Summary
Colorectal cancer (CRC) presents a considerable disease burden worldwide. The human colon is also an anatomical location with the largest number of microbes. It is natural therefore to anticipate a role for microbes, particularly bacteria, in colorectal carcinogenesis. The increasing accessibility of microbial meta’omics is fueling a surge in our understanding of the role that microbes and the microbiota play in CRC. In this review, we will discuss recent insights into contributions of the microbiota to CRC and explore conceptual frameworks for evaluating the role of microbes in cancer causation. We also highlight new findings on candidate CRC-potentiating species and current knowledge gaps. Finally, we explore the roles of microbial metabolism as it relates to bile acids, xenobiotics, and diet in the etiology and therapeutics of CRC.
Introduction
The human large bowel is a common site for adenocarcinomas and also one of the most densely populated microbial ecosystems on our planet. Colorectal cancers (CRC) affect over a quarter of a million people each year. In industrialized nations, the lifetime risk of developing CRC is approximately 5% and the lifetime risk of developing an adenoma, a non-cancerous colon tumor that can develop into CRC, is 20%. When the disease is local or confined cure rates range from 70–90%, however, advanced colorectal cancer has a high mortality rate, consistently ranking in the top 3 causes of cancer-related death around the globe. There has been long standing curiosity about the role of bacteria in colorectal carcinogenesis, because of the large disease burden of CRC and the microbial load of the colon; and recent heightened interest in the gut microbiome in CRC, because of the increasing accessibility of microbial meta’omics.
Sequencing technologies have vastly expanded our understanding of the human genetic landscape of colorectal cancer. Similarly, efforts at sequencing CRC microbiomes are providing leads into how a microbe’s interactions with an individual’s entire colonic microbial community, clades within that community, or the human holobiont (Gordon et al 2013), the entirety of the assemblage of both human and microbe, may be associated with colorectal carcinogenesis. Studies of candidate species in model systems have been useful in evaluating cancer causality and are in keeping with reductionist scientific experimental paradigms. However, an equally plausible concept is that consortia of microbes contribute to colorectal cancer risk over time, which can be a far more challenging concept to observationally or experimentally interrogate. This concept is well-aligned with human genetic-based models of colorectal carcinogenesis, namely that, molecular alterations in multiple genes underlie the development of a hyperplastic epithelium and propel progression onto adenoma and then towards adenocarcinoma. Mutations in human genes that influence adenoma and adenocarcinoma development may shape the growth rate of colonic epithelial cells, reduce their susceptibility to cell death, endow them with metabolic specializations, and confer on them abilities to commandeer immune cells to further promote growth and spread. Similarly, microbes can be viewed as collections of gene networks that affect cancer genomic stability, metabolism, and immune responsiveness. In turn, it is possible that the characteristics of transformed colonic epithelial cells render them more sensitive to microbially-influenced carcinogenesis.
Herein, we will discuss recent insights into contributions of the microbiota to CRC. We explore conceptual frameworks for evaluating the role of microbes in cancer causation as we highlight new findings on candidate CRC-potentiating species and knowledge gaps. We will not summarize how microbially-elicited inflammation or host microbial-sensing pathways affect carcinogenesis as these topics have been the subject of several recent reviews (Dejea et al., 2013; Kostic et al., 2013; Jobin, 2012; Schwabe and Jobin, 2013 and see review by Trinchieri and Goldzmid in this issue). Instead, we concentrate on microbial metabolism in colorectal carcinogenesis with a focus on bile acids and also touch upon xenobiotics and food, all of which are areas where the microbiota has the potential to explicate observations about host gene-environmental interactions in carcinogenesis.
Causality Theory
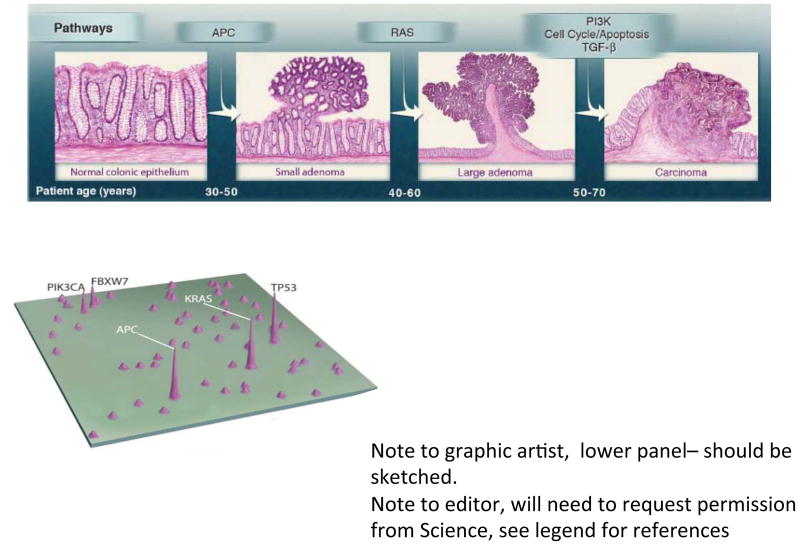
CRC is essentially a genetic disease (Figure 1). Gradual accumulation of oncogenic gene mutations leads to autonomous colonic epithelial cell (CEC) proliferation that slowly progresses typically over 10 to 40 years, resulting in first colon adenomas then, in a minority of individuals, cancers. Initiation of colon tumors (adenomas, adenocarcinomas) refers to events yielding biologic changes fostering CEC proliferation; progression refers to the subsequent events that liberate growth of the incipient colon tumor and, ultimately, transformation to cancer. Yet, it remains unknown with any precision what events precipitate either the initial, disease-initiating mutation(s) or foster the subsequent disease progression. The microbiome, however, is a prime suspect for triggering the initiation and/or progression of colonic carcinogenesis. Certainly, in murine disease models, colon mucosal inflammation, often induced by mucosal irritants (dextran sulfate sodium, 2,4,6-trinitrobenzenesulfonic acid) combined with administration of a carcinogen (often azoxymethane, a compound in engine fuel), yields colon tumorigenesis, substantiating the intersection of inflammation and exposure to carcinogens in colon tumorigenesis. Certain engineered murine gene knockouts, with potential mucosal inflammation sequelae, also yield colon carcinogenesis that is ameliorated in germ-free animals or sometimes merely by a vivarium change, often considered a proxy for acquisition of a new microbiota. While these models support the hypothesis that the microbiota contributes to colon carcinogenesis, they mimic human disease development poorly. Until recently, the contribution of the human colon environment, home to the largest and most complex microbial mass of human ecology, was not integral to the analytical framework of translational CRC research.
