasseri) and features of the technology used to produce them. Caciocavallo Pugliese is manufactured from raw cows' milk in the Apulia region. The cheese is sold after 2 weeks of ripening or, more commonly, after 2 to 4 months or longer (>12 months), only if it has to be grated. Natural whey or commercial thermophilic cultures are used as primary starters (1, 3).
Previously, culture-dependent approaches were used to determine the microbiological features of ripened Caciocavallo Pugliese cheese (4), even after the addition of autochthonous nonstarter lactic acid bacteria (NSLAB) or attenuated adjunct cultures (5, 6). The microbial dynamics which lead from raw cows' milk to ripened pasta filata cheese, including all typical manufacturing steps, had never been thoroughly described, and deep-sequencing approaches had never been used.
The microbial contribution to cheese flavor is mainly determined by the protocol of cheese making. During the manufacture of pasta filata cheeses, the main role of starter cultures is to synthesize enough lactic acid to demineralize and transform the curd into the state that undergoes stretching in hot water at the target pH. Primary starters provide the most significant contribution to the microbial biomass in young curd, typically attaining densities of ≥108 CFU g−1 within 1 day and declining throughout ripening (7). This decline marks the beginning of a presumptive microbial succession, which should involve the appearance of adventitious microorganisms, mainly represented by NSLAB (7, 8). NSLAB derive from raw milk (9) or the dairy environment and equipment surfaces (10), and their growth may be affected by primary starters (11). Although an extensive body of literature (7, 9, 12) has described the sources and factors affecting the growth of NSLAB, the transition between primary starters and NSLAB during ripening of pasta filata caciocavallo cheese had not been examined through deep-sequencing approaches.
The most recent literature (13, 14) shows how deep-sequencing approaches are successful in describing the structure and evolution of the cheese microbiota. None of these studies established the influence of the successive microbial populations on proteolysis and synthesis of volatile components during cheese ripening. Recently, one study proposed a polyphasic approach to find the in situ causal relationship between microbiota composition and proteolysis of raw ewes' milk pecorino cheese (15). When used for the pasta filata cheese Caciocavallo Pugliese, this approach might provide new insights regarding (i) raw cows' milk as the source of microbial diversity; (ii) the cheese manufacture, including the addition of primary starters, as the main factor influencing microbial diversity; (iii) the unpredictable and dynamic occurrence of NSLAB; and (iv) the causal relationship among technology, microbiota, and features of pasta filata cheese.
This study used a polyphasic approach, which was based on pyrosequencing of the 16S rRNA targeting RNA, community-level physiological profiles obtained with Biolog EcoPlates, and proteolysis and volatile-component analyses to characterize the pasta filata cheese Caciocavallo Pugliese during manufacture and ripening.
MATERIALS AND METHODS
Manufacture of cheese.
Caciocavallo Pugliese was manufactured with raw cows' milk (Friesian cow breed), using commercial freeze-dried (Sacco, Cadorago, Como, Italy) primary starter Streptococcus thermophilus (initial cell count of ca. 7.0 ± 0.2 log CFU ml−1 of milk). The manufacture was carried out at the industrial plant Ignalat (vat of about 200 liters), located in Noci, Bari (Apulia region), Italy. Cheese making was carried out on 3 consecutive days (total of 3 batches), using cows' milk from 2 daily milkings. All the results were the averages for 3 batches, which were analyzed in triplicate (total of 9 samples analyzed). Raw cows' milk was heated at 37°C and inoculated with primary starter, and liquid calf rennet (10 ml 100 l−1) was added. Coagulation took place within 30 min. The coagulum was first cut coarsely by hand, held under whey at 37°C for 2 h, and then reduced mechanically to particles of 1.5 to 2 cm. When the curd reached a pH of 5.25 (ca. 5 h, at room temperature), it was stretched in hot water (80°C). Cheeses were salted in brine (30% [wt/vol] NaCl) for 12 h. Ripening was at ca. 10°C and a relative humidity of 83% for 90 days. The weight of the cheese was approximately 1.5 kg. Raw cows' milk, curd immediately after coagulation, curd after 5 h of incubation (when the pH reached ca. 5.25), curd after stretching, and cheeses after 1 (C1), 3 (C3), 7 (C7), 15 (C15), 30 (C30), 45 (C45), 60 (C60), 75 (C75), and 90 (C90) days of ripening (after brine treatment) were collected from each batch. All samples were transported to the laboratory in thermal plastic bags under refrigerated conditions (ca. 4°C) and analyzed immediately (microbiological analysis and community-level catabolic profiles) or frozen (−80°C) (biochemical analysis and extraction of total bacterial genomic RNA).
Compositional, microbiological, and biochemical analyses.
Samples of milk, curd, and cheese were analyzed for protein (16), fat (17), moisture (oven drying at 102°C) (18), and salt (19). The pH was measured by a Foodtrode electrode (Hamilton, Bonaduz, Switzerland). The raw cows' milk used had the following composition: pH 6.7; fat, 3.8%; protein, 3.7%; lactose, 4.9%; and salt, 0.09%. No significant (P > 0.05) differences were found between the 3 batches.
Microbiological analyses were carried out as described previously (20). Twenty grams of sample was homogenized with 180 ml of sterile sodium citrate (2% [wt/vol]) solution. Presumptive mesophilic lactobacilli and lactococci were enumerated on MRS and M17 agar (Oxoid), respectively, under anaerobiosis at 30°C for 48 h. Presumptive thermophilic streptococci were enumerated on M17 agar (Oxoid, Basingstoke, Hampshire, United Kingdom) under anaerobiosis at 42°C for 48 h. Enterococci were counted on Slanetz and Bartley agar (Oxoid) at 37°C for 48 h. The number of yeasts was estimated at 30°C for 48 h on Sabouraud dextrose agar (SDA) (Oxoid) medium supplemented with chloramphenicol (0.1 g liter−1). The number of molds was estimated on wort agar (Oxoid) at 25°C for 5 days. Total coliforms were counted using violet red bile lactose (Oxoid) at 37°C for 24 h. Except for enterococci, the media for plating bacteria were supplemented with cycloheximide at 0.1 g liter−1.
The pH 4.6-insoluble and -soluble nitrogen fractions of the cheeses were analyzed by urea polyacrylamide gel electrophoresis (urea-PAGE) and reverse-phase high pressure liquid chromatography (RP-HPLC), as described by Andrews (21) and Gobbetti et al. (4), respectively. Total and individual free amino acids (FAA) from the water-soluble extracts were determined by a Biochrom 30 series amino acid analyzer (Biochrom Ltd., Cambridge Science Park, United Kingdom), as described by Di Cagno et al. (22).
Determinations of volatile components and volatile free fatty acids.
Neutral volatile components (VOC) were determined by a purge-and-trap method (PT) coupled with gas chromatography-mass spectrometry (PT–GC-MS). Prior to PT, 3 g of cheese was mixed with 27 g of ultra-high-quality (UHQ) deionized water with an Ultra-Turrax 4 times for 40 s each time, separated by 10-s rests, in a glass flask with a narrow neck plunged into ice. Ten milliliters of this suspension was placed in a glass extractor connected to the PT apparatus (Tekmar 3000; Agilent Instruments, NY, USA). Extraction was performed with helium at a flow rate of 40 ml min−1 on a Tenax trap at 37°C. The trap was desorbed at 225°C, and injection used a cryoconcentrator at −150°C. The chromatograph (model 6890; Agilent Instruments) was equipped with a DB5-like capillary column (RTX5 Restek; Agilent Instruments) with a 60-m length, 0.32-μm internal diameter, and 1-μm thickness. The helium flow rate was 2 ml min−1; the oven temperature was 40°C during the first 6 min and then was increased at 3°C min−1 to 230°C. The mass detector (MSD5973; Agilent Instruments) was used in electronic impact at 70 eV and in scan mode, at an atomic mass from 29 to 206. Quantification of compounds was expressed in arbitrary units of area.
For solid-phase microextraction (SPME) extraction of volatile free fatty acids (VFFA), each sample was analyzed twice with two different dilutions. A 400- or 1,000-μl portion of the suspension described above was poured into a 10-ml flask with 100 μl of 2 N sulfuric acid and 1,200 μl of UHQ water. The flask was sealed and placed in a bath at 60°C for 15 min. An SPME Carboxen-polydimethylsiloxane 75-μm fiber (Supelco, L'Isle d'Abeau, France) was introduced into the flask and held in the headspace for 30 min at 60°C. It was then removed and desorbed for 5 min in a splitless chromatograph injector at 240°C. The chromatograph (CE8160; Thermoquest, Les Ulis, France) was equipped with an FFAP column with a 30-m length, 0.53-μm diameter, and 1-μm thickness (Stabilwax DA; Restek, Lisses, France) and a flame ionization detector (FID). The helium flow rate was 6 ml min−1; the oven temperature was 120°C for 1 min, was increased to 162°C at 1.8°C min−1 and to 240°C at 10°C min−1, and lastly was held at 240°C for 5 min. Concentrations of VFFAs were calculated from calibration curves established with external standards (Sigma, L'Isle d'Abeau, France).
Extraction of total bacterial genomic RNA.
Total RNA was extracted using the RiboPure bacterial kit (Ambion RNA; Life Technologies Co., Carlsbad, CA, USA), according to the manufacturer's instructions. The quality of the RNA was checked by agarose gel electrophoresis. The RNA concentration was measured in a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Rockland, DE). The purified RNA (100 ng; final volume, 20 μl) was incubated at 42°C for 2 min in 2 μl of genomic DNA (gDNA) wipeout buffer (7×; QuantiTect reverse transcription kit; Qiagen srl, Milan, Italy) and RNase-free water (final volume, 14 μl). The cDNA was obtained with the QuantiTect reverse transcription kit (Qiagen), according to the manufacturer's instructions. All reactions were set up in a Rotor Gene 6000 instrument (Corbett Life Science, New South Wales, Australia) equipped with a 36-well reaction rotor.
Pyrosequencing and data analyses.
