Abstract
Human genetic diversity has long been studied both to understand how genetic variation influences risk of disease and infer aspects of human evolutionary history. In this article, we review historical and contemporary views of human genetic diversity, the rare and common mutations implicated in human disease susceptibility, and the relevance of genetic diversity to personalized medicine. First, we describe the development of thought about diversity through the 20th century and through more modern studies including genome-wide association studies (GWAS) and next-generation sequencing. We introduce several examples, such as sickle cell anemia and Tay–Sachs disease that are caused by rare mutations and are more frequent in certain geographical populations, and common treatment responses that are caused by common variants, such as hepatitis C infection. We conclude with comments about the continued relevance of human genetic diversity in medical genetics and personalized medicine more generally.
Genetic diseases cannot be fully understood by studying only one human population. Rare gene variants (e.g., mutations causing Tay–Sachs disease) can show markedly different patterns across groups.
We all differ at the level of our DNA sequence, and geneticists obsess over trying to understand the significance of this genetic diversity. This is an important goal, as by understanding human genetic diversity we can learn about the evolutionary history of our species, where we have come from, and perhaps where we are headed. More practically, understanding human genetic diversity is essential to understanding the biology of our diseases of various kinds, from the genetically more simple to more complex, and how we respond to treatment at both the population and individual levels (Torkamani et al. 2012). Indeed, improving our knowledge of human disease biology is the primary driver behind the largest and most systematic studies of human genetic diversity today. These studies, and the population- and disease-specific investigations made possible by them, are essential for reducing health disparities and improving health outcomes for the species as a whole. Unfortunately, largely because of which DNA samples are most easily accessible, most genomics research programs have concentrated their discovery efforts in populations of European ancestry (Need and Goldstein 2009; Bustamante et al. 2011). As we discuss in this essay, this approach is myopic and carries with it untoward consequences for both the scientific and public health enterprises.
The successful completion of the Human Genome Project in 2003 was the first in a series of large multinational public efforts that began to move the field of medical genetics away from purely descriptive documentation of patients’ physical features coupled with laborious one-by-one examination of a small subset of their genes for potentially pathogenic changes. For example, the International HapMap Project’s collection of millions of genotypes from four global populations was indispensable to the pursuit of hereditary changes in genes that contribute to disease by providing the platform for so called “genome-wide association studies” (GWAS). GWAS gave us the ability to efficiently and comprehensively assay genetic variants that are common in a population and identify those that appear more commonly in patients with a given disease than they do in controls without the disease. Such variants can sometimes provide clues to the genetic basis of human disease (Manolio et al. 2009).
In parallel, researchers have capitalized on our improved understanding of population history to identify disease-causing genes. Population-specific studies of disease, from myocardial infarction in Icelanders to prostate cancer in African-Americans, have cleverly exploited the enrichment of specific disease-susceptibility alleles in more genetically homogeneous populations (Torkamani et al. 2012).
Along the way, advances in our understanding of patterns of human genetic variation have also informed our view of the history of modern human populations. Our interpretation of the scientific data, however, has been influenced by the constantly evolving sociopolitical milieu. During the early part of the 20th century, two schools of thought emerged on how natural selection influenced the frequency and distribution of genetic variation. The “classical” school believed that most genetic variation was rare and that variants present in the population are almost always deleterious. In the very occasional cases in which new mutations are advantageous, they quickly became “fixed” (cases in which the new advantageous allele replaced the ancestral one). The “balanced” school, on the other hand, believed that genetic variation was quite common and often actively maintained by selection favoring multiple forms of a gene in the population. This might be because of so-called overdominance, in which selection favored the heterozygote or other forms of selection-maintaining diversity. In fact, either of these perspectives could readily be, and were, marshaled in support of eugenic perspectives that were common before and after the Second World War. For example, under the classical school, it was easy to postulate a genetic underclass that carried a greater-than-average load of deleterious mutations. Because natural selection pressures can be assumed to have differed among diverse human populations, it was possible to imagine that populations from some geographic regions would have a “superior” complement of variants in terms of key phenotypic characteristics as compared with other geographic regions.
Early “modern” approaches to quantifying biological differences were based on physical measurements that were heavily biased in the ways they were deployed. The distinguishing characteristics used to construct racial classification were those to which human perception is most finely tuned (skin color, eye shape and color, hair color and texture, etc.). Direct, objective methods of quantifying “genetic” variation (as opposed to “physical” characteristics) simply did not exist. Further, physical measurements were typically prone to environmental (nongenetic) influences, blurring the relationship between the measurement and genetic makeup of the individual.