Figure 1. Genetic alterations and the progression of CRC (Vogelgram).
The major signaling pathways that drive the development of CRC are shown at the transitions between each tumor stage. One of several driver genes in each signaling pathway can be altered in an individual tumor. Patient age indicates the time interval during which the driver genes are usually mutated. The classic ‘vogelgram’ shown in the upper panel is adapted from Vogelstein et al. (2013). A map of genes mutated in CRC is shown in the lower panel with peak height indicating that a large percentage of human colorectal tumors harbor such mutations (adapted from Wood et al. 2007).
What can guide us as we seek to determine if and how the colon microbiome contributes to the pathogenesis of sporadic human CRC? One clear limitation in seeking a microbe as the cause of a chronic disease is the possibility that the inciting microbe is no longer present at the time the disease is identified, perhaps gradually eliminated by changes in tumor microenvironment no longer hospitable to the microbe or, alternatively, because the microbe acts by a ‘hit and run’ mechanism whereby limited microbial exposure is sufficient to incite disease. For example, in the case of H. pylori and gastric cancer, isolation of H. pylori declines with advancing gastric cancer although detection of H. pylori exposure is usually still possible by serology (Ota H et al., 1998). This reinforces the importance of utilizing multiple approaches in seeking to link a microbe to disease ‘causation’.
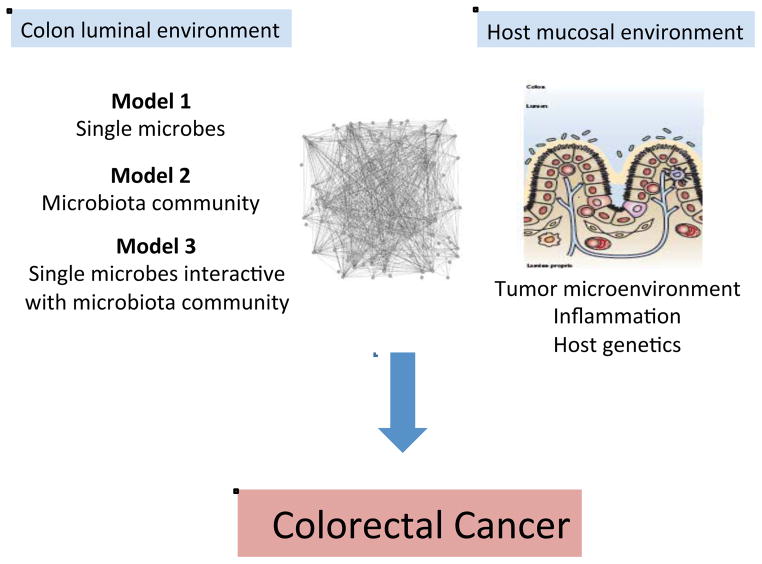
Figure 2 provides a framework for considering the microbiota and specific members of the microbiota as either primary (initiators) or secondary (fostering progression) contributors to human CRC pathogenesis (Sears and Pardoll, 2011). We consider 3 models by which specific microbes (Model 1), a microbial community (Model 2) or the two acting sequentially and/or in synergy (Model 3) influence colon carcinogenesis. With respect to the first model, we well understand that individual microbes such as the pneumococcus, the meningococcus, Helicobacter pylori or hepatitis viruses are established etiologies of host pathology. We consider that these microbial pathogens possess sufficient virulence mechanisms enabling them to act alone in disease causation. In contrast, as a second model, inflammatory bowel disease stands as the best prototype for microbial community disease causation (Sears and Pardoll, 2011). In this disease, host genetics is the presumed initiator permissive to development of a dysbiotic (implying dysfunctional, disease-initiating or -amplifying) microbiome with an ensuing cycle of host gene-microbiota interactions causing intestinal and possibly extraintestinal disease. Experimental work definitively supports this pathogenetic sequence in that dysbiotic colonic microbiota develop in, at least select, mice with gene knockouts (e.g., Tlr5, Il10, T-bet and Rag2) and that this emergent dysbiotic microbiome alone possesses the capacity to transmit to a healthy mouse (without any gene mutations) the disease of interest (Garrett et al., 2007, 2010; Vijay-Kumar et al., 2010). Importantly, these murine experiments strongly support the idea that the dysbiotic microbiota can, as a community, encode tissue-specific (e.g., colitis) as well as systemic disease (e.g., metabolic syndrome, obesity) (Ridaura et al., 2013; Vijay-Kumar et al., 2010).
Figure 2. Microbial contributions to the pathogenesis of colorectal cancer.
Complex microbiota:host interactions are considered probable primary or secondary contributors to the pathogenesis of colorectal cancer. From the microbiota perspective, several hypotheses are actively under investigation including disease instigation or promotion through individual microbes (Model 1), the collective microbiota (Model 2) or an interactive model in which single microbes drive the emergence of a modified, disease-generating microbiota (Model 3). From the host perspective, the microbiota may alter tumor-associated inflammation with consequences for tumor biology or, conversely, the tumor microenvironment or associated inflammation may induce microbiota shifts with the potential to further inhibit or promote tumor biology. Host genetic polymorphisms that modify immune and metabolic responses are predicted to play a key role in host:microbiota interactions during colonic carcinogenesis. See text for details.