Three cDNA samples, corresponding to the three batches, were pooled and used for 16S based bacterial diversity analysis. Bacterial diversity was assessed via pyrosequencing on a 454 FLX sequencer (454 Life Sciences, Branford, CT, USA) and was performed by Research and Testing Laboratories (Lubbock, TX), according to standard laboratory procedures. Primers targeting the V1-V3 region (Escherichia coli positions 27 to 519; forward, 28F [GAGTTTGATCNTGGCTCAG], and reverse, 519R [GTNTTACNGCGGCKGCTG]) of the 16S rRNA gene (23) were used. Pyrosequencing procedures were carried out based upon protocols from Research and Testing Laboratories.
Bioinformatics.
Sequence data were processed using Research and Testing Laboratory's in-house pipeline, described at http://www.researchandtesting.com/. Briefly, sequences were grouped using their barcodes, and any sequence that contained a low-quality barcode or that failed to be at least half the expected amplicon length (or 250 bp, whichever was shorter) was removed from the data pool. Sequences that passed the quality filter were denoised using an algorithm based on USEARCH (24) and then checked for chimeras using UCHIME (25). Finally, sequence data were separated into operational taxonomic units (OTUs) and annotated using USEARCH and a distributed. NET algorithm that utilizes BLASTN+. The output from BLASTN+ was then parsed, and the taxonomic information for each sequence was resolved using the following rules: sequences with more than 97% identity to well-characterized database sequences (<3% divergence) were resolved at the species level, those with 95% to 97% at the genus level, those with 90% to 95% at the family level, those with 85% to 90% at the order level, those with 80 to 85% at the class level, and those with 77% to 80% at the phylum level. Any match below this percent identity was discarded. In addition, the highest-scoring pair had to be at least 75% of the query sequence or it was discarded, regardless of identity.
Community level catabolic profiles (CLCPs).
Biolog Eco-Microplates (Biolog, Inc., Hayward, CA, USA) were used to determine bacterial CLCPs (26). Microplates contain 31 carbon sources grouped by chemical class (carbohydrates, carboxylic acids, polymers, amino acids and amines) and the control, without a carbon source, in triplicate. Ten grams of sample was homogenized with 90 ml of sterile sodium chloride (0.9% [wt/vol]) solution and centrifuged at 10,000 × g for 15 min at 4°C. The pellet was washed with sterile 50 mM Tris-HCl (pH 7.0) and again with sterile sodium chloride solution and then centrifuged again. The cell suspension was diluted (1:1,000) in sterile sodium chloride solution and dispensed (150 μl) into each of the 96 wells of the Biolog Eco-Microplates. Incubation was at 30°C in the dark, and color development was measured at 590 nm with a microplate reader (Biolog Microstation), every 24 h up to 120 h. Three indices were determined (27). Shannon's diversity (H′), indicating the substrate utilization pattern, was calculated as −∑pi ln(pi), where pi is the ratio of the activity of a particular substrate to the sums of activities of all substrate activity at 120 h. Substrate richness (S), measuring the number of different substrates used, was calculated as the number of wells with a corrected absorbance greater than 0.25. Substrate evenness (E) was defined as the equitability of activities across all utilized substrates: E = H′/log(S).
Statistical analyses.
Data were subjected to one-way analysis of variance (ANOVA), and pairwise comparison of treatment means was achieved by Tukey's procedure at a P value of <0.05, using the statistical software Statistica 7.0 for Windows. Data for VOC and VFFA analyses were subjected to permutation analysis using PermutMatrix (28).
RESULTS
Compositional and microbiological analyses.
Table S1 in the supplemental material shows the main chemical composition of Caciocavallo Pugliese during manufacture and ripening. As expected, the pH decreased from the curd immediately after coagulation (6.40 ± 0.04) to the curd after ca. 5 h of incubation (5.28 ± 0.02). An increase (5.62 ± 0.03) was found after curd stretching, followed by a decrease at the beginning of ripening (5.24 ± 0.01) until 30 days of ripening (5.09 ± 0.03). Then, the pH increased over time, reaching 5.36 ± 0.04 at 90 days and varying only slightly during ripening. The highest increase in the NaCl concentration was found at the beginning of ripening, even though it progressively rose over time. As expected, the cheese moisture decreased progressively during ripening, reaching a value of 38.1 ± 1.5% at 90 days. The concentration of fat and protein inversely followed the moisture trend. At 90 days, the cheese had 33.1% ± 1.5% fat and 23.8% ± 1.5% protein.
Presumptive mesophilic lactobacilli were already present in the raw cows' milk (4.2 ± 0.2 log CFU g−1) (Table 1). Their numbers increased by ca. 1.4 log cycles after curd incubation and decreased after curd stretching (4.1 ± 0.2 log CFU g−1). During ripening, presumptive mesophilic lactobacilli progressively increased, especially after 30 days (7.2 ± 0.1 log CFU g−1), and attained the highest number during late aging (average value of ca. 8.5 log CFU g−1). Compared to mesophilic lactobacilli, raw cows' milk contained a slight but significantly (P < 0.05) higher number of presumptive mesophilic lactococci, which decreased after curd stretching (4.1 ± 0.1 log CFU g−1) and remained almost constant throughout ripening. As expected, a high number of presumptive thermophilic streptococci was found after coagulation and incubation of the curd (average value of ca. 8.7 log CFU g−1). Although the number decreased by ca. 1 log cycle after stretching of the curd, it progressively increased up to 15 days (9.0 ± 0.3 log CFU g−1). From 30 days onwards, the number decreased and attained the lowest value at the end of ripening (7.3 ± 0.1 log CFU g−1). Enterococci were found in the raw cows' milk (2.6 ± 0.1 log CFU g−1), and the number increased after 1 day of ripening (4.1 ± 0.2 log CFU g−1) and remained almost constant for 15 days but decreased during late ripening (from 30 to 90 days). Yeasts and molds disappeared during ripening and manufacture, respectively. Total coliforms were detected in the raw cows' milk but progressively disappeared.
TABLE 1.
Cell numbers, determined by a culture-dependent method, of various microbial groups during manufacture and ripening of Caciocavallo Pugliese
| Microbial group | Log CFU g−1 ina: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Milk | Curd-cgb | Curd-ic | Curd-std | Cheese at indicated day of ripening |
|||||||||
| 1e | 3 | 7 | 15 | 30 | 45 | 60 | 75 | 90 | |||||
| Mesophilic lactobacilli | 4.2 ± 0.2 F | 5.2 ± 0.1 DE | 5.6 ± 0.1 D | 4.1 ± 0.2 F | 5.0 ± 0.1 E | 5.1 ± 0.3 E | 6.2 ± 0.3 C | 6.6 ± 0.3 C | 7.2 ± 0.1 B | 7.5 ± 0.5 B | 8.4 ± 0.2 A | 8.6 ± 0.2 A | 8.5 ± 0.3 A |
| Mesophilic lactococci | 5.2 ± 0.1 A | 5.3 ± 0.3 A | 5.5 ± 0.2 A | 4.1 ± 0.1 C | 4.2 ± 0.4 C | 4.3 ± 0.4 C | 4.2 ± 0.2 C | 4.6 ± 0.1 B | 4.5 ± 0.3 B | 4.4 ± 0.4 B | 4.6 ± 0.3 B | 4.2 ± 0.2 C | 4.0 ± 0.1 B |
| Thermophilic streptococci | 4.7 ± 0.1 E | 8.6 ± 0.2 B | 8.8 ± 0.2 B | 7.9 ± 0.2 C | 8.8 ± 0.4 B | 8.9 ± 0.2 B | 9.3 ± 0.2 A | 9.0 ± 0.3 A | 8.2 ± 0.1 C | 8.1 ± 0.2 C | 8.0 ± 0.2 C | 7.7 ± 0.3 CD | 7.3 ± 0.1 D |
| Enterococci | 2.6 ± 0.1 E | 3.9 ± 0.2 B | 4.3 ± 0.2 A | 3.5 ± 0.2 C | 4.1 ± 0.2 AB | 4.0 ± 0.1 AB | 4.3 ± 0.2 A | 4.1 ± 0.2 AB | 3.0 ± 0.1 D | 2.5 ± 0.1 E | 2.0 ± 0.2 F | 1.7 ± 0.2 G | 1.5 ± 0.2 G |
| Yeasts | 3.4 ± 0.2 B | 4.5 ± 0.1 A | 5.4 ± 0.3 A | 3.7 ± 0.1 B | 3.7 ± 0.1 B | 2.6 ± 0.1 C | 2.4 ± 0.2 C | 2.3 ± 0.1 C | 1.8 ± 0.2 D | <1 | <1 | <1 | <1 |
| Molds | 2.3 ± 0.0 A | 2.2 ± 0.1 B | 2.1 ± 0.1 C | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 | <1 |
| Total coliforms | 3.4 ± 0.2 CD | 4.4 ± 0.1 B | 5.2 ± 0.3 A | 3.2 ± 0.1 C | 2.7 ± 0.2 CD | 2.5 ± 0.2 CD | 2.3 ± 0.1 D | 2.9 ± 0.1 CD | 2.4 ± 0.2 D | 2.4 ± 0.1 D | <1 | <1 | <1 |
Data are means ± standard deviations for two batches of each type of cheese, analyzed in triplicate. Data in the same row with different letters are significantly different (P < 0.05).
Curd after coagulation.
Curd after 5 h of incubation, when the pH reached a value of ca. 5.25.
Curd after stretching.
Curd after brine treatment.
Proteolysis.