The population characterization of the ABO blood group system by the Hirszfelds in the early 1900s, therefore, was seminal. It provided a biochemical marker that was closely aligned to underlying genetic variation, and, in so doing, provided the first major system for exploring patterns of human genetic diversity in an unbiased manner. Indeed, when tested in soldiers in armies of World War I, the pattern of A and B blood types showed frequency gradients that correlated with the geographic origin of the soldiers (Hirszfeld and Hirszfeld 1919). Soldiers from Western Europe (English and French) had a lower frequency of the “B” blood group, which appeared to gradually increase as one moved east toward Eastern European (Greeks, Turks, Russians) and Asian groups, suggesting that gene frequencies changed gradually across geographically defined populations.
A decisive break with the tradition of assuming sharp genetic divisions among ethnic groups came with the work of Richard Lewontin in 1972. Using multiple different polymorphic genetic markers, comprising blood group systems and serum protein markers that were ascertained in an unbiased manner, Lewontin generated data from more than 100 populations sampled across seven socially constructed “racial” groups (Caucasians, Africans, East Asians, South Asians, Amerindians, Oceanians, and Australians). Lewontin showed that the vast majority of human genetic diversity (∼85%) is caused by individual differences that are shared across “all” populations and races. Only a small percentage (∼15%) was because of differences “between” populations and a smaller percentage, again (∼6%), was caused by differences between “racial” groups (Lewontin 1972). Lewontin’s data suggested that although there is substantial genetic variation within the human population, such variation has accumulated over time; most of this variation appeared before the expansion of Homo sapiens out of Africa and the resulting isolation of populations within continents. Put another way, the considerable genetic differences we see between individuals has very little to do with so-called “racial” boundaries. Rather, it is merely the variation that was present in the original human population that seeded all the current human populations. Therefore, the same polymorphic alleles (genetic variants) are found in most populations, although their frequencies may differ substantially. Broadly speaking, there has been too little time for the accumulation of substantive divergence in a young species such as ours. The fact that the classical model predicted extensive genetic differentiation between populations was explained by the molecular evolution pioneer Motoo Kimura, who hypothesized that most variation was selectively neutral (neither enhancing nor retarding human survival) and, therefore, largely free from the influence of Darwinian selective forces.
At the time of publication, Lewontin’s findings were controversial, but consensus gradually emerged that genetic differences among populations are modest (Nei and Roychoudhury 1972; Cavalli-Sforza et al. 1994). Before Lewontin, the general consensus was that genetic diversity would be structured according to racial labels and, thus, the labels were scientifically justified. The observation that patterns in human genetic variation were largely gradual according to geographic boundaries and not subject to sudden population-specific changes that followed preconceived racial notions removed the biological argument for race (or, we would argue, it should have).
Lewontin illustrated that genetic variation was extensive and largely shared across populations. But, it was not until the sequencing of the first human genome (actually, a consensus of several human genomes) in 2003 that we appreciated just how extensive genetic variation really was in the human genome. Any two randomly selected individuals of European descent will differ at ∼3 million points in their genome, or ∼0.1% of their >3 billion bases of DNA. The fact that most of this variation is, in effect, selectively neutral presents an enormous challenge for characterizing those alleles that contribute to our common diseases in substantive ways. In other words, the challenge is to identify the few trait-altering variants that lie in an ocean of irrelevant ones.
A major breakthrough in this challenge was the development of GWAS. The basic framework used in these studies is to select key variants that inform about virtually all common variations in the human genome. These specially selected variants are often called tagging single-nucleotide polymorphisms (SNPs) because they are near perfect surrogates for variants not directly assayed. These variants could be tested easily by newly developed technologies using specially designed genotype chips. GWAS chips are also relatively inexpensive; one can now genotype a million variants for <$50 a sample. Applied to large studies involving thousands of disease (case) and nondisease (control) individuals, the GWAS approach provided the framework to associate specific genetic variants and their cognate genomic regions with diseases, even if the study design was not well suited to identifying the actual genetically causal variants. The GWAS approach was successful in that it provided much needed momentum in the push to identify disease genes. Nevertheless, in most cases, even when applied to studies involving hundreds of thousands of participants, the approach failed to explain the majority of the presumed genetic component of any given trait (Manolio et al. 2009).