Human CRC provides an opportunity to consider individually the above disease causation scenarios as well as a novel hypothesis in which these theories blend with individual or limited bacterial species acting in concert with a locally modified microbiome to cause CRC (Figure 2, Model 3). Nuances of a limited microbial consortium inducing colon tumorigenesis include the possibilities of sequential microbial exposure or polymicrobial disease causation. For this third disease model, we lack clear, clinically relevant historical examples to guide us except for the requirement of the hepatitis B virus for replication and disease induction by the hepatitis D virus. For example, while we understand that the pneumococcus or the meningococcus invade the host from among a complex microbiota in the respiratory tree or nasopharynx, we have no clear data suggesting that the composition of these microbial communities are required for or contribute to the disease causation potential of these bacteria. Similarly, the intestinal microbiota has been shown to hinder or exacerbate viral infections in murine models but the responsible bacteria are not known (Wilks et al., 2013).
It will not be easy to discern among these potential disease models that provide a framework for defining the microbe contributions to human CRC pathogenesis. Carefully designed studies that consider the Bradford-Hill criteria (1965) are needed to link the microbiota and/or select microbes with CRC initiation and progression. A view from the colon lumen may be insufficient. Rather, microbial data must be considered in the context of key host parameters such as the host immunologic response (including the tumor microenvironment) and host gene polymorphisms that influence the host immune response as well as host susceptibility to CEC gene mutations.
The Microbiome Community as Protagonist
The seminal work of Eckburg et al. clarified the complexity of the fecal and mucosal colon microbiota, importantly illustrating two key points relevant to the colon microbiome as causal in colonic carcinogenesis (Eckburg et al. 2005). First, the majority of microbes, predominantly bacteria, within the colon microbial community are ‘non-cultivatible’. While this concept has been challenged by subsequent work, it remains clear that a complete cultured or genome sequence catalog of the colon microbiome with strain-level resolution is still far from our reach (Lagier et al., 2012). Thus, at this time, associations between the microbiota and CRC rely on approaches to broadly define the composition or function of the colon microbiome using various ‘omic’ approaches (16S rRNA gene sequencing, metagenomics, transcriptomics, proteomics, metabolomics). Second, while the colon mucosal community may not vary substantially along the axis of the colon, the mucosa-associated community differs from the intraluminal microbiome. These data raise the important and yet unanswered question of whether the fecal microbiome alone will sufficiently mirror mucosal events to allow ‘causation’ to be established.
The dataset to address microbial community associations in human CRC is limited and has focused on defining bacterial communities associated with colon tumorigenesis. The available studies yield several worthy, albeit preliminary, observations (Dejea et al., 2013). First, the bacterial community composition in colon adenoma or CRC patients, both in mucosal samples and feces, differs from the examined control samples although consistent associations of bacterial groups with tumor samples or tumor hosts is not yet discernible. Second, ‘on tumor’ and ‘off tumor’ mucosa bacterial populations differ within the tumor host. Third, the microbiota of CRC tissue samples and luminal samples differ consistent with prior studies in healthy individuals. Lastly, while considerable study-to-study differences in detected bacterial groups by sample are reported (including nearly polar opposite findings), studies have identified enrichment of Fusobacterium spp. (most often identified as F. nucleatum) associated with CRC mucosa compared to non-tumor, histologically normal colon tissues from the same cancer-bearing host (Castellarin et al., 2012; Kostic et al., 2012). Additional work has further identified enrichment of Fusobacterium spp. in fecal samples from CRC hosts compared to healthy individuals (Ahn et al., 2013; Kostic et al., 2013). These observations are discussed in more detail below. Several of the available colon tumor microbiome studies have limitations including small sample sizes, undefined tissue sampling sites, limited or absence of control samples (including healthy controls and/or control tissues within the tumor host), limited or no metadata (subject clinical data, e.g. patient age, family history, past medical history, current and past medications, dietary patterns, cancer stage, or tumor KRAS, PIK3CA, BRAF, NRAS mutational status) and/or poorly described analyses. Differing study designs including types of samples analyzed and populations included combined with, for example, unknown dietary habits or other risk factors for CRC that are associated in other datasets with modified colon microbiomes, make it difficult to glean themes across these studies.
When considered within the Bradford-Hill framework for establishing causality, our knowledge of the community microbial associations in CRC remains quite limited. For example, as yet, no study has examined in parallel fecal and mucosal samples from non-tumor hosts compared to the fecal and mucosal communities present in colon tumor hosts (‘on-tumor’ and ‘off-tumor’). This approach might help us discern specific microbial associations and could provide the basis for prospective testing for disease identification. Prospective, longitudinal study of individuals at high risk for development of CRC will need to be done to discern if shifts of the colon microbiome occur coincident with disease development consistent with the epidemiologic principle that exposure must antecede disease expression. Serologic, proteomic or metabolomic studies of human CRC and controls to identify potential fecal, mucosal or serum colon tumor specific molecular markers for disease prediction that might simplify screening for this highly preventable disease are also outstanding. Further, although the right and left colonic mucosal communities appear similar in the healthy host, the molecular features of right and left CRC can differ (Yamauchi et al., 2012), raising the question of whether regional microbial associations may distinctly affect the genesis of colon tumors, a point not yet addressed by available data. Lastly, there is uncertainty about the population sizes required to identify the ‘correct’ causal associations, if these exist. For example, the population size to detect a single microbe as causal will likely be much smaller than if a particular community exposure or polymicrobial, even sequential, exposure is critical. Certainly, large populations were necessary to identify the effect of KRAS mutations, aspirin or diet on colorectal carcinogenesis.
Individual Microbes as Protagonists
There is a long history of attempting to associate individual microbes, mostly bacteria, with human CRC (Aries et al., 1969; Hill et al., 1971). As examples to guide our thinking, herein we discuss data on select bacteria that may serve as protagonists of human CRC. Table 1 summarizes how the data on colon carcinogenesis promoted by these individual bacteria align with criteria supportive of disease causality as classified by Evans AS (1976) and Fredricks DA and Relman DA (1996).
Table 1.