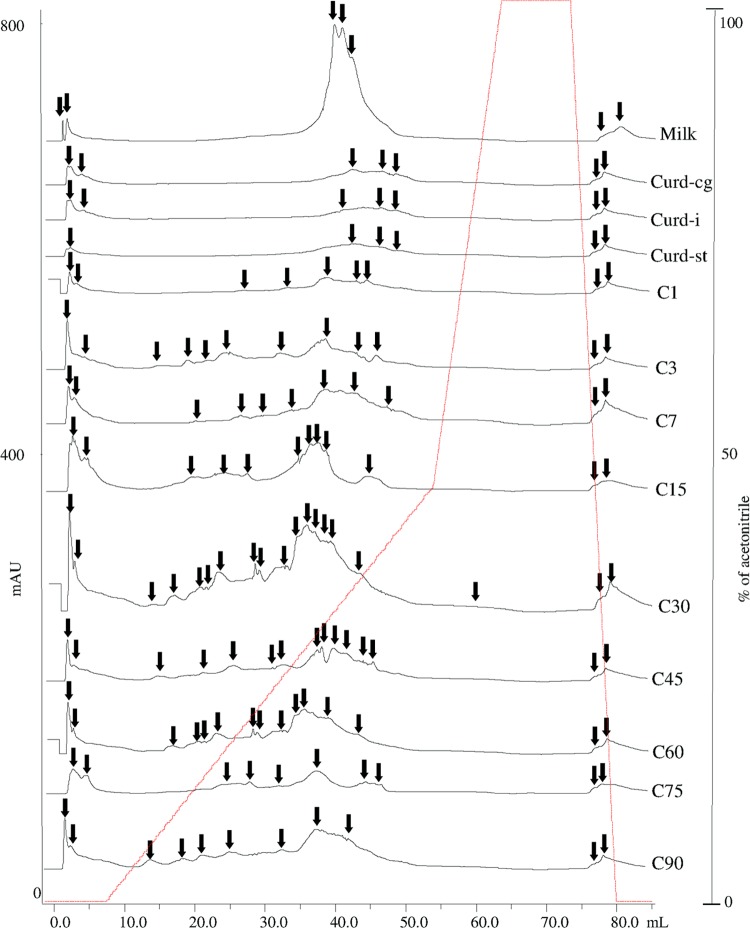
The pH 4.6-insoluble and -soluble nitrogen fractions of the cheese were analyzed by urea-PAGE (Fig. 1A and B, respectively). αs1-Casein (αs1-CN) persisted to the end of ripening, and its main degradation probably began at 7 days. β-CN also persisted. The formation of protein bands with low electrophoretic mobility, which presumably corresponded to γ-CN, was evident. The urea-PAGE electrophoretogram of the pH 4.6-soluble fraction showed differences over time. Characteristic polypeptide bands appeared at 7 days of ripening and persisted during late ripening. Complementary information emerged with RP-HPLC analysis (Fig. 2). The number of peaks which were recognized and matched visually with the Unicorn program (Amersham Biosciences) varied during manufacture and ripening of Caciocavallo Pugliese. Seven (raw cows' milk) to 19 (30 days) peaks were detected, which decreased to 11 at 90 days. An increase in the area of hydrophobic and especially hydrophilic peptide peaks was found up to 30 days. These results agreed with the concentration of total free amino acids (FAA) (Table 2). As expected, raw cows' milk (105.4 ± 14 mg kg−1) and curd after coagulation, incubation, and stretching had the lowest concentrations of FAA (140.4 ± 29 to 218.2 ± 18 mg kg−1). Subsequently, the concentration of FAA markedly (P < 0.05) increased, showing the highest rate from 30 days (1,343 ± 42 mg kg−1) to 75 days (2,335 ± 63 mg kg−1). The FAA found at the highest concentrations (>100 mg kg−1) were Ser, Cys, Val, Ile, Leu, Phe, His, Trp, Lys, Arg, and Pro.
FIG 1.
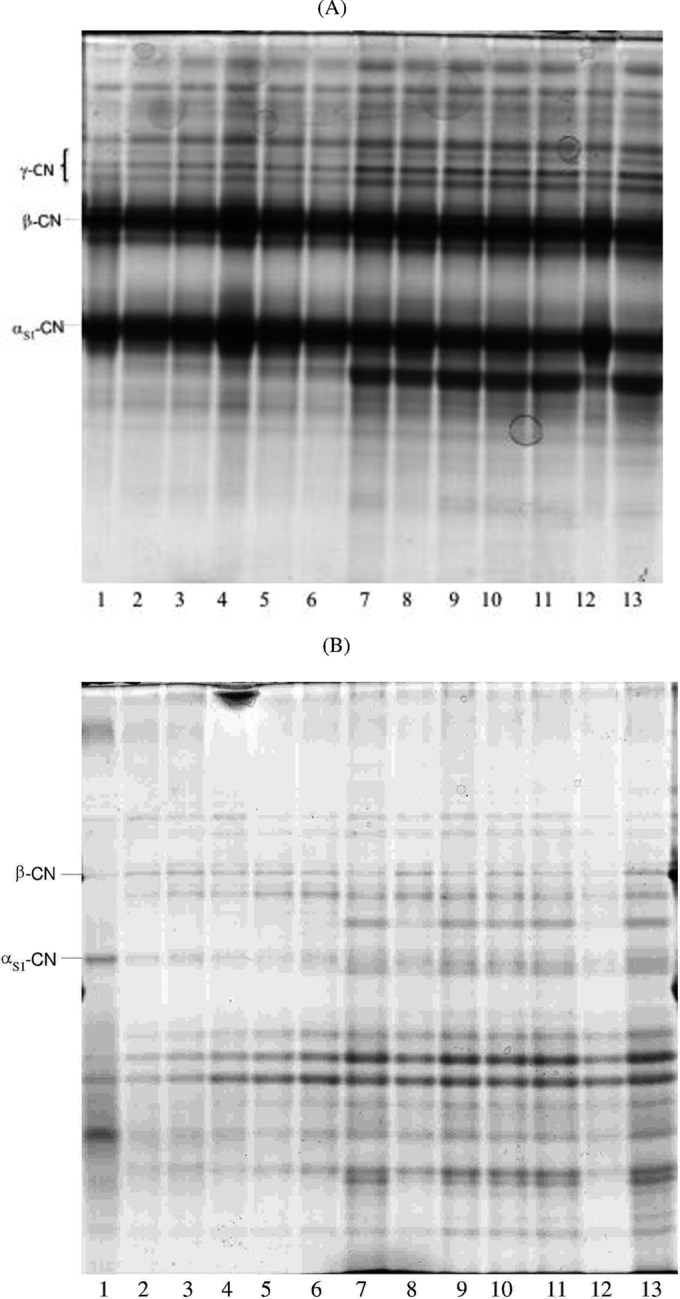
Urea-polyacrylamide gel electrophoresis (PAGE) of pH 4.6-insoluble (A) and -soluble (B) nitrogen fractions during manufacture and ripening of Caciocavallo Pugliese. Lanes: 1, bovine casein (CN) standard; 2, curd after coagulation; 3, curd after 5 h of incubation (when the pH reached a value of ca. 5.25); 4, curd after stretching and molding; 5 to 13, cheese (after brine treatment) after 1, 3, 7, 15, 30, 45, 60, 75, and 90 days of ripening, respectively.
FIG 2.
Reverse-phase fast protein liquid chromatography (RP-FPLC) of the pH 4.6-soluble nitrogen fraction during manufacture and ripening of Caciocavallo Pugliese. Data for raw cows' milk, curd after coagulation (Curd-cg), curd after 5 h of incubation (when the pH reached a value of ca. 5.25) (Curd-i), curd after stretching (Curd-st), and cheese (C) after brine treatment (C1) and during ripening (3, 7, 15, 30, 45, 60, 75, and 90 days; C3 to C90) are shown. Arrows indicate hydrophilic and hydrophobic peptide peaks. The gradient of acetonitrile is reported (red line). mAU, milli-absorbance units (UV, 214 nm).
TABLE 2.
Levels of FAA in Caciocavallo Pugliese during manufacture and ripening
| FAA | Mean concn of FAA (mg kg−1) ina: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Milk | Curd-cgb | Curd-ic | Curd-std | Cheese at indicated day of ripening |
|||||||||
| 1e | 3 | 7 | 15 | 30 | 45 | 60 | 75 | 90 | |||||
| Asp | 0.0 | 0.0 | 0.0 | 4.5 | 9.4 | 11.3 | 11.3 | 23.0 | 40.0 | 47.6 | 55.1 | 56.9 | 59.6 |
| Thr | 0.0 | 4.9 | 5.7 | 5.0 | 5.0 | 5.9 | 6.0 | 7.3 | 7.8 | 8.5 | 9.4 | 12.6 | 16.4 |
| Ser | 11.0 | 16.5 | 16.9 | 17.2 | 17.5 | 17.9 | 18.5 | 20.2 | 28.1 | 39.6 | 66.4 | 122.5 | 133.3 |
| Glu | 10.0 | 15.5 | 15.6 | 21.7 | 24.0 | 25.0 | 24.7 | 30.5 | 31.3 | 29.3 | 37.6 | 38.9 | 39.9 |
| Gly | 2.4 | 4.5 | 4.9 | 6.4 | 6.7 | 16.6 | 16.8 | 24.5 | 17.6 | 22.5 | 31.2 | 42.0 | 42.2 |
| Ala | 6.1 | 7.2 | 7.8 | 9.2 | 15.5 | 12.0 | 15.8 | 27.5 | 26.1 | 28.8 | 48.6 | 44.4 | 47.3 |
| Cys | 2.1 | 3.2 | 4.9 | 11.5 | 17.9 | 17.7 | 18.8 | 49.2 | 122.6 | 130.8 | 138.3 | 190.1 | 190.5 |
| Val | 14.7 | 18.6 | 22.3 | 4.3 | 7.2 | 8.0 | 7.3 | 12.9 | 66.5 | 70.5 | 103.1 | 144.6 | 149.6 |
| Met | 3.1 | 4.2 | 4.8 | 0.0 | 3.0 | 3.5 | 4.5 | 7.2 | 16.6 | 13.2 | 29.9 | 55.5 | 57.7 |
| Ile | 2.8 | 5.4 | 7.6 | 10.5 | 15.6 | 14.2 | 15.6 | 45.0 | 141.0 | 141.5 | 141.8 | 175.3 | 185.1 |
| Leu | 10.3 | 11.0 | 11.3 | 6.7 | 10.3 | 9.6 | 13.6 | 20.4 | 161.7 | 191.1 | 267.9 | 361.0 | 372.1 |
| Tyr | 4.5 | 8.8 | 8.9 | 10.3 | 18.8 | 19.3 | 24.9 | 38.7 | 106.7 | 132.7 | 120.0 | 45.5 | 53.8 |
| Phe | 8.2 | 9.9 | 13.8 | 18.9 | 16.3 | 17.5 | 14.8 | 28.9 | 113.7 | 133.4 | 174.4 | 242.9 | 239.6 |
| His | 2.3 | 2.7 | 4.2 | 5.9 | 15.6 | 21.1 | 25.4 | 35.2 | 71.4 | 85.4 | 95.6 | 121.7 | 138.7 |
| Trp | 0.0 | 2.2 | 2.0 | 1.7 | 12.6 | 32.1 | 18.4 | 62.4 | 90.8 | 98.1 | 99.3 | 176.3 | 183.6 |
| Orn | 1.0 | 3.1 | 3.4 | 10.5 | 14.3 | 17.5 | 21.3 | 26.4 | 28.8 | 47.0 | 49.6 | 134.9 | 126.6 |
| Lys | 9.8 | 10.3 | 15.5 | 25.5 | 29.1 | 34.8 | 32.6 | 49.7 | 80.7 | 91.1 | 117.3 | 132.9 | 148.5 |
| Arg | 0.0 | 0.0 | 2.1 | 6.4 | 13.6 | 14.8 | 13.3 | 35.3 | 103.6 | 119.9 | 153.8 | 109.5 | 106.9 |
| Pro | 17.1 | 12.4 | 24.3 | 42.0 | 44.1 | 45.6 | 43.8 | 54.9 | 88.3 | 111.2 | 119.2 | 127.5 | 134.6 |
| Total | 105.4 ± 14 H | 140.4 ± 29 G | 175.9 ± 24 G | 218.2 ± 18 F | 296.5 ± 19 F | 344.4 ± 31 E | 347.4 ± 32 E | 599.2 ± 19 D | 1343 ± 42 C | 1542.2 ± 51 C | 1858.5 ± 58 B | 2335 ± 63 A | 2426 ± 51 A |
Data are means ± standard deviations for two batches of each type of cheese, analyzed in triplicate. Data in the same row with different letters are significantly different (P < 0.05).