One explanation for this problem of “missing heritability” lies in the fact that the GWAS approach only tests for genetic variants that are common in a population, that is to say, those that Lewontin first observed as shared across individuals and populations. The reason for this is that the research community (through the International HapMap Project) had a good understanding of the nature and extent of common variation; it was, after all, “common” and therefore easy to find and test in large populations. Thus, it was a logical starting point for genome-wide studies. Further, it was not until the development of novel DNA-sequencing techniques in the last few years that the study of rare variants became logistically and financially feasible (Cirulli and Goldstein 2010). As a result, geneticists using GWAS in the late 2000s were akin to the drunken man who would only look for his lost keys under the streetlamp; he looked there because that is where the light was.
THE PATTERN OF GEOGRAPHIC VARIATION FOR COMMON VARIATION MAY BE QUITE DIFFERENT FROM THAT OF VARIANTS INFLUENCING DISEASE RISK AND DRUG RESPONSE
Although most common variants are indeed common among most human populations, it has long been known that rarer gene variants can show markedly different patterns across human groups. Perhaps the best evidence of this comes from the successor to the HapMap Project, the 1000 Genomes Project (The 1000 Genomes Project Consortium 2010), whose goal is to sequence the genomes of a large number of humans to provide a comprehensive survey of human genetic variation (Via et al. 2010). Investigators in the 1000 Genomes Project discovered that 63% of novel variants (that is, those that have never before been observed in humans) are found in African ancestry populations as compared with 33% with European ancestry.
In the same study, several hundred thousand SNPs with large allele-frequency differences were found across geographically distinct populations. Within these variants, there was enrichment for so-called “nonsynonymous” variants, which are characterized by important changes in the DNA sequence that lead to structural and functional changes in the proteins produced by these genes. This observation suggests that local populations adapted to their specific environments and the genetic changes that allowed this to happen were selected for by evolution (The 1000 Genomes Project Consortium 2010). These results also illustrate the fact that Lewontin’s assessment related specifically to common variants because those are the ones most important to overall variation present in an individual. If you look at one individual, most of the variants that individual has are common variants, and those are the ones that follow Lewontin’s pattern; they are mostly derived from the common human ancestral population. But, if the variants that are most important to phenotype variation are more rare, then this assessment that Lewontin provides does not apply to those most responsible for phenotypic variation.
MENDELIAN MUTATIONS ARE HIGHLY POPULATION SPECIFIC FOR A NUMBER OF REASONS
Because the Moravian monk Gregor Mendel was the first one to work out the basic laws of heredity, we refer to diseases within a family that “obviously” follow the rules of inheritance described by Mendel as Mendelian diseases. Typically, these mutations have a major effect on disease risk (and gene function) and relatively few genes can carry mutations that cause a given disease and still allow the organism to survive. Some of the mutations responsible for Mendelian diseases have long been known to show a high degree of population specificity. In some exceptional cases, this is clearly because of positive, as opposed to negative, natural selection. The autosomal recessive disease sickle cell anemia (that is caused by two defective copies of the β hemoglobin gene and, thus, producing a hemoglobin protein with reduced function), for example, is largely restricted to African, Mediterranean, and South Asian ancestry populations. In African-Americans, the allele frequency of the sickle hemoglobin (Hb S) mutation is ∼4% (Ashley-Koch et al. 2000). Why? Because although carrying two mutant Hb S alleles causes the devastating condition sickle cell anemia, carrying a single copy of Hb S does not usually cause health problems. However, it does protect against malarial infection. For this reason, it has been selected for in regions of the world where malaria has been endemic: Africa, the Mediterranean, and South Asia. Its frequency is significantly higher among populations that originate from these regions. In other words, carriers of one defective copy of the hemoglobin gene are at an evolutionary advantage in regions of the world where malaria is common and, therefore, this version of the hemoglobin gene has become more common in those areas (Aidoo et al. 2002).
Carrier advantage is not the principal reason why many Mendelian mutations can be thought of as more or less population specific. Most fundamentally, mutations that have a major impact on risk are rare because of natural selection against them. That means that they have been relatively recently introduced into the population by mutation, and the specific mutations are, therefore, usually geographically quite restricted. This, by itself, would mean that the mutations tend to be very different in different geographic regions, but not the total burden of diseases they cause. In fact, the collective frequency of disease-causing mutations in specific populations at specific genes can be quite different from global averages because small population size and demographic history can also be important.