Criteria for Disease Causation: Human Colorectal Cancer & Putative Bacterial Protagonists
| Criteria1 | S. gallolyticus | ETBF | E. faecalis | E. coli | F. nucleatum |
|---|---|---|---|---|---|
| Epidemiology2 | + | + | − | + | + |
| Measurable immunological responses3 | + | − | − | − | − |
| Experimental disease reproduction4 | − | + | + | +5 | + |
| Biological plausability6 | +/− | + | +/− | + | + |
| Elimination or modification of agent prevents disease7 | − | − | − | − | − |
Presence or absence of data is noted by + (present) or − (absent); +/− denotes overall data is variable.
Adapted from Evans AS 1976 and Fredricks DN and Relman DA, 1996.
Epidemiology encompasses several types of evidence including prevalence, exposure or incidence of disease significantly higher in those exposed to the putative cause than controls; data comparing cases and controls should show consistency and strength of association; a range of controls should be evaluated to assess specificity of the epidemiologic association; temporality (exposure antedates disease development)
Only data assessing human immunologic responses are considered.
Experimental disease induction refers to animal models demonstrating increased colon carcinogenesis by the listed bacterium.
Experimental model data are only available for E. coli possessing the pks island
Biologic plausibility reflects the authors’ judgment of the strength of the data available at present regarding the potential role of the bacterial protagonist in human colorectal cancer.
Elimination or modification of agent prevents disease refers to human studies such as use of antibiotics, probiotics or vaccines to prevent disease. As yet, no such studies are reported for these bacteria and CRC.
Streptococcus gallolyticus
Identification of S. gallolyticus subspecies gallolyticus (the former S. bovis, biotype I) in the bloodstream has been a strong and consistent predictor of colon pathology, often colon adenomas or cancers, for nearly 40 years (Boleij and Tjalsma, 2013; Boleij et al., 2011b). In fact, positive blood cultures for S. gallolyticus mandate clinical examination of the colon for pathology. While co-occurrence of S. gallolyticus bloodstream infection and colon neoplasia is infrequent, estimated at less than 1%, molecular approaches have detected S. gallolyticus DNA in ~20–50% of colon tumor or non-tumor colon mucosal tissues from tumor hosts compared to <5% of control tissues (Abdulamir et al., 2010). Despite evidence of S. gallolyticus colonization in colon tumor hosts, it is uncertain whether S. gallolyticus is mere opportunist invader in the setting of a breach in the colon mucosal barrier or if it contributes, through specific virulence mechanisms, to transformation in the colon. S. gallolyticus, compared to other members of the S. bovis complex, possesses a pilus protein (encoded by the pil1 locus) with a collagen-binding domain, exhibits a growth advantage under metabolic conditions associated with colon tumors, can translocate efficiently through a paracellular epithelial route and is associated with enhanced inflammatory signals including Ptgs2 (COX-2) (Abdulamir et al., 2010; Boleij et al., 2012; 2011a). These observations suggest that S. gallolyticus is poised to colonize and invade colon tumors and may enhance tumor growth through inflammatory signals. In vivo detection studies as above and serologic assays suggest that exposure to S. gallolyticus does not characterize the majority of individuals with CRC (Boleij and Tjalsma, 2013). Thus, the S. gallolyticus contribution to the pathogenesis of human CRC is likely restricted to a subset of individuals, in whom it may enhance tumor growth.
Enterococcus faecalis
E. faecalis strains differ in their capacity to produce radical oxygen species capable of inducing DNA damage and genomic instability and those E. faecalis strains producing extracellular superoxide anions have been proposed as initiators of CRC. Experimental studies using an oral commensal E. faecalis strain suggest that E. faecalis-triggered carcinogenesis is mediated by inducing mucosal macrophages to produce diffusible clastogens (chromosomal-breaking factors) such as 4-hydroxy-2-nonenal (a breakdown product of ω-6 polyunsatured fatty acids) that mediate DNA damage through a bystander effect (Wang et al., 2008; 2012; Yang et al., 2013). The potential oncogenicity of certain E. faecalis strains is further supported by observations that superoxide-producing E. faecalis induce marked distal colitis, DNA damage and cancer in germ-free Il10−/− mice whereas superoxide-deficient E. faecalis induce inflammation but not tumor formation (Wang et al., 2012). Human data testing these concepts are limited to a prospective case cohort study of consecutive colonoscopy patients in which fecal E. faecalis populations were identified as unstable over >1 year and an association of superoxide-producing E. faecalis with detection of large colon adenomas or cancer was not found (Winters et al., 1998).
Enterotoxigenic Bacteroides fragilis (ETBF)
ETBF, a cause of human diarrheal illnesses, was initially proposed as a potential microbial instigator of human CRC based on studies of the mechanism of action of the organism’s only recognized virulence factor, the B. fragilis toxin (BFT; also known as fragilysin). BFT, a zinc-dependent metalloprotease toxin, rapidly alters CEC structure and function including cleavage of the tumor suppressor protein, E-cadherin, resulting in enhanced nuclear Wnt/β–catenin signaling that yields increased colonic carcinoma cell proliferation and expression of the protooncogene, MYC. Loss of E-cadherin increases the permeability of polarized CEC monolayers and an increase in colonic permeability prior to tumor development is an early pathophysiologic change associated with incipient colorectal cancer (Soler et al., 1999). BFT also triggers NF-κB signaling that induces CEC secretion of cytokines that likely contribute to the development of mucosal inflammation; further NF-κB signaling may contribute to CEC carcinogenesis. Collectively, these in vitro studies of the mechanism of action of BFT suggested the toxin, and hence the organism, may be oncogenic (Sears, 2009).