Curd after coagulation.
Curd after 5 h of incubation, when the pH reached a value of ca. 5.25.
Curd after stretching.
Cheese after brine treatment.
Volatile components.
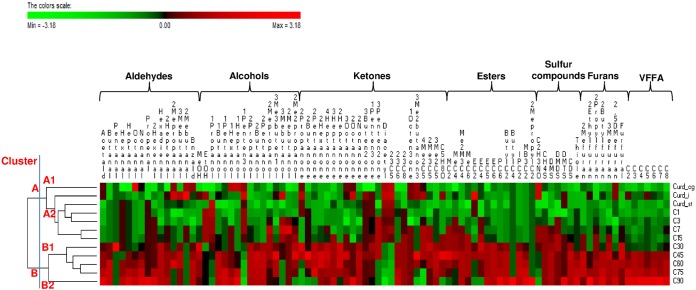
Volatile components (VOC) (95 in total) were identified by PT-GC/MS. VOC belonged to several chemical classes: aldehydes (17), alcohols (19), ketones (26), esters (14), sulfur compounds (7), and furans (8). The levels of 80 VOC significantly (P < 0.05) differentiated samples during manufacture and ripening (Fig. 3; also, see Table S2 in the supplemental material). Overall, aldehydes were the VOC found at the lowest levels. Acetaldehyde, octanal, nonanal, 2-methyl-propanal, 3- and 2-methyl-butanal, and benzaldehyde, which were found in the curd after coagulation and incubation, decreased after curd stretching, and their levels remained almost constant up to 15 days. Slight increases were found later. The levels of all the other aldehydes increased from 30 days onwards (45 days for heptanal and 60 for hexanal). The levels of 9 alcohols (methanol, 1-butanol, 1-pentanol, 1-hexanol, 1-heptanol, 2-propanol, 2-butanol, 2-pentanol, and 3-methyl-3-buten-1-ol) increased after 30 or 45 days of ripening and remained almost constant throughout ripening. 2-Methyl-1-propanol and 1-hexanol increased earlier. 1-Penten-3-ol was the only alcohol found at the highest level before ripening (curd after coagulation). Regarding the number of compounds identified, ketones (26 compounds identified) were the most abundant VOC. Most of them (e.g., 2-pentanone, 2-heptanone, 2-octanone, and 2-nonanone) were found at the highest levels from 30 to 90 days of ripening. The highest levels of 2-butanone and 2,3-octanedione and of 2,3-butanedione (diacetyl) and 2,3-pentanedione were found during manufacture and early ripening, respectively. Except for butyl butanoate and 2-methyl propyl-acetate, which were identified at the highest levels at the end of ripening, all the other esters increased from 15 (e.g., methyl hexanoate) and 30 (e.g., propyl hexanoate) days onwards. Almost all sulfur compounds increased after 7 days of ripening. Except for 2-ethyl- and 2,5-dimethyl-furans, which were identified at the highest levels before ripening, all the others furans increased after 15 (e.g., 2-methyl furan) and 30 (e.g., furfural) days onwards.
FIG 3.
Concentrations of volatile components (log arbitrary units of area) and volatile free fatty acids (ppm) identified during manufacture and ripening of Caciocavallo Pugliese. Euclidean distance and McQuitty's criterion (weighted pair group method with averages) were used for clustering. The colors correspond to normalized mean data levels from low (green) to high (red). The color scale, in terms of units of standard deviation, is shown at the top. Acetald, acetaldehyde; 2Hexenal, 2-hexenal; Hexadienal, 2,4-hexadienal; 2Heptenal, 2-heptenal; 2Mepropanal, 2-methyl-propanal; 3Mebutanal, 3-methyl-butanal; 2Mebutanal, 2 methyl-butanal; Bnzald, benzaldehyde; MeOH, methanol; EtOH, ethanol; 1Propanol, 1-propanol; 1Butanol, 1-butanol; 1Pentanol, 1-pentanol; 1Hexanol, 1-hexanol; 1Heptanol, 1-heptanol; 1Penten3ol, 1-penten-3-ol; 2Propanol, 2-propanol; 2Butanol, 2-butanol; 2Pentanol, 2-pentanol; 2Mepropanol, 2-methyl-1-propanol; 3Me3buten1ol, 3-methyl-3-buten-1ol; 3Mebutanol, 3-methyl-1-butanol; 2Mebutanol, 2-methyl-1-butanol; 2Me2propanol, 2-methyl-2-propanol; 2Propanone, 2-propanone; 2Butanone, 2-butanone; 2Pentanone, 2-pentanone; 2Hexanone, 2-hexanone; 4Heptanone, 4-heptanone; 3Heptanone, 3-heptanone; 2Heptanone, 2-heptanone; 3Octanone, 3-octanone; 2Octanone, 2-octanone; 2Nonanone, 2-nonanone; 3Buten2one, 3-buten-2-one; 1Penten3one, 1-penten-3-one; 3Penten2one, 3-penten-2-one; Diacetyl, 2,3-butanedione; 2–3C5, 2,3-pentanedione; 2–3C6, 2,3-hexanedione; 2–3C8, 2,3-octanedione; 1Octen3one, 1-octen-3-one; 3Me2butanone, 3-methyl-2-butanone; 4Me2C5, 4-methyl-2-pentanone; 2Me3C5, 2-methyl-3-pentanone; 3Me2C5, 3-methyl-2-pentanone; C5H8O, cyclopentanone; MC2, methyl acetate; MeC4, methyl butanoate; Me2M3C4, methyl 2-methyl-butanoate; MeC6, methyl hexanoate; EC2, ethyl acetate; EC4, ethyl butanoate; EC5, ethyl pentanoate; EC6, ethyl hexanoate; PC6, propyl hexanoate; ButylC2, butyl acetate; ButylC4, butyl butanoate; IPC2, isopropyl acetate; MB3C2, 3-methyl-butyl acetate; 2MepropylC2, 2-methyl propyl acetate; C2H3NO, methyl isocyanate; CH4S, methanethiol; DMS, dimethyl-sulfide; DMDS, dimethyl-disulfide; DMTS, dimethyl-trisulfide; CDS, carbon disulfide; Thiole, thiophene; 2Mefuran, 2-methyl furan; 2Ethylfuran, 2-ethyl-furan; 2Propylfuran, 2-propyl-furan; 2Butylfuran, 2-butyl-furan; 3Mefuran, 3-methyl-furan; 2,5DMefuran, 2,5-dimethyl-furan; Furfural, furfural; C2, acetic acid; C3, propionic acid; C4, butyric acid; C5, valeric acid; C6, hexanoic acid; C7, heptanoic acid; C8, octanoic acid.
The levels of all volatile free fatty acids (VFFA) increased throughout ripening (Fig. 3; also, see Table S2 in the supplemental material). In particular, acetic (8.1 ± 0.4 to 26.7 ± 1.1 ppm) and hexanoic (8.9 ± 0.2 to 23.4 ± 0.7 ppm) acids were found at the highest concentrations, followed by butyric acid (3.7 ± 0.1 to 16.9 ± 0.5 ppm). The permutation analysis based on VOC and VFFA (Fig. 3) distributed the samples into two major clusters (A and B). Cluster A included curds after the various manufacturing steps and cheeses from 1 to 15 days of ripening, which contained the lowest levels of most of the VOC and VFFA. Cluster B contained cheeses from 30 days of ripening onwards. Within cluster B, subclusters B1 and B2 further differentiated the cheeses based on the time of ripening (from 30 to 75 and at 90 days, respectively).
Structure and changes of the microbiota.