Consider the Ashkenazi Jews, who are statistically more likely to carry mutations that cause autosomal recessive Tay–Sachs disease in which affected children die at an early age because their mutations deprive them of a particular enzyme. In all likelihood, mutations causing Tay–Sachs increased in frequency during a time when the Ashkenazi Jewish population was small. Perhaps when the Ashkenazim were beginning to establish themselves in Europe during the early Middle Ages, one or more Tay–Sachs mutations arose by chance and the small breeding population led to a “founder effect,” that is, persistence of particular alleles because those alleles were overrepresented when the population in question first emerged. Similarly, perhaps Tay–Sachs mutations were overrepresented after the Ashkenazi Jewish population underwent a “population bottleneck,” that is, experienced a sharp contraction. For example, the European Jewish population declined precipitously following persecution of Jews during the First Crusade in the late 11th century and the subsequent spread of Black Death in the mid-14th century. It may well be that, by chance, Tay–Sachs mutations were present in surviving members of the Ashkenazim following these events and those mutations were, therefore, preferentially transmitted to subsequent generations (Slatkin 2004). Ashkenazi Jews’ historical propensity to preferentially choose mates within their group has also served to keep Tay–Sachs alleles within their community at a relatively high frequency. Today, the carrier frequency of Tay–Sachs disease is on the order of 1 in 30 in self-identified Ashkenazi Jews, 10 times higher than in other populations. Before widespread population carrier screening of this disorder, 1 in 3600 children born to Ashkenazi parents had Tay–Sachs disease (Fernandes Filho and Shapiro 2004; Bray et al. 2010). Screening has since reduced Tay–Sachs births among Ashkenazim by some 90% (Ostrer and Skorecki 2013).
In other cases in which specific genetic diseases appear to be more common in a certain population, it is not clear whether the high frequency of rare disease-causing mutations is caused by chance, selective mating among carriers within the population, carrier selection advantage, or some combination of these factors. Cystic fibrosis, for example, is most common in European ancestry populations. In Caucasians, the frequencies of cystic fibrosis mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene are significantly higher than in other populations and cause the autosomal recessive disease in 1 of 2500 newborns (Ratjen and Doring 2003). Over the years, geneticists have speculated as to why this is the case, often focusing on Darwinian selection as an explanation: perhaps carriers of CFTR mutations were more resistant to cholera and other dehydrating intestinal diseases (Bertranpetit and Calafell 1996). Or perhaps they were more resistant to contracting tuberculosis (Poolman and Galvani 2007). Another hypothesis suggested that carrier frequencies rose in Europe after farmers on the continent began raising dairy cattle, which led to the transmission of various pathogens from livestock to humans, perhaps via cow’s milk (Alfonso-Sanchez et al. 2010). Although these ideas are intriguing, none have been proven to the extent of the implication of malaria as the selective force accounting for the rise of the Hb S allele. A particularly provocative hypothesis was promulgated by Harpending and Cochran that some of the mutations causing Tay–Sachs and other lysosomal storage diseases, several of which also occur at increased frequency in Ashkenazi Jews, were the result of positive selection; the idea was that somehow being a carrier for these diseases was associated with greater intelligence (Cochran et al. 2006). However, there remains no real evidence to support this speculation.
Whatever the reason for the emergence of disease-causing alleles at relatively high frequencies in specific populations, their existence suggests the possibility that rarer variants are also important in common diseases, and there may be more population specificity than anticipated by Lewontin’s analysis of common variation. Consequently, we would do well to pay attention to the population frequencies of various human diseases and traits to better understand their genetic underpinnings.
COMMON VARIATION INFLUENCING DISEASE RISK AND DRUG RESPONSE
Even among common variants, some show relatively greater differentiation (frequency differences) among population groups because of genetic drift or selection, with clinically important consequences even at the level of the population average. One of the most well-known diseases for which common genetic variation affects both the spontaneous clearance of an infectious agent and treatment response is hepatitis C virus (HCV) infection. Treatment response refers to medical treatment with the combination of peginterferon-α (PegIFN-α) and antiviral therapies to induce viral clearance, whereas spontaneous clearance is the automatic viral clearance without exogenous drug administration. It was already well known that African ancestry individuals respond more poorly to HCV drug treatment than Caucasian and Asian individuals. In 2009, GWAS discovered a SNP (also known as rs12979860) in the IL28B locus (abbreviated as IL28B polymorphism below) that is highly associated with patient drug responses to medicines designed to treat HCV (Ge et al. 2009). Allele frequencies of IL28B polymorphism were found to differ largely among these ethnic populations, and explain the differences of treatment success rate among those populations. IL28B encodes interferon-λ-3, which is an important cytokine for innate immunity and one of the first responders to the invasion of foreign pathogens. Some believe the allele frequency of IL28B has been selected among different populations by one or more pathogen and, thus, evolved at different stages of human history. However, the exact natural selection pressure that causes the distinct pattern of allele frequency is unclear. Overall, the discovery of IL28B polymorphism illustrates that the frequency distribution of certain risk alleles is sufficient to affect the disease progression and drug responses. Below, we will discuss in detail how this common variant was discovered and its impact on both treatment-induced and spontaneous HCV clearance.