This hypothesis was tested using multiple intestinal neoplasia (ApcMin/+) mice that are heterozygous for the Apc gene and considered a classic model for CRC pathogenesis given that Apc mutations are present in most human colorectal cancers. In ApcMin/+ mice, persistent colonization with ETBF (a piglet isolate) markedly and rapidly increased colon adenoma formation (Wu et al., 2009). ETBF accelerated both the time course and altered the distribution of colon tumor formation in ApcMin/+ mice. By histology, colon microadenomas were detectable as early as 1–2 weeks after colonization and visible colon tumors by one month post-colonization, a time line that is markedly accelerated compared to ApcMin/+ mice not colonized with ETBF. Further, the majority of adenomas in ApcMin/+ mice are detected in the small bowel with limited adenoma formation scattered through the colon. Upon ETBF colonization, distal colon tumorigenesis, but not small bowel adenoma formation, is augmented. Mechanistically, specific Stat3 activation with induction of a mucosal IL17 response was shown to mediate, at least in part, ETBF colon carcinogenesis, the first demonstration of the contribution of Th17 adaptive immune responses to carcinogenesis. Subsequently, development of an IL17 immune response was linked to a worse prognosis in human CRC (Tosolini et al., 2011). ETBF in vivo and BFT in vitro also induce DNA damage in CECs (Goodwin et al., 2011). Thus, the in vitro and in vivo biologic basis for considering ETBF as a potential contributor to colon carcinogenesis is strong.
In contrast, human data to directly link ETBF to CRC is currently limited although the framework for considering ETBF as an instigator of CRC is more promising. Exposure to ETBF begins in early childhood where this bacterium was first associated with human diarrheal disease and ETBF acquisition appears to be relatively common, at least, in some locales (Sears, 2009). Subsequent data suggest that asymptomatic adult colonization with ETBF is also common, occurring in up to 40% of individuals (Nitomersky et al., 2011). However, whether ETBF is a persistent colon colonizer throughout life and/or is associated with subclinical, potentially CRC-promoting, colonic inflammation in humans remains unknown. Consistent with this idea, a persistent, subclinical IL17-dominant colitis is identified in mice with long-term ETBF colonization (Wick et al., in press). Further, chronic ETBF colonization is associated with subclinical, inflammatory foci in which coincident Stat3 activation in inflammatory cells and overlying CECs occurs. These murine data suggest that long-term ETBF colonization may yield foci at risk for CEC transformation. Overall, the data suggest that ETBF may exhibit the correct temporal relationship for consideration as an instigator of CRC, a disease estimated to require 10 to 40 years from inception to clinical detection. A single study from Turkey has examined ETBF in a CRC population so far (Toprak et al., 2006). Using bft as the marker, ETBF was detected significantly more often in the stools of consecutive cases of CRC compared to concurrent hospital-based, age-and gender-matched patients without CRC (38%, ETBF in 73 cases of CRC and 12%, ETBF in 59 controls, P<0.01). Importantly, B. fragilis was isolated in similar percentages of both CRC cases and controls. Together in vitro and in vivo experimental models and early human data support the concept that ETBF may act to promote colon carcinogenesis.
Escherichia coli
One of the earliest studies to employ molecular methods to examine the microbiota of adenomas, CRCs and healthy colonoscopy control biopsies identified the versatile E. coli as disproportionately associated with tumor samples. Using the classic gentamicin protection assay, intracellular E. coli could be recovered from 81% of 16 colon tumor (adenoma or cancer) samples examined compared to none of 25 control biopsies (Swidsinski et al., 1998). Subsequently, two groups of E. coli have been of particular interest with respect to the pathogenesis of CRC, genotoxic E. coli and tightly adherent E. coli. Among potential genotoxic E. coli, phylogenetic group B2 E. coli induce double-strand DNA breaks through the polyketide synthase (pks) island containing the genotoxin, colibactin (Cuevas-Ramos et al., 2010; Nougayrède et al., 2006). The concept that E. coli through the pks island and colibactin promotes colon carcinogenesis was strengthened by data revealing that deletion of the pks island reduced DNA damage, tumor numbers and bacterial invasion, but not inflammation, in a murine colonic oncogenesis model (Il10−/− mice treated with azoxymethane) colonized with a murine E. coli strain possessing the pks island (Arthur et al., 2012). Thus, carcinogenesis in this model required inflammation plus a specific bacterial virulence factor, a concept further enhanced by observations that a strain of E. faecalis also induced inflammation but not colon tumorigenesis in this murine model. However, other data indicate that the type of inflammation is critical in facilitating carcinogenic biologic events with Th1 and Th17 inflammatory responses generally being anti- and pro-carcinogenic, respectively (Yu et al., 2007). The character of the colonic inflammation induced by E. coli possessing or not the pks island was not determined in this model and, thus, it is possible that deletion of the pks island resulted in a shift in the inflammatory environment from pro-carcinogenic to anti-carcinogenic. Limited human data suggest that colibactin-positive E. coli are more common in CRC and inflammatory bowel disease patients and even are identified in fecal samples from infants where persistent colonization, at least to 18 months of age, is frequent (Arthur et al., 2012; Buc et al., 2013; Nowrouzian and Oswald, 2012). Other E. coli with defined (e.g., cytotoxic necrotizing factor or cytolethal distending toxin) or undefined genotoxic factors have also been isolated from the colonic mucosa although there is little data on their associations with human CRC (Buc et al., 2013). Enteropathogenic E. coli (EPEC) that exhibit characteristic tight intestinal epithelial cell adherence (also known as attaching and effacing lesions) and are well-known as a common cause of acute and persistent diarrhea in children have also been proposed as potentially carcinogenic via downregulation of DNA mismatch repair proteins (Maddocks et al., 2009). EPEC were detected in 25% of 20 formalin-fixed adenocarcinoma tissues but not in normal colon tissues from the same individuals. Overall, E. coli possessing the pks island are the current strongest E. coli candidate for being a contributor to colon carcinogenesis.