Pyrosequencing of 16S rRNA was used to characterize metabolically active bacteria. No significant (P > 0.05) differences were found between the three batches analyzed. After pyrosequencing, a total of 197,385 raw sequence reads of 16S rRNA gene amplicons were obtained (average length, 515 bp). The calculated alpha diversity parameters are reported in Table S3 in the supplemental material. The highest values of Chao1 richness and the Shannon diversity index were found in raw cows' milk (50.2 and 2.15, respectively). Further, the indices fluctuated according to technology steps. After these variations, Chao1 richness and the Shannon diversity index slightly increased from day 7 onwards. Good's estimated sample coverage (ESC) was above 99% for all the samples, indicating a satisfactory description of the microbial diversity. The community structure was also analyzed using three phylogeny-based beta-diversity measures (see Fig. S1 in the supplemental material). The two principal coordinate analyses (PCoA) clearly differentiated raw cows' milk, which was characterized by higher variability and was spread in the right part of the plot. Curds at various technological steps (coagulation, incubation, and stretching) and cheeses at early ripening (1 to 15 days) tended to form a separate subgroup. As the time of ripening proceeded (30 days onwards), cheeses grouped together.
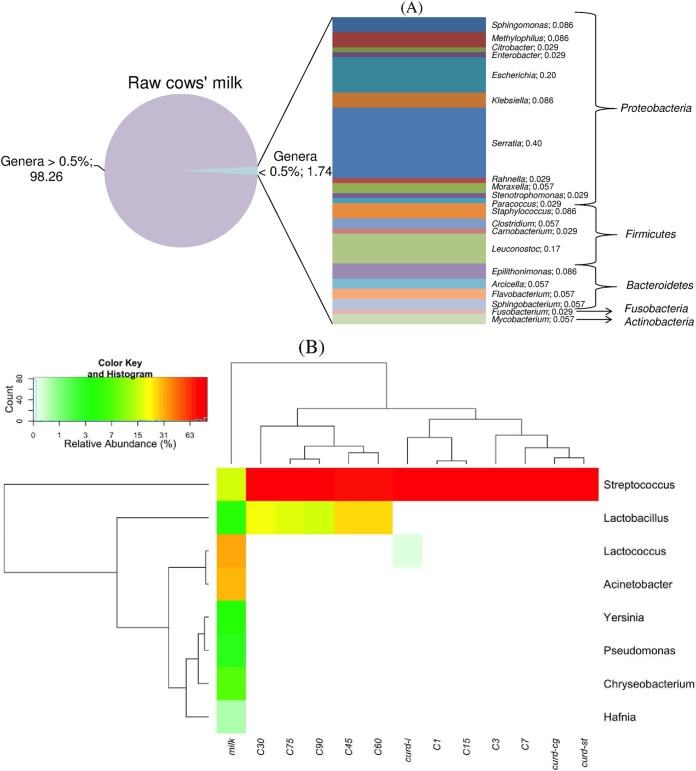
The bacterial sequences from RNA which were assigned to bacterial phyla and their relative abundance varied during cheese manufacture and ripening (data not shown). RNA from raw cows' milk included mainly Firmicutes (53%) and Proteobacteria (39%), followed by Bacteroidetes (7.8%) and, at low frequency, Actinobacteria and Fusobacteria (0.06% and 0.03%, respectively). From curd after coagulation onwards, the phylum Firmicutes dominated (ca. 99 to 100%). The phylum Proteobacteria appeared at a very low frequency only after 30 and 75 days of ripening (0.04 to 0.03%, respectively).
The distribution of the OTUs that were classified at genus level is reported in Fig. 4. Although 35 OTUs were identified in raw cows' milk (Fig. 4A and B), only 8 had a relative abundance higher than 0.5% in at least one sample (Fig. 4B). Lactococcus (31.9%), Acinetobacter (30.5%), Streptococcus (16.2%), Chryseobacterium (7.5%), Lactobacillus (4.6%) Yersinia (4.2%), Pseudomonas (2.9%), and Hafnia (0.55%) were the main genera found in raw cows' milk. Subdominant genera belonging to the phyla Proteobacteria (Sphingomonas, Methylophilus, Citrobacter, Enterobacter, Escherichia, Klebsiella, Serratia, Rahnella, Moraxella, Paracoccus, and Stenotrophomonas) (0.40 to 0.029%), Firmicutes (Staphylococcus, Clostridium, Carnobacterium, and Leuconostoc) (0.17 to 0.029%), Bacteroidetes (Epilithonimonas, Arcicella, Flavobacterium, and Sphingobacterium) (0.086 to 0.057%), Fusobacteria (Fusobacterium) (0.029%), and Actinobacteria (Mycobacterium) (0.057%) were also found at very low frequency (Fig. 4A). As expected, the bacterial profile of the curd after the inoculum of the primary starter (St. thermophilus) and coagulation markedly changed and became dominated by Streptococcus (100%) (Fig. 4B). At this manufacturing step, all the other genera were not detectable. Only in the curd after incubation was Streptococcus accompanied by Lactococcus (0.22%) and Lactobacillus at very low frequency (0.04%). From the curd after stretching to 15 days of ripening, Streptococcus still dominated (ca. 99%) and Lactobacillus was present in low numbers. In agreement with the numbers of presumptive thermophilic streptococci and mesophilic lactobacilli (Table 1), Streptococcus and Lactobacillus, respectively, decreased (80.7%) and increased (19.2%) at 30 days of ripening. This trend maintained throughout ripening, and at 90 days the bacterial profile of the Caciocavallo Pugliese was characterized by Streptococcus (79.2%), followed by Lactobacillus (20.1%). Other genera, occasionally and in low abundance, were also present. In particular, Paracraurococcus, Pseudoalteromonas, and Rhodococcus (after 30 days) and Azospirillum and Gelria (after 75 days) were found.
FIG 4.
Distribution of the OTUs assigned at genus level occurring in raw cows' milk at a relative abundance lower than 0.5% (A) and pseudo-heatmap depicting the distribution (percent) of bacterial genera during manufacture and ripening of Caciocavallo Pugliese (B). Only OTUs occurring at 0.5% abundance in at least one sample were included in the pseudo-heatmap. Clustering of samples and taxa was obtained using hierarchical clustering of unweighted UniFrac distances between samples or Euclidean distance measure between taxa. Data for raw cows' milk, curd after coagulation (Curd-cg), curd after 5 h of incubation (when the pH reached a value of ca. 5.25) (Curd-i), curd after stretching (Curd-st), and cheese (C) after brine treatment (C1) and during ripening (3, 7, 15, 30, 45, 60, 75 and 90 days; C3 to C90) are shown.
The abundance of OTUs from RNA samples (Firmicutes only) is reported in Table 3, with taxonomic details up to species level when such assignment was possible. Lactococcus lactis (25.6%) and St. thermophilus (11.5%) were the most abundant species in raw cows' milk, followed by Lactococcus sp. (5.62%), Streptococcus sp. (3.2%), Streptococcus parauberis (1.34%), Lactobacillus kefiranofaciens (1.28%), and Lactobacillus plantarum (1.2%). Lactobacillus paracasei, L. casei, and Lactobacillus sp. were also found at very low frequency. After the addition of the primary starter, St. thermophilus (82.6%) and Streptococcus sp. (16.8%) dominated the curd. They persisted at the same ratio up to 15 days of ripening. After 30 days, the microbiota changed. The relative abundance of St. thermophilus decreased (ca. 65%) and remained almost constant. At the same time, Streptococcus sp. also decreased slightly, whereas L. paracasei became the dominant species of mesophilic lactobacilli, which persisted up to 90 days of ripening (11%). Although less abundant (ca. 3%), the L. casei group and Lactobacillus sp. accompanied L. paracasei and stably persisted.
TABLE 3.
Incidences of OTUsa assigned to the Firmicutes species level during manufacture and ripening of Caciocavallo Pugliese
| Taxon | Incidence of OTU (%) in: |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Milk | Curd-cdb | Curd-ic | Curd-std | Cheese at indicated day of ripening |
|||||||||
| 1e | 3 | 7 | 15 | 30 | 45 | 60 | 75 | 90 | |||||
| Lactobacillus sp. | 0.54 | 0 | 0 | 0 | 0 | 0 | 0.035 | 0.036 | 2.34 | 3.69 | 3.78 | 2.61 | 2.13 |
| Lactobacillus buchneri | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.013 | 0.12 | 0.018 | 0 | 0.026 |
| Lactobacillus casei | 0.31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.65 | 4.51 | 3.42 | 3.435 | 3.28 |
| Lactobacillus kefiranofaciens | 1.28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lactobacillus paracasei | 0.94 | 0 | 0 | 0 | 0 | 0.028 | 0 | 0 | 13.15 | 16.48 | 18.53 | 11.77 | 10.86 |
| Lactobacillus pentosus | 0.20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lactobacillus plantarum | 1.20 | 0 | 0.013 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Leuconostoc mesenteroides | 0.17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lactococcus sp. | 5.62 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lactococcus lactis | 25.58 | 0 | 0.22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Lactococcus piscium | 0.71 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Streptococcus sp. | 3.17 | 16.86 | 11.26 | 15.0 | 14.58 | 18.96 | 13.9 | 12.94 | 14.90 | 13.39 | 10.57 | 14.20 | 16.60 |
| Streptococcus parauberis | 1.34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Streptococcus salivarius | 0.057 | 0.55 | 0.88 | 0.74 | 0.66 | 0.72 | 0.63 | 0.78 | 0.73 | 0.34 | 0.54 | 0.49 | 0.59 |
| Streptococcus thermophilus | 11.57 | 82.59 | 87.58 | 84.23 | 84.73 | 80.24 | 85.44 | 86.20 | 65.08 | 61.30 | 63.03 | 67.33 | 66.42 |
Based on 16S rRNA gene pyrosequencing analysis of all RNA samples directly from Caciocavallo Pugliese cheese during manufacture and ripening. Only OTUs occurring at 0.1% abundance in at least one sample are included.
Curd after coagulation.
Curd after 5 h of incubation (pH 5.25).
Curd after stretching.
Cheese after brine treatment.
Changes in the community level catabolic profiles.