IL28B DISCOVERY FOR HCV TREATMENT RESPONSES
HCV is a positive-strand RNA virus belonging to the family Flaviviridae. HCV transmission is mainly through blood-to-blood contact and chronic infection usually results in fibrosis, cirrhosis, liver carcinoma, and even liver failure. It is estimated that 170 million people are chronically infected by HCV worldwide, and it is the major cause for liver transplants in the United States. Because HCV has been a serious public health problem in the United States and worldwide, there have been efforts to develop treatments for chronic HCV infection. However, the treatment success rate has been unsatisfactory. PegINF-α combined with ribavirin (RBV) therapy has been widely used to treat chronically infected HCV patients since 2002. The treatment success rate is moderate (from 20% to 70%) and is dependent on a patient’s ancestry. Treatment success is defined as reaching sustained virological response (SVR), when the blood viral load is suppressed below the detectable level for 24 wk after 48 wk of combination treatment (Ghany et al. 2009). In East Asian populations, the PegIFN-α plus RBV treatment for chronically infected HCV patients has been shown to reach 76% of the overall SVR rate, which is dramatically higher than the 56% SVR rate of European-Americans and 24% of African-Americans (Liu et al. 2008; Ge et al. 2009). Before the genetic discovery of IL28B, the reason for the differences observed among major ethnic groups was unclear, and race had been used as a profiling feature to predict HCV treatment response.
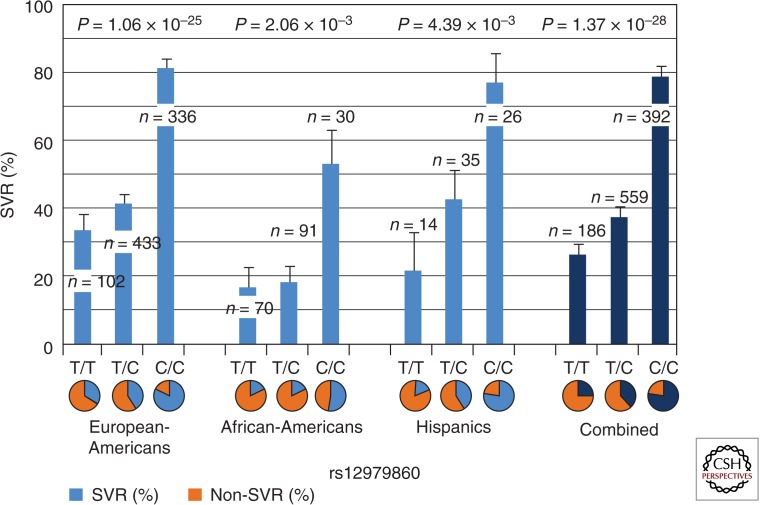
The GWAS performed by Ge and colleagues, as well as studies performed by two other groups, identified a SNP (rs12979860) on the IL28B locus associated with the response of PegIFN-α plus RBV therapy. This genetic variant (rs12979860) is a C-to-T substitution with C being the major allele in Europeans and East Asians. The relative risk for SVR (chance to reach treatment success) is around threefold higher in C/C than non-C/C patients (including C/T and T/T), and is statistically highly significant (Fig. 1). Similar results were also found in several other studies; patients with the homozygous C/C genotype at IL28B generally have a two to three times higher treatment success rate than patients with C/T or T/T genotypes (Ge et al. 2009; Suppiah et al. 2009; Tanaka et al. 2009). European-American patients with C/C genotypes under different treatment regimens show ∼80% SVR, compared with 30% and 40% SVR rates of C/T and T/T genotypes, respectively. In African-Americans, patients with the C/C genotype show ∼50% of the SVR rate compared with <20% of the SVR rate for C/T and T/T patients (Fig. 1). The overall effect of the IL28B polymorphism is, therefore, substantial in predicting HCV treatment response. In general, regardless of ethnicity, the C/C genotype has higher SVR rate than non-C/C genotypes (twofold higher in European-Americans and Hispanics, and threefold in African-Americans). This result suggests that C/C universally favors treatment success versus non-C/C, although in African-Americans, the same C/C genotype shows a lower SVR rate than in European-Americans (50% in African-Americans vs. 80% in European-Americans). The factors that cause this success rate difference in C/C genotype among individuals of different ethnicities are still unclear.
Figure 1.