Fusobacterium spp
Fusobacterium spp. initially arose as potential bacteria contributing to the pathogenesis of CRC through complementary unsupervised genomic methods analyzing the microbial associations of CRCs versus matched normal tissues from the same host. Metagenomic, 16S rDNA and RNA-sequence analyses supported an enrichment of Fusobacterium sequences associated with tumor samples relative to the normal colon tissue from the same cancer-bearing host (Castellarin et al., 2012; Kostic et al., 2012). The results were further supported by visualization of excess Fusobacterium by fluorescent in situ hybridization (FISH) on tumors compared to parallel host normal colon tissue and quantitative PCR (qPCR). F. nucleatum appeared to be the dominant phylotype although multiple species were detected. A single cancer-associated F. nucleatum isolate was invasive in tissue culture, consistent with the known clinical spectrum of fusobacteria that act as invasive anaerobes in oral and endometrial infections as well as appendicitis and inflammatory bowel disease (Castellarin et al., 2012; Strauss et al., 2011). Subsequent work further confirmed the tumor-association of Fusobacterium spp. as well as identified fusobacteria as more abundant in the normal rectal mucosa of adenoma patients compared to non-adenoma controls; more abundant in adenoma tissue than patient-matched normal colon tissues; and more abundant by qPCR in the stool of patients with adenomas or adenocarcinomas compared to the stools of healthy individuals without colon tumors (Ahn et al., 2013; Kostic et al., 2013; McCoy et al., 2013; Warren et al. 2013). Although Fusobacterium was not associated with particular tumor characteristics, it was more abundant on colon tumors from Spain when compared to tumors from the United States and Vietnam. This suggests that Fusobacterium colonization may vary regionally although the reasons for this such as diet, for example, are unknown (Kostic et al., 2012).
Recent data provide experimental support for a tumor-inducing role of F. nucleatum. Chronic exposure of ApcMin/+ mice to a F. nucleatum strain isolated from an inflammatory bowel disease patient induced a modest, but significant, increase in colon adenomas as well as small bowel tumors. Again excess F. nucleatum was detected on the tumor compared to normal murine colonic tissues (Kostic et al., 2013). In contrast to most murine models of intestinal carcinogenesis, mucosal inflammation was not detected in non-tumorous colon tissue of the F. nucleatum-infected ApcMin/+ mouse. However, F. nucleatum induced an expansion of myeloid-derived immune cells in the tumor microenvironment of small bowel tumors as well as upregulated inflammatory genes in both small intestinal and colon tumors. These results were further supported by a correlation between the abundance of Fusobacterium, but not other bacterial genera, and NF-κB p65 nuclear translocation as well as expression of myeloid-associated and NF-κB-driven inflammatory genes in human CRCs.
In experimental work using a periodontal disease-derived F. nucleatum strain, the invasive and carcinogenic properties of F. nucleatum were suggested to be mediated by the activated complex of the FadA adhesin (FadAc) of F. nucleatum (Rubinstein et al., 2013). In vitro colon carcinoma cell line studies and tumor xenograft models revealed that FadAc binds to a select extracellular domain of E-cadherin triggering invasion of the organism as well as activation of β-catenin/Wnt signaling with stimulation of cell proliferation or tumor growth, respectively. Of note, E-cadherin binding appeared to be sufficient to trigger β-catenin/Wnt signaling with oncogene transcription but internalization of E-cadherin was required for activation of additional NF-κB signaling. Evaluations of tumor tissues from adenoma and adenocarcinoma patients compared to normal colon tissue from non-tumorous individuals revealed fadA gene copy number was elevated in tumor tissues. The highest fadA gene copies were detected in cancer tissues in association with increased fadA transcripts and concomitant increases in expression of representative Wnt and NF-kB genes consistent with the results of in vitro FadAc studies.
Collectively, F. nucleatum and possibly other Fusobacterium spp. are more abundant in some CRC-bearing hosts, found particularly in association with tumor tissues, and experimental data provide support for carcinogenesis being mediated through Wnt signaling with coincident skewing towards myeloid-derived and NF-κB inflammation in the tumor microenvironment. The data also reveal that there is a gradient in the fecal abundance of F. nucleatum across healthy and tumor hosts suggesting that Fusobacterium detection, in and of itself, may not be a sufficiently robust biomarker for identifying patients at increased risk for colon tumors (Kostic et al., 2013). Similarly, and consistent with data on other individual bacterial CRC protagonists, only a subset of tumors display enhanced abundance of Fusobacterium and the relative abundance differs markedly between tumors [e.g., 2-fold to >10,000-fold compared to normal tissues within the tumor host, (Castellarin et al., 2012)]. Data to support a correlation between Fusobacterium abundance and tumor oncogenic properties or disease stage remains outstanding. The epidemiology of acquisition or colonic colonization of Fusobacterium spp. is also unknown. In particular, whether the oral fusobacteria commonly associated with periodontal disease are in fact related to the Fusobacterium spp. detected in the colon requires further investigation.
Overall, two common themes emerge regarding the mechanisms by which individual bacteria may contribute to human CRC pathogenesis. The first theme, based on studies on ETBF, E. faecalis, E. coli and F. nucleatum suggest that members of the colonic microbial community capable of triggering Wnt signaling and/or select types of inflammation may be human CRC protagonists. This concept is further supported by the near universal detection of gene mutations promoting Wnt signaling in human CRC and our understanding that all CRC exhibits enhanced inflammatory tone with evolving human data suggesting that the features of this inflammatory response are linked to disease prognosis. The second theme supported by data on ETBF, E. faecalis and E. coli is that bacterial members of the colon microbiota capable of inducing DNA damage and/or of interfering with DNA repair processes may be critical to tumor initiation in the colon. Further, it is easy to speculate, based on an understanding of bacterial pathogenesis, that the capacity to breach the colonic mucus layer and persistently adhere to the colonic mucosa is necessary for members of the microbiota to initiate oncogenic CEC signaling and/or to deliver specific oncogenic virulence proteins or molecules in the colon. In this context, the early CEC barrier changes associated with CRC (Soler et al., 1999 and Grivennikov et al., 2012) likely enhance the uptake of bacterial molecules contributing to inflammatory signals and colon carcinogenesis.