The relative utilization of carbon sources (amino acids, carbohydrates, carboxylic acids, amines, and polymers) by the bacterial community varied during manufacture and ripening of Caciocavallo Pugliese (Fig. 5). Carbohydrates and amino acids followed by carboxylic acids were the chemical classes mainly used over time. After curd incubation, the most intense degradation of all the chemical classes occurred. The ability to use the carbon sources progressively decreased after curd stretching until 7 days of ripening. After 15 and especially 30 days, the catabolic activity increased and almost stabilized over time. Catabolic profiles were also determined using the indices H′, S, and E (see Table S4 in the supplemental material). Raw cows' milk showed the lowest values of substrate utilization (H′) (2.75 ± 0.2) and substrate richness (S) (13.33 ± 1.6). These indices significantly (P < 0.05) increased during cheese manufacture and ripening, and the highest values were found after curd incubation (3.08 ± 0.1 and 16.87 ± 1.9, respectively). The index E, a measure of the statistical significance (equitability) of the values of H′ and S, confirmed the significant (P < 0.05) differences over time.
FIG 5.
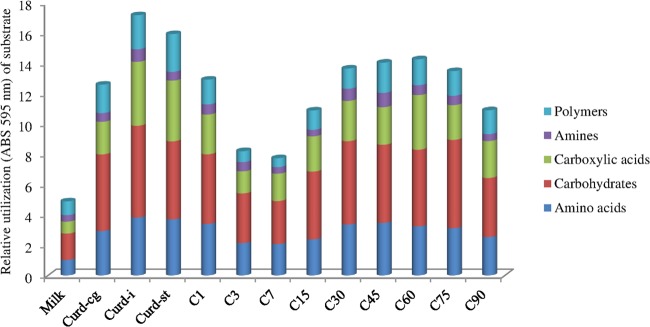
Relative utilization of different carbon sources, grouped by chemical class (carbohydrates, carboxylic acids, polymers, amino acids, and amines), during manufacture and ripening of Caciocavallo Pugliese. Data for raw cows' milk, curd after coagulation (Curd-cg), curd after 5 h of incubation (when the pH reached a value of ca. 5.25) (Curd-i), curd after stretching (Curd-st), and cheese (C) after brine treatment (C1) and during ripening (3, 7, 15, 30, 45, 60, 75 and 90 days; C3 to C90) are shown.
Correlations among microbiota, proteolysis, catabolic profiles, and volatile components.
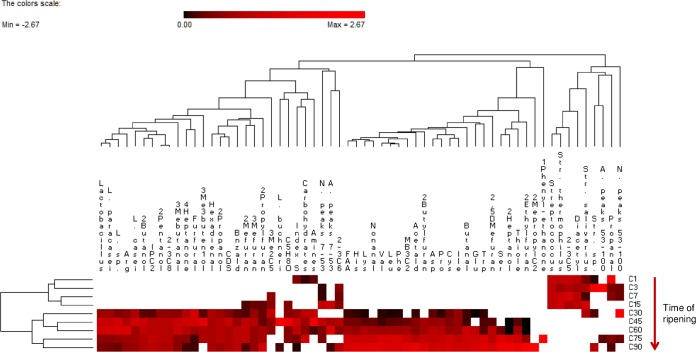
All the OTUs were used to find correlations. Only positive correlations (P < 0.05; r > 0.6) were further detailed. The concentration of total FAA and the abundance of Lactobacillus sp. and L. paracasei were correlated (P < 0.05). Also, the concentration of Asp, Glu, Leu, Phe, and Val was mainly correlated with L. paracasei (r > 0.80). The level of Lactobacillus sp. and L. paracasei was also correlated with the area of hydrophilic peptide peaks, whereas the abundance of St. thermophilus was positively correlated with the area of hydrophobic peptide peaks. St. thermophilus was also positively correlated with the substrate richness (S index) and with the utilization of amines and carbohydrates (P < 0.05). Regarding VOC, the abundance of L. paracasei was positively correlated with several aldehydes (e.g., propanal, butanal, nonanal, 2,4-hexadienal, 3-methyl-butanal, and benzaldehyde), alcohols (2-propanol, 2-butanol, 2-pentanol, 2-heptanol, and 3-methyl-3-buten-ol), ketones (2-hexanedione, 2-methyl 3-pentanone, cyclopentanone, 1-phenyl-ethanone, and 4-heptanone), esters (isopropyl acetate and 3-methyl-butyl acetate), sulfur compounds (carbon disulfide and thiophene), and all furans. St. thermophilus was correlated only with acetaldehyde, 2,3-butanedione (diacetyl), and 2,3-pentanedione. Permutation analysis based on the above parameters clustered cheeses throughout ripening (Fig. 6).
FIG 6.
Relative abundance of OTUs assigned to genus (Lactobacillus and Streptococcus) and species (Lactobacillus paracasei, Lactobacillus casei, Lactobacillus buchneri, Streptococcus thermophilus and Streptococcus salivarius) levels, total free amino acids (FAA, mg kg−1), amino acids found at the highest concentration (>100 mg kg−1) (Arg, His, Lys, Val, Leu, Phe, Asp, Pro, Cys, Ile, Glu, Trp, and Ser), substrate richness index (S index), relative utilization of carbon sources (carbohydrates and amines), number (N) and area (A) of hydrophilic (peaks 7 to 53) and hydrophobic (peaks 53 to 100) peptide peaks, and concentrations of volatile components (arbitrary units of area) positively correlated with the abundance of L. paracasei during ripening of Caciocavallo Pugliese. Euclidean distance and McQuitty's criterion (weighted pair group method with averages) were used for clustering. The colors correspond to normalized mean data levels from low (white) to high (red). The color scale, in terms of units of standard deviation, is shown at the top. Data for cheese (C) after brine treatment (C1) and during ripening (3, 7, 15, 30, 45, 60, 75 and 90 days; C3 to C90) are shown. L., Lactobacillus; Str., Streptococcus; Hexadienal, 2–4 hexadienal; 3Mebutanal, 3-methyl-butanal; Bnzald, benzaldehyde; 2Propanol, 2-propanol; 2Butanol, 2-butanol; 2Pentanol, 2-pentanol; 2Heptanol, 2-heptanol; 3Me3buten1ol, 3-methyl-3-buten-1-ol; 2–3C8, 2-hexanedione; 3Me2C5, 2-methyl-3-pentanone; C5H8O, cyclopentanone; 4Heptanone. 4-heptanone; IPC2, isopropyl acetate; MB3C2, 3-methyl-butyl acetate; CDS, carbon disulfide; Thiole, thiophene; 2Mefuran, 2-methyl-furan; 2Ethylfuran, 2-ethyl-furan; 2Propylfuran, 2-propyl-furan; 2Butylfuran, 2-butyl-furan; 3Mefuran, 3-methyl-furan; 2,5DMefuran, 2,5-dimethyl-furan; Acetald, acetaldehyde; Diacetyl, 2,3-butanedione; 2–3C5, 2,3-pentanedione.
DISCUSSION
First, this study explained how the microbial ecology evolved and affected proteolysis and synthesis of volatile components during the manufacture and ripening of Caciocavallo Pugliese.
Mean values for the gross composition approached those previously found for Caciocavallo Pugliese (4, 6, 29) and other semihard pasta filata cheeses (1, 3). Due to curd stretching in hot water (80°C) and addition of thermophilic starter, the highest decreases in pH and moisture, and, consequently, the highest increase in NaCl took place within 3 days. As shown by plating, raw cows' milk harbored adventitious lactic acid bacteria, especially mesophilic lactococci. The use of mature milk (storage for 24 h at 4°C before use) certainly favored the growth of mesophilic and psychrotrophic bacteria (8, 30). As expected, thermophilic streptococci markedly increased during curd incubation (pH ca. 5.25). The cell density of all microbial groups decreased by 1.5 to 2 log cycles after curd stretching. The lowest decrease (0.9 log cycle) was found for thermophilic streptococci, which better tolerated the temperature of stretching (3). After cheese manufacture, the low cell density of mesophilic lactococci remained almost constant, whereas thermophilic streptococci increased up to 15 days and then decreased. The number of mesophilic lactobacilli increased through ripening, remarkably after 30 to 45 days of ripening. As in other artisan raw milk cheeses (9, 31–34), enterococci were not negligible components of the adventitious microbiota.
A residual activity of chymosin toward αs1-CN was assumed, especially from 7 days of ripening onwards. Chymosin activity toward β-CN is usually limited, mainly because of hydrophobic interactions between salt and proteins (35). Usually, plasmin is active in pasta filata cheeses, since it withstands stretching conditions (3). Plasmin activity toward β-CN was suggested due to the concomitant formation of γ-CN. As shown by electrophoretic and chromatographic analyses of the pH 4.6-soluble fraction, and by liberation of FAA, peptides, generated through primary proteolysis, were substrates for the marked secondary proteolysis by lactic acid bacteria (3). The number and area of hydrophilic and hydrophobic peptide peaks increased up to 30 days of cheese ripening. At this time, the concentration of FAA became significantly high, further increasing and approaching the values usually found for other Italian cheeses (6, 36, 37). Ser, Val, Leu, Phe, His, Trp, and Pro were found at the highest concentrations, which is typical for semihard pasta filata cheeses (e.g., kashkaval) (1).
According to secondary proteolysis, the levels of most VOC increased from 30 days onwards. Only several aldehydes (e.g., acetaldehyde, octanal, nonanal, and 2-methyl propanal) were identified at highest levels during cheese manufacture and early ripening. Aldehydes are unstable compounds which are reduced to alcohols or oxidized to acids, indicating progressive cheese maturation (38, 39). Straight-chain aldehydes result from the oxidation of fatty acids, whereas branched-chain ones result from amino acid catabolism. Acetaldehyde may derive from different sources, but during early ripening it may result mainly from lactate metabolism, after the sudden fermentation of residual lactose (40). Primary, secondary, and branched-chain alcohols were identified, especially after 30 days of ripening. Secondary alcohols derive from reduction of methyl ketones, through the reductase activities of autochthonous lactic acid bacteria (41). Methyl-branched alcohols are synthesized from reduction of aldehydes, which are formed from branched FAA microbial activity (42). Ketones, the most abundant chemical class of VOC identified, are formed by enzymatic oxidation of free fatty acids to keto acids and subsequent decarboxylation to methyl ketones (40). Ketones are intermediate compounds which are further reduced to alcohols. Indeed, the levels of 3-penten-2-one, 2,3-butanedione, and 2,3-pentanedione decreased throughout ripening. Diacetyl, which mainly originates from the unstable precursor α-aceto-lactate via citrate metabolism (43), was mainly found during early ripening. As shown for the Turkish pasta filata cheese kashar (39), esters (e.g., ethyl acetate and ethyl butanoate) increased throughout ripening. VFFA contribute to the cheese flavor but also serve as precursors for the synthesis of methyl-ketones, alcohols, and esters (40). As in other pasta filata cheeses, acetic, hexanoic, and butyric acids were the most abundant (1, 3, 39). A relationship between autolysis of primary starters (e.g., Lc. lactis subsp. lactis and St. thermophilus) and levels of VFFA during cheese ripening may be hypothesized (44, 45).