The SVR% as a function of ancestry and IL28B genotype. Figure based on data from Ge et al. (2009).
IL28B has received a great deal of attention since the GWAS discovery for its ability to predict the pretreatment drug response outcome, and the potential for its biological antiviral activities. Before the GWAS, the reason behind the link between ethnicity and drug responses was elusive, but we now clearly know that the IL28B allele frequencies show very different distributions across populations. Using random controls with unknown hepatitis C status, 90% of the East Asian population carried the IL28B C allele versus 70% in European-Americans. However, in the African-American population, the C allele has become the minor allele (smaller allele frequency) at 40%. Strikingly, according to the study performed by Ge and colleagues (2009), the C allele frequency showed linear correlation with the SVR rate in four distinct populations (Table 1). This concordance strongly suggests that the difference observed in HCV treatment response can be mostly explained by the allele frequency distribution among populations. In a subsequent study by Thomas et al. (2009), 51 geographical subpopulations were examined for the IL28B polymorphism. The results were similar: the C allele frequency was highest in Asian populations, modest in European populations, and the lowest in African ancestry populations. This result showed the IL28B allele frequency distribution in higher resolution and corroborated the initial observations.
Table 1.
Correlation between SVR rate and IL28B C allele frequency
| SVR% | IL28B C allele frequency | Sample size | |
|---|---|---|---|
| African-Americans | 24% | 0.40 | 191 |
| Hispanics | 51% | 0.58 | 75 |
| European-Americans | 56% | 0.63 | 871 |
| East Asians | 76% | 0.95 | 154 |
Linear regression r2 = 0.93. Data adapted from Ge et al. (2009).
This correlation substantially explains the reason why different populations have significantly different treatment success rates. Up until now, the only gene for which there is strong evidence of an influence on HCV treatment response has been IL28B. An extensive search for other genetic factors that might contribute to HCV treatment response has been performed, but no statistically significant result for other genes that modify the effect of IL28B has been found thus far.
The profile based on race to predict treatment success rate in the past is now proven to be overly simplified. It is actually the IL28B genotype that plays a major role in determining treatment response, not ethnicity, and the differences observed among ethnicity can be explained merely by the allele frequency differences among geographic populations. HCV treatment response is a great example of how allele frequency can affect treatment outcomes among populations, and it seems highly likely that there will be other examples like this to be found in the future.
VARIATION OF IL28B ALSO AFFECTS SPONTANEOUS CLEARANCE OF HCV
Spontaneous clearance is the clearance of virus by the immune system without the administration of additional drugs. Based on studies of the natural history of HCV, 20%–30% of infected patients can spontaneously clear the virus, whereas the other 70%–80% become chronically infected and require drug therapy. The spontaneous clearance rate was estimated to be 36% in patients of non-African ancestry and 9% in patients of African ancestry (Thomas et al. 2000). Soon after the discovery of genetic association with treatment response for HCV, IL28B again was shown to be associated with the spontaneous clearance of HCV. Thomas and colleagues examined the IL28B polymorphism in six independent patient cohorts with the diagnosis of HCV infection. Patients were categorized as being chronically infected or having spontaneously cleared HCV by at least two blood tests separated by an interval of at least 6 months. Strikingly, the C allele of IL28B (rs12979860) also favors HCV clearance in these cohorts consisting of both European- and African-Americans. Individuals with the IL28B C/C genotype were, once again, two to three times more likely to clear the virus than the non-C/C patients. This result was similar to what had been observed in drug-induced HCV clearance. This finding suggests that IL28B has a universal effect on HCV resolution in natural settings without the administration of drugs, an important biological clue.
Because there is clear evidence of IL28B association with both treatment-induced and spontaneous viral clearance, it would be intriguing to know whether IL28B is also associated with the geographic distribution of HCV prevalence. However, the prevalence of HCV in major continents and IL28B frequency do not seem to be highly correlated. Although in most African countries, the prevalence rates are >3% of the total population (>3% is considered high prevalence for HCV), many East Asian countries (Asian populations have the highest rate of protective IL28B C allele) also comprise the majority of HCV chronic infections worldwide. For example, there is a 3.2% seroprevalence rate in China, which accounts for a major global HCV-infected population (Shepard et al. 2005). Many believe that country-specific features of the health care system itself may play a major role in determining the likelihood of HCV exposure. For example, the availability of safe injections dramatically decreases the chance of exposure. Nevertheless, because HCV was first discovered in 1989, it has been impossible to obtain actual data of global HCV prevalence before industrialization.