A Consortium as Protagonist
The concept of a keystone species was introduced into environmental science in 1969 and is defined as a species that plays a critical role in maintaining the structure of an ecological community and whose impact on the community is greater than would be expected based on its relative abundance or total biomass (http://www.washington.edu/research/pathbreakers/1969g.html). Consistent with this idea, a number of studies seeking to catalog and categorize the complex microbiota of humans have suggested that minority microbiota members may be prime contributors to microbiota function; in some cases, single species or a limited set of species may serve to distinguish the populations under study (Arumugam et al., 2011; Koren et al., 2013; Qin et al., 2012). Further, the field of microbial communication through quorum sensing and secretion of hormones or anti-bacterial factors such as bacteriocins strongly suggests that individual bacteria possess the capacity to modify, if not commandeer, their nearby microbial community. Given these data and the complexity of the colonic microbiota, the concept of a microbial leader that recruits a consortium of disease-facilitating microbes to initiate the biologic events causing CRC is appealing (Hajishengallis et al., 2012; Sears and Pardoll, 2011; Tjalsma et al., 2012). After all, given the complexity of the colonic microbiota, could all but one microbe be simply passengers not contributing to disease development? Despite its appeal, as yet, no study has specifically tested this idea in the causation of CRC and, thus, this is an area where carefully designed experimental work might help further our concepts of how the microbiota contribute to the pathogenesis of colon carcinogenesis.
Over the decades it takes for human colorectal cancers to develop, several microbial actors may be primary or secondary contributors. Their biomolecular activities in relation to host physiology and in response to a host’s diet or ingested pharmaceuticals may be the factors that explicate environment and human gene interactions in cancer causation and offer up new cancer biomarkers and therapeutic targets. While well-designed microbial discovery efforts are still warranted in human CRC, preclinical models and other approaches which consider the contribution of microbial metabolism to cancer prevention, development, and treatment merit further investigation. Given our evolving understanding of the microbiome, longstanding clinical observations, such as the connections between bile acids or dietary components and gastrointestinal cancers, should be re-examined with a fresh perspective. With this in mind, we will explore select aspects of microbial metabolism in colorectal carcinogenesis and treatment that are more speculative in terms of their mechanistic roles, but represent areas warranting reexamination and further investigation.
High fat diet and CRC: a microbial link?
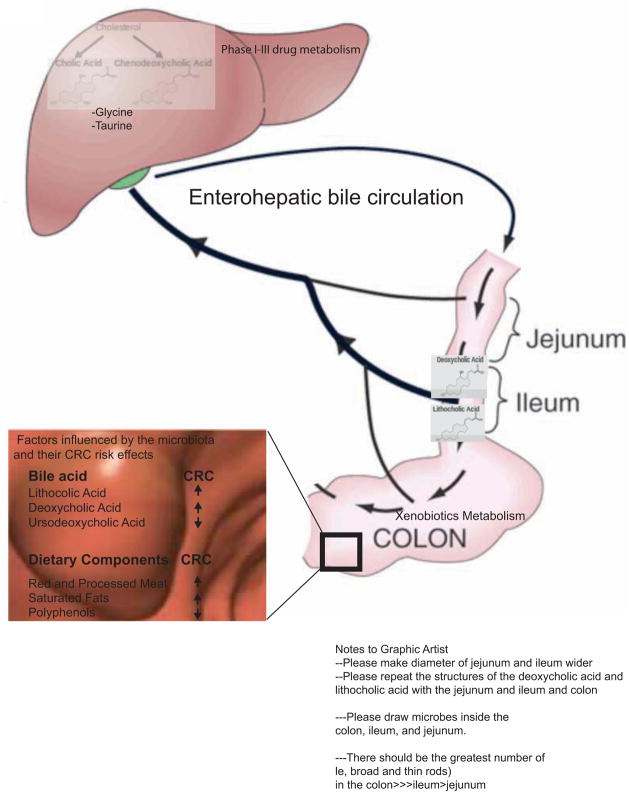
Diets rich in saturated fats increase bile acid production, and numerous studies have identified associations between diets high in saturated fats and CRC (Williams et al., 2010 and Reddy 2002). Gut bacteria are important contributors to bile acid metabolism and thus may play a role in the biology linking bile acids to CRC. As illustrated in Figure 3, there is significant interplay between host and microbe in bile acid metabolism. The liver secretes glycine and taurine conjugates of two bile acids, cholic and chenodeoxycholic acid. Conjugated bile acids can be deconjugated by bacteria to produce the secondary bile acids, lithocholic and deoxycholic acid, the two dominant fecal bile acids.
Figure 3. Host and microbial metabolism affect CRC risk.
Host and microbiota co-metabolism influence colonic bile pool exposure, drug metabolism, and the breakdown of ingested foodstuffs with significant consequences for CRC. The enterohepatic circulation of bile acids is shown from the generation of the primary bile acids from cholesterol in the liver to the generation of secondary bile acids by the intestinal microbiota. Bile acids linked to decreased or increased colorectal cancer risk are highlighted. Both the liver and the gut microbiota play critical roles in drug metabolism with significant effects on drug toxicity and response in CRC. Specific dietary components have been implicated in increasing or decreasing CRC risk, e.g. saturated fats, red and processed meats, and polyphenols. An individual’s gut microbial metabolism may play a role in the beneficial or detrimental effects of certain foods.
Both deoxycholic acid and lithocholic acid may contribute to carcinogenesis. In studies of dietary fat intake and CRC risk, elevated fecal lithocholic and deoxycholic acid have been found in CRC patients relative to healthy controls (Gill and Rowland, 2002). Vancomycin-based perturbations of the gut microbiota support that bacteria are key drivers of high-fat diet-mediated increases in deoxycholic acid in mice (Yoshimoto et al., 2013). Both lithocholic acid and deoxycholic acid function can enhance mutagenesis in classic mutagenesis test, like the Ames test, (Kawalek et al., 1983; Shibuya et al., 1997). In rodent models of colorectal carcinogenesis employing chemical carcinogens, deoxycholic acid and lithocholic acid potentiated tumor number and invasivity, when their endogenous levels were altered by surgery, high-fat diet, or by delivery of supraphysiologic doses to the gastrointestinal tract (Reddy, 1975; Summerton et al., 1985). Focusing on carcinogen-induced liver cancer in mice, Yoshimoto et al. found that a high-fat diet or genetic susceptibility to obesity resulted in deoxycholic acid-mediated induction of the senescence-associated secretory phenotype (secretion of pro-inflammatory and tumor permissive factors by fibroblast-type cells) in hepatic stellate cells and potentiated the development of liver cancer. Notably, perturbing gut bacterial-mediated deoxycholic acid production was sufficient to blunt the development of liver cancer in this model.