The culture-independent analysis was carried out using RNA as the template, and the OTUs found during processing were active members of the microbial population (46). According to plating, the alpha diversity analysis demonstrated that the microbial community fluctuated during curd incubation and stretching. Further, diversity enlarged and varied during ripening. Raw cows' milk contained bacterial phyla that resulted from environmental contamination. Bacteroidetes, Actinobacteria, and especially Proteobacteria and Firmicutes dominate different areas throughout the farm, which include teat surfaces, milking parlors, hay, air, and dust (30). Genera belonging to Proteobacteria (e.g., Acinetobacter, Pseudomonas, and Hafnia), Firmicutes (e.g., Lactococcus, Streptococcus, and Lactobacillus), and Bacteroidetes (Chryseobacterium) were mainly identified. Psychrotrophic Chryseobacterium spp. was previously detected in raw cows' milk through culture-dependent and -independent approaches (30). Overall, Acinetobacter spp. and Pseudomonas spp., which dominated the raw cows' milk, may constitute 70 to 90% of the microbial population during milk storage at low temperature (47). Cows' milk also contains a population of lactic acid bacteria, including Lactococcus spp., Streptococcus spp., and Lactobacillus spp., that also adapt well to low temperatures (30). A large subpopulation of taxa, corresponding to <1% of the total reads, was also highlighted. Almost the same taxonomic structure characterized the microbiota of raw ewes' milk used for making pecorino cheese (15). Most of the above genera were inhibited after the thermophilic starter was grown. Only after incubation and stretching of the curd did Lactococcus and Lactobacillus recur at low frequency. The Gram-negative organisms Paracraurococcus, Pseudoalteromonas, and Azospirillum and the Gram-positive organisms Rhodococcus and Gelria were part of the subdominant population at the end of manufacture. The distribution of these low-abundance OTUs was variable among the samples, indicating sporadic contamination. The microbial succession reflected the overall physiological diversity (Fig. 3 and Fig. 6). The lowest utilization pattern substrate was found in raw cows' milk. The greatest ability to degrade carbon sources and the highest H′ index were observed almost at the end of cheese manufacturing, which coincided with environmental conditions becoming hostile. The starter organism St. thermophilus dominated the population of the curd. It was accompanied by Streptococcus sp., and both persisted at an almost constant ratio up to 15 days. At 30 days, a microbial succession was clearly highlighted. The relative abundances of Streptococcus sp. and especially of St. thermophilus decreased, while those of L. casei, Lactobacillus sp., and especially L. paracasei increased consistently. Mesophilic lactobacilli persisted at high levels up to the end of ripening. Previous studies (3, 4, 7) showed the coexistence of thermophilic primary starters and NSLAB (Lactobacillus parabuchneri, L. casei, and L. paracasei subsp. paracasei) during ripening of Caciocavallo Pugliese, but none clearly described the succession and diversity of the microbiota. Compared to the previous pyrosequencing analyses of the microbiota of the raw ewes' milk cheese pecorino (15), a lower diversity was found during ripening, and mesophilic lactobacilli were noticeably more abundant. Middle ripening (ca. 30 to 45 days) was the pivotal phase, where microbiota, secondary proteolysis, and synthesis of VOC concomitantly switched to confer the typical features of the semihard Caciocavallo Pugliese, which was completed at 90 days of ripening (Fig. 3 and 6). Despite the lower relative abundance of mesophilic lactobacilli with respect to St. thermophilus, only these bacteria were positively correlated with the total and individual (Ser, Val, Leu, and Phe) concentrations of FAA and the area of hydrophilic peptide peaks. L. paracasei was also positively correlated with several VOC (e.g., aldehydes, alcohols, ketones, esters, and all furans), which confirms the role of raw milk microbiota, especially that of NSLAB, during ripening of most cheeses (48, 49). St. thermophilus was correlated only with acetaldehyde, diacetyl, and 2,3-pentanedione. The synthesis of diketones and acetaldehyde by St. thermophilus from different substrates, including amino acids, was well documented (50, 51). All these compounds are common constituents of several foods (52), and the amounts commonly found are too low to be detrimental to health. However, many compounds were sufficiently concentrated in the mature cheeses to be perceived and contribute to flavor.
This study described the microbial ecology dynamics during manufacture and ripening of the pasta filata cheese Caciocavallo Pugliese through a deep-sequencing approach. Technology (e.g., curd stretching and use of thermophilic primary starter) affects the microbial diversity of raw cows' milk. A core microbiota was naturally selected during middle ripening, which seemed to be the main factor responsible for cheese ripening.
Supplementary Material
ACKNOWLEDGMENTS
We thank the industrial plant Ignalat, located in Noci, Bari (Apulia region), Italy, for the supply of cows' milk, cheese manufacture, and technical support.
This work was funded by Ministero dell'Istruzione, dell'Università e della Ricerca, Ministero dello Sviluppo Economico and Fondo Europeo di Sviluppo Regionale (PON02_00186_3417037, project PROINNO_BIT).
Footnotes
Published ahead of print 1 August 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02097-14.
REFERENCES
- 1.Kindstedt P, Carić M, Milanović S. 2004. Pasta-filata cheeses, p 251–277 In Fox PF, McSweeney PLH, Cogan TM, Guinee TP. (ed), Cheese: chemistry, physics and microbiology, 3rd ed, vol 2 Elsevier Academic Press, London, United Kingdom. 10.1016/S1874-558X(04)80047-2 [DOI] [Google Scholar]
- 2.Salvadori del Prato O. 1998. Trattato di tecnologia casearia. Edagricole—Edizioni Agricole della Calderini s.r.l. Press, Bologna, Italy [Google Scholar]
- 3.De Angelis M, Gobbetti M. 2011. Cheese: pasta-filata cheeses: traditional pasta-filata cheese, p 745–752 In Fuquay JW, Fox PF, McSweeney PLH. (ed), Encyclopedia of dairy sciences, 2nd ed, vol 1 Elsevier Academic Press, San Diego, CA [Google Scholar]
- 4.Gobbetti M, Morea M, Baruzzi F, Corbo MR, Matarante A, Considine T, Di Cagno R, Guineee T, Fox PF. 2002. Microbiological, compositional, biochemical and textural characterization of Caciocavallo Pugliese cheese during ripening. Int. Dairy J. 12:511–523. 10.1016/S0958-6946(02)00042-0 [DOI] [Google Scholar]
- 5.Morea M, Matarante A, Di Cagno R, Baruzzi F, Minervini F. 2007. Contribution of autochthonous non-starter lactobacilli to proteolysis in Caciocavallo Pugliese cheese. Int. Dairy J. 17:525–534. 10.1016/j.idairyj.2006.05.010 [DOI] [Google Scholar]
- 6.Di Cagno R, De Pasquale I, De Angelis M, Gobbetti M. 2012. Accelerated ripening of Caciocavallo Pugliese cheese with attenuated adjuncts of selected non-starter lactobacilli. J. Dairy Sci. 9:4784–4795. 10.3168/jds.2011-5283 [DOI] [PubMed] [Google Scholar]
- 7.Gobbetti M, De Angelis M, Di Cagno R, Rizzello CG. 2007. Relative contributions of starter cultures and non-starter bacteria to flavour of cheese, p 121–156 In Weimer BC. (ed), Improving the flavour of cheese. CRC Press, Boca Raton, FL [Google Scholar]
- 8.Quigley L, O'Sullivan O, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. 2011. Molecular approaches to analyzing the microbial composition of raw milk and raw milk cheese. Int. J. Food Microbiol. 150:81–94. 10.1016/j.ijfoodmicro.2011.08.001 [DOI] [PubMed] [Google Scholar]
- 9.Berthier F, Beuvier E, Dasen A, Grappin R. 2001. Origin and diversity of mesophilic lactobacilli in Comtè cheese, as revealed by PCR with repetitive and species-specific primers. Int. Dairy J. 11:293–305. 10.1016/S0958-6946(01)00059-0 [DOI] [Google Scholar]
- 10.Somers EB, Johnson ME, Wong ACL. 2001. Development of amino acids and organic acids in Norvegia, influence of milk treatment and adjunct Lactobacillus. J. Dairy Sci. 84:1926–1936. 10.3168/jds.S0022-0302(01)74634-6 [DOI] [PubMed] [Google Scholar]
- 11.Di Cagno R, De Angelis M, Upadhyayd VK, McSweeney PLH, Minervini F, Gallo G, Gobbetti M. 2003. Effect of proteinases of starter bacteria on the growth and proteolytic activity of Lactobacillus plantarum DPC2741. Int. Dairy J. 13:145–157. 10.1016/S0958-6946(02)00143-7 [DOI] [Google Scholar]
- 12.Fox PF, Guinee TP, Cogan TM, McSweeney PLH. 2000. Fundamentals of cheeses sciences. Aspen Publishers, Gaithersburg, MD [Google Scholar]