Thanks to advances in tools for human genetic research, GWAS methods provide us with insight on common variants and infectious disease. In many carefully controlled clinical trials for HCV treatments, clear and consistent correlation between treatment success and the presence of the IL28B polymorphism has been shown. The genetic discovery shatters the long-lasting myth that race plays a role in HCV clearance. In fact, most of the difference in SVR rate can be explained solely by the frequency differences of IL28B alleles among populations. HCV infection, therefore, provides a salutary example of how common variation affects disease susceptibility and drug response.
IMPLICATIONS FOR HOW TO THINK ABOUT DISCOVERY AND ITS CLINICAL USE
The relative importance of rare and common variants in the traits that impose the greatest public health burden in the developed world remains unclear. Many believe that most cases of common disease are influenced by variants distributed across many genes, each with small effect, interacting with the environment in ways we do not yet understand. But we also know that sometimes cases of relatively common and certainly complex diseases can be caused by rare genetic changes of large effect. Among the best examples of the latter is neuropsychiatric disease, including conditions such as autism, epilepsy, and schizophrenia, in which rare, large genetic rearrangements (so-called “copy-number” variants) collectively account for a small but significant fraction of cases (Murdoch and State 2013). Another illustrative example of rare variants with large effects is epilepsy. In a recent report on two classical epileptic encephalopathies (infantile spasms and Lennox–Gastaut syndrome), researchers have discovered statistically significant enrichment of de novo mutations, that is, new variants that arise in the germline of the patient’s parents, in specific gene sets. Some of these genes have significantly more de novo mutations in the patient cohort than would be expected by chance. This finding demonstrates that de novo mutations (occurring at one of several different genes) can have a strong influence on the risk of epilepsy (Epi4K Consortium, Epilepsy Phenome/Genome Project 2013).
A related question concerns the proportion of the functional genetic variation that is present in the human population as a result of some form of carrier advantage, as is clearly important in sickle cell anemia, or some sort of mutation-selection balance (in which the arisal of de novo mutations is balanced by their loss because they reduce the fitness of the mutation bearer) as is clearly responsible for the “copy number” variants mentioned above. In the latter, genetic rearrangements can lead to changes in human cognitive potential, but in ways that we cannot yet predict with a high degree of confidence.
Despite the clear similarity of human populations as described by Lewontin, the few examples listed here show that no matter what sort of evolutionary tradeoffs existed in the past of the human species, the genetic bases of medically relevant traits can be profoundly different at both the individual and population levels. The nature of these differences will obviously depend, in large part, on what sort of genetic variation causes most diseases. More generally, we still do not exactly know whether the majority of important variants is generally deleterious and present because of mutation-selection balance, or whether the important ones are more nuanced in their effects in that they are sometimes helpful, sometimes harmful.
It is clear that geneticists cannot assume that aspects of human genetic disease and other medically relevant traits can be understood by studying only one population because human groups are neither homogeneous nor genetic studies fruitful unless they are population comparative. As such, research programs that concentrate their discovery efforts in populations of European ancestry alone, as most genomics efforts have performed to date (Need and Goldstein 2009; Bustamante et al. 2011), are inefficient and incomplete. If we are to fulfill the promise of the Human Genome Project, enhance biological discovery, and begin to bring our knowledge of population genetics to bear on long-standing health disparities, then we must understand and appreciate the enormous range of variation within our species. As the poet Audra Lorde wrote, “It is not our differences that divide us. It is our inability to recognize, accept, and celebrate those differences.”