- 13.Alegria A, Szczenny P, Mayo B, Bardowski J, Kowalczyk M. 2012. Biodiversity in Oscypek, a traditional Polish cheese, determined by culture-dependent and independent approaches. Appl. Environ. Microbiol. 78:1890–1898. 10.1128/AEM.06081-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Filippis F, La Storia A, Stellato G, Gatti M, Ercolini D. 2014. A selected core microbiome drives the early stages of three popular Italian cheese manufactures. PLoS One 9:1–8. 10.1371/journal.pone.0089680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Pasquale I, Calasso M, Mancini L, Ercolini D, La Storia A, De Angelis M, Di Cagno R, Gobbetti M. 2014. Causal relationship between microbial ecology dynamics and proteolysis during manufacture and ripening of Canestrato Pugliese PDO cheese. Appl. Environ. Microbiol. 80:4085–4094. 10.1128/AEM.00757-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Dairy Federation. 1964. Determination of the protein content of processed cheeses products. Standard 25. International Dairy Federation, Brussels, Belgium [Google Scholar]
- 17.Institute for Industrial Research and Standards. 1955. Determination of the percentage of fat in cheese. Irish standard 69. Institute for Industrial Research and Standards, Dublin, Ireland [Google Scholar]
- 18.International Dairy Federation. 1982. Cheese and processed cheese. Determination of the total solid content. IDF standard 4A. International Dairy Federation, Brussels, Belgium [Google Scholar]
- 19.Fox PF. 1963. Potentiometric determination of salt in cheese. J. Dairy Sci. 46:744–745. 10.3168/jds.S0022-0302(63)89134-1 [DOI] [Google Scholar]
- 20.Di Cagno R, Quinto M, Corsetti A, Minervini F, Gobbetti M. 2006. Assessing the proteolytic and lipolytic activities of single strains of mesophilic lactobacilli as adjunct cultures using a Caciotta cheese model system. Int. Dairy J. 16:119–130. 10.1016/j.idairyj.2005.01.012 [DOI] [Google Scholar]
- 21.Andrews AT. 1983. Proteinases in normal bovine milk and their action on caseins. J. Dairy Res. 50:45–55. 10.1017/S0022029900032519 [DOI] [PubMed] [Google Scholar]
- 22.Di Cagno R, Buchin S, de Candia S, De Angelis M, Fox PF, Gobbetti M. 2007. Characterization of Italian cheeses ripened under nonconventional conditions. J. Dairy Sci. 90:2689–2704. 10.3168/jds.2006-654 [DOI] [PubMed] [Google Scholar]
- 23.Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods 7:668–669. 10.1038/nmeth0910-668b, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 25.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siragusa S, Di Cagno R, Ercolini D, Minervini F, Gobbetti M, De Angelis M. 2009. Taxonomic structure and monitoring of the dominant population of lactic acid bacteria during wheat flour sourdough type I propagation using Lactobacillus sanfranciscensis starters. Appl. Environ. Microbiol. 75:1099–1109. 10.1128/AEM.01524-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon CE. 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27:379–423, 623–656. 10.1002/j.1538-7305.1948.tb01338.x [DOI] [Google Scholar]
- 28.Minervini F, Di Cagno R, Lattanzi A, De Angelis M, Antonielli L, Cardinali G, Cappelle S, Angelis M, Gobbetti M. 2012. Lactic acid bacterium and yeast microbiotas of 19 sourdoughs used for traditional/typical Italian breads: interactions between ingredients and microbial species diversity. Appl. Environ. Microbiol. 78:1251–1264. 10.1128/AEM.07721-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ottogalli G. 2001. Atlante dei Formaggi. Hoepli Editore, Milan, IT [Google Scholar]
- 30.Quigley L, O'Sullivan O, Beresford TP, Ross RP, Fitzgerald GF, Ross RP, Stanton C, Cotter PD. 2013. The microbial content of raw and pasteurised cow's milk as determined by molecular approaches. J. Dairy Sci. 96:1–10. 10.3168/jds.2012-5409 [DOI] [PubMed] [Google Scholar]
- 31.Aquilanti L, Dell'Aquila L, Zannini E, Zocchetti A, Clementi F. 2006. Resident lactic acid bacteria in raw milk Canestrato Pugliese cheese. Lett. Appl. Microbiol. 43:161–167. 10.1111/j.1472-765X.2006.01935.x [DOI] [PubMed] [Google Scholar]
- 32.Cogan TM, Barbosa M, Beuvier E, Bianchi-Salvadori B, Cocconcelli PS, Fernandes I, Gomez J, Gomez R, Kalantzopoulos G, Ledda A, Medina M, Rea MC, Rodriguez E. 1997. Characterization of the lactic acid bacteria in artisanal dairy products. J. Dairy Res. 64:409–421. 10.1017/S0022029997002185 [DOI] [Google Scholar]
- 33.Prodromou K, Thasitou P, Haritonidou E, Tzanetakis N, Litopoulou-Tzanetaki E. 2001. Microbiology of “Orinotyri,” a ewe's cheese from the Greek mountains. J. Food Microbiol. 18:319–328. 10.1006/fmic.2001.0403 [DOI] [Google Scholar]
- 34.Dolci P, Alessandria V, Rantsiou K, Rolle L, Zeppa G, Cocolin L. 2008. Microbial dynamics of Castelmagno PDO, a traditional Italian cheese, with a focus on lactic acid bacteria ecology. Int. J. Food Microbiol. 122:302–311. 10.1016/j.ijfoodmicro.2007.12.018 [DOI] [PubMed] [Google Scholar]
- 35.Fox PF. 1989. Proteolysis during cheese manufacture and ripening. J. Dairy Sci. 72:1379–1383. 10.3168/jds.S0022-0302(89)79246-8 [DOI] [Google Scholar]
- 36.Coda R, Brechany E, De Angelis M, De Candia S, Di Cagno R, Gobbetti M. 2006. Comparison of the compositional, microbiological, biochemical, and volatile profile characteristics of nine Italian ewes' milk cheeses. J. Dairy Sci. 89:4126–4143. 10.3168/jds.S0022-0302(06)72458-4 [DOI] [PubMed] [Google Scholar]
- 37.Di Cagno R, Banks J, Sheehan L, Fox PF, Brechany EY, Corsetti A, Gobbetti M. 2003. Comparison of the microbiological, compositional, biochemical, volatile profile and sensory characteristics of three Italian PDO ewes' milk cheeses. Int. Dairy J. 13:961–972. 10.1016/S0958-6946(03)00145-6 [DOI] [Google Scholar]
- 38.Carbonell M, Nunez M, Fernandéz-Garcìa E. 2002. Evolution of the volatile components of ewe raw milk La Serena cheese during ripening. Correlation with flavour characteristics. Lait 82:683–698. 10.1051/lait:2002042 [DOI] [Google Scholar]
- 39.Hayaloglu AA. 2007. Comparisons of different single-strain starter cultures for their effects on ripening and grading of Beyaz cheese. Int. J. Food Sci. Technol. 42:930–938. 10.1111/j.1365-2621.2006.01312.x [DOI] [Google Scholar]
- 40.McSweeney PLH, Sousa MJ. 2000. Biochemical pathways for the production of flavour compounds in cheese during ripening: a review. Lait 80:293–324. 10.1051/lait:2000127 [DOI] [Google Scholar]
- 41.Molimard P, Spinnler HE. 1996. Compounds involved in the flavor of surface mold-ripened cheese: origins and properties. J. Dairy Sci. 79:169–184. 10.3168/jds.S0022-0302(96)76348-8 [DOI] [Google Scholar]
- 42.Yvon M, Rijnen L. 2001. Cheese flavor formation by amino acid catabolism. Int. Dairy J. 11:185–201. 10.1016/S0958-6946(01)00049-8 [DOI] [Google Scholar]
- 43.Garde S, Carbonell M, Fernandez-Garcia E, Medina M, Nunez M. 2002. Volatile compounds in Hispanico cheese manufactured using a mesophilic starter, a thermophilic starter and bacteriocin-producing Lactococcus lactis subsp. lactis INIA 415. J. Agric. Food Chem. 50:6752–6757. 10.1021/jf020577v [DOI] [PubMed] [Google Scholar]
- 44.Chich JF, Marchessau K, Gripon JC. 1997. Intracellular esterase from Lactococcus lactis subsp. lactis NDCO763: purification and characterization. Int. Dairy J. 7:169–174. 10.1016/S0958-6946(97)00001-0 [DOI] [Google Scholar]
- 45.Collins YF, McSweeney PLH, Wilkinson MG. 2003. Evidence for a relationship between autolysis of starter bacteria and lipolysis in Cheddar cheese. J. Dairy Res. 70:105–113. 10.1017/S0022029902005915 [DOI] [PubMed] [Google Scholar]
- 46.Ercolini D. 2013. High-throughput sequencing and metagenomics: moving forward in the culture-independent analysis of food microbial ecology. Appl. Environ. Microbiol. 79:3148–3155. 10.1128/AEM.00256-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raats D, Offek M, Minz D, Halpern M. 2011. Molecular analysis of bacterial communities in raw cow milk and the impact of refrigeration on its structure and dynamics. Food Microbiol. 28:465–471. 10.1016/j.fm.2010.10.009 [DOI] [PubMed] [Google Scholar]
- 48.Beuvier E, Buchin S. 2004. Raw milk cheeses, p 319–346 In Fox PF, McSweeney PLH, Cogan T, Guinee T. (ed), Cheese: chemistry, physics and microbiology, 3th ed, vol 1, Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 49.Montel MC, Buchin S, Mallet A, Delbes-Paus C, Vuitton DA, Desmasures N, Berthier F. 2014. Traditional cheeses: rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 177:136–154. 10.1016/j.ijfoodmicro.2014.02.019 [DOI] [PubMed] [Google Scholar]
- 50.Ott A, Germond JE, Chaintreau A. 2000. Vicinal diketone formation in yogurt: 13C-labeled precursors and effect of branched-chain amino acids. J. Agric. Food Chem. 48:724–731. 10.1021/jf990487z [DOI] [PubMed] [Google Scholar]
- 51.Ott A, Germond JA, Chaintreau A. 2000. Origin of acetaldehyde during milk fermentation using 13C-labeled precursors. J. Agric. Food Chem. 48:1512–1517. 10.1021/jf9904867 [DOI] [PubMed] [Google Scholar]
- 52.Maarse H, Visscher CA. 1989. In Maarse H, Visscher CA, Willemsens LC, Boelens MH. (ed), Volatile compounds in food, qualitative and quantitative data. TNO-CIVO, Zeist, The Netherlands [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.