Footnotes
Editor: Aravinda Chakravarti
Additional Perspectives on Human Variation available at www.perspectivesinmedicine.org
REFERENCES
- Aidoo M, Terlouw DJ, Kolczak MS, McElroy PD, ter Kuile FO, Kariuki S, Nahlen BL, Lal AA, Udhayakumar V 2002. Protective effects of the sickle cell gene against malaria morbidity and mortality. Lancet 359: 1311–1312 [DOI] [PubMed] [Google Scholar]
- Alfonso-Sanchez MA, Perez-Miranda AM, Garcia-Obregon S, Pena JA 2010. An evolutionary approach to the high frequency of the ΔF508 CFTR mutation in European populations. Med Hypotheses 74: 989–992 [DOI] [PubMed] [Google Scholar]
- Ashley-Koch A, Yang Q, Olney RS 2000. Sickle hemoglobin (HbS) allele and sickle cell disease: A HuGE review. Am J Epidemiol 151: 839–845 [DOI] [PubMed] [Google Scholar]
- Bertranpetit J, Calafell F 1996. Genetic and geographical variability in cystic fibrosis: Evolutionary considerations. Ciba Found Symp 197: 97–114; discussion 114–118 [DOI] [PubMed] [Google Scholar]
- Bray SM, Mulle JG, Dodd AF, Pulver AE, Wooding S, Warren ST 2010. Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. Proc Natl Acad Sci 107: 16222–16227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Burchard EG, De la Vega FM 2011. Genomics for the world. Nature 475: 163–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A 1994. The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Cirulli ET, Goldstein DB 2010. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genets 11: 415–425 [DOI] [PubMed] [Google Scholar]
- Cochran G, Hardy J, Harpending H 2006. Natural history of Ashkenazi intelligence. J Biosoc Sci 38: 659–693 [DOI] [PubMed] [Google Scholar]
- Epi4K Consortium, Epilepsy Phenome/Genome Project. 2013. De novo mutations in epileptic encephalopathies. Nature 501: 217–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes Filho JA, Shapiro BE 2004. Tay–Sachs disease. Arch Neurol 61: 1466–1468 [DOI] [PubMed] [Google Scholar]
- Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, et al. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461: 399–401 [DOI] [PubMed] [Google Scholar]
- Ghany MG, Strader DB, Thomas DL, Seeff LB, American Association for the Study of Liver Diseases. 2009. Diagnosis, management, and treatment of hepatitis C: An update. Hepatology 49: 1335–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirszfeld L, Hirszfeld H 1919. Essai d’application des methodes au problem des races [Testing the application of methods to the question of race]. Anthropologie 29: 505–537 [Google Scholar]
- Lewontin R 1972. The apportionment of human diversity. In Evolutionary biology (ed. Dobzhansky T, Hecht M, Steere W), pp. 381–398 Appleton Centuary Crofts, New York [Google Scholar]
- Liu CH, Liu CJ, Lin CL, Liang CC, Hsu SJ, Yang SS, Hsu CS, Tseng TC, Wang CC, Lai MY, et al. 2008. Pegylated interferon-α-2a plus ribavirin for treatment-naive Asian patients with hepatitis C virus genotype 1 infection: A multicenter, randomized controlled trial. Clin Infect Dis 47: 1260–1269 [DOI] [PubMed] [Google Scholar]
- Lorde A 2007. Sister outsider: Essays and speeches. Ten Speed Press, New York [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch JD, State MW 2013. Recent developments in the genetics of autism spectrum disorders. Curr Opin Genet Dev 23: 310–315 [DOI] [PubMed] [Google Scholar]
- Need AC, Goldstein DB 2009. Next generation disparities in human genomics: Concerns and remedies. Trends Genet 25: 489–494 [DOI] [PubMed] [Google Scholar]
- Nei M, Roychoudhury AK 1972. Gene differences between Caucasian, Negro, and Japanese populations. Science 177: 434–436 [DOI] [PubMed] [Google Scholar]
- Ostrer H, Skorecki K 2013. The population genetics of the Jewish people. Hum Genet 132: 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman EM, Galvani AP 2007. Evaluating candidate agents of selective pressure for cystic fibrosis. J R Soc Interface 4: 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratjen F, Doring G 2003. Cystic fibrosis. Lancet 361: 681–689 [DOI] [PubMed] [Google Scholar]
- Shepard CW, Finelli L, Alter MJ 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis 5: 558–567 [DOI] [PubMed] [Google Scholar]
- Slatkin M 2004. A population-genetic test of founder effects and implications for Ashkenazi Jewish diseases. Am J Hum Genet 75: 282–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E, et al. 2009. IL28B is associated with response to chronic hepatitis C interferon-α and ribavirin therapy. Nat Genet 41: 1100–1104 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, et al. 2009. Genome-wide association of IL28B with response to pegylated interferon-α and ribavirin therapy for chronic hepatitis C. Nat Genet 41: 1105–1109 [DOI] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DL, Astemborski J, Rai RM, Anania FA, Schaeffer M, Galai N, Nolt K, Nelson KE, Strathdee SA, Johnson L, et al. 2000. The natural history of hepatitis C virus infection: Host, viral, and environmental factors. JAMA 284: 450–456 [DOI] [PubMed] [Google Scholar]
- Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, et al. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461: 798–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkamani A, Pham P, Libiger O, Bansal V, Zhang G, Scott-Van Zeeland AA, Tewhey R, Topol EJ, Schork NJ 2012. Clinical implications of human population differences in genome-wide rates of functional genotypes. Front Genet 3: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via M, Gignoux C, Burchard EG 2010. The 1000 Genomes Project: New opportunities for research and social challenges. Genome Med 2: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]