Numerous mechanisms underpinning lithocholic and deoxycholic role in carcinogenesis are likely at play. Both lithocholic and deoxycholic acid can be pro-inflammatory in that they can elicit reactive oxygen and nitrogen species production and NF-κB activation in intestinal epithelial cells (Da Silva et al., 2012; Lee et al., 2004; Mühlbauer et al., 2004; Payne et al., 2007). Chronic exposure to deoxycholic acid in vitro results in DNA adducts as well as enhanced epithelial cell proliferation and decreased apoptosis (Barrasa et al., 2013). Thus, lithocholic and deoxycholic acid may represent pro-carcinogenic bacterial metabolites and promising therapeutic targets.
Devkota et al., identified a mechanism that may explain the observations about diets high in saturated fats and escalations in inflammatory diseases in industrialized nations (Devkota et al., 2012). Using Il10−/− mice, the investigators found that a diet enriched in milk-derived saturated fat versus polyunsaturated-fat altered the composition and function of the gut microbiota. A milk fat-enriched diet enhanced taurine (2-aminoethanesulfonic acid) conjugation of primary bile acids and resulted in a bloom of the sulfur-reducing bacterium Bilophila wadsworthia, which in turn exacerbated colitis in inflammation-prone Il10−/− mice. A recent human feeding intervention study also supports that a high-fat diet may result in blooms in B. wadsworthia (David et al., 2013). Examining the intersections between dietary fat intake, gut microbiota composition and function, and both primary and secondary bile acids may hold promise for not only mitigating inflammatory disease like colitis but also for attenuating the smoldering inflammation that contributes to sporadic CRC.
Beyond dietary modification, there are other aspects of bile acid microbial metabolism that may offer opportunities for cancer prevention. In contrast to lithocholic and deoxycholic acid, ursodeoxycholic acid has a promising safety and health benefit profile in humans and CRC7 pre-clinical models. Two enzymes, α-hydroxysteroid dehydrogenase and 7β-hydroxysteroid dehydrogenase, catalyze the transformation of chenodeoxycholic acid to ursodeoxycholic acid. In vitro many human intestinal bacteria, including Clostridium, Ruminococcus, and Eubacterium strains, can convert chenodeoxycholic acid to ursodeoxycholic acid. However, it is important that a given strain not also express 7β-dehydroxylase, which would convert the ursodeoxycholic acid to the potentially toxic lithocholic acid. Ursodeoxycholic acid is an approved therapy for primary biliary cirrhosis and is generally well-tolerated. A few small, retrospective studies examined if ursodeocycholic acid was useful for CRC prevention in patients with a history of adenomas and inflammatory bowel diseases, however none of these studies were sufficiently powered to demonstrate efficacy (Carey and Lindor, 2012; Serfaty, 2012). Screening, selection, and identification of human intestinal bacteria that can generate ursodeoxycholic acid from chenodeoxycholate in the small intestine could represent a microbe-based approach for optimizing bile acid metabolism for CRC prevention (Lepercq et al., 2004).
Xenobiotic Metabolism, the Gut Microbiota, and Colon Cancer
Many compounds from pharmaceuticals to chemical carcinogens, collectively referred to as xenobiotics, are metabolized in the liver and undergo further metabolism by the gut microbiota (see Figure 3). The liver tends to engage in metabolism via oxidation and conjugation reactions while the gut microbiota favor reduction and hydrolytic processes (Sousa et al., 2008). In the liver some compounds are conjugated to glucuronic acid, which makes them more water-soluble and facilitates their excretion in the urine and feces. Colonic bacterial β-glucuronidases can hydrolyze these conjugates, resulting in their release within the colonic lumen. This joint host and microbe co-metabolism is necessary for the chemical carcinogen azoxymethane to induce intestinal tumors in mice (Fiala, 1977; Takada et al., 1982) and can also contribute to the toxicity of many over-the-counter and prescription drugs (Saitta et al., 2013).
Irinotecan, a commonly used intravenously-delivered CRC chemotherapy, is a topoisomerase-1 inhibitor. Like many chemotherapies, irinotecan can cause both immune suppression and diarrhea. However, in some patients, irinotecan can cause a severe and refractory diarrhea that requires hospitalization and limits the drug’s subsequent dosing and usage. Irinotecan is a prodrug and carboxylesterases, present in the serum and throughout the body, convert it to SN-38. SN-38 is a topoisomerase-1 inhibitor 1000X more potent that irinotecan, and it is glucuronidated in the liver (Kawato et al., 1991). However, within the intestinal lumen, bacterial β-glucuronidase can liberate SN-38 (Roberts et al., 2013). Thus the levels of intestinal bacterial β-glucuronidase and subsequent degree of intestinal epithelial SN-38 exposure influence the drug toxicity for patients. In an elegant study, the Redinbo laboratory identified a potent β-glucuronidase inhibitor that alleviated irinotecan toxicity in mice (Wallace et al., 2010) and in subsequent studies they have identified a number of compounds that are both microbially-selective and have EC50s in the nanomolar range (Roberts et al., 2013). The identification of compounds that can improve drug efficacy and reduce toxicity represents an exciting direction for microbiota-based oncology therapeutics.
The gut microbiota may also contribute to chemotherapy responsiveness via its influence on immune system function. Recently Trinchieri and Goldzmid examined how oxaliplatinum, a drug used to treat CRC, depends on the gut microbiota-immune system interactions for the host to reap full benefits of its anti-cancer effects (Iida et al., 2013 and see review by Trinchieri and Goldzmid in this issue). While chronic exposure to reactive oxygen species can increase the risk of developing CRC, tumor exposure to reactive oxygen species coincident with exposure to DNA adduct-forming platinum-based chemotherapies can be the death knell for cancer cells. Iida et al. found that the gut microbiota influence the expression of several enzymes that myeloid cells require to make reactive oxygen species in the tumor microenvironment. These reactive oxygen species were important for optimal response to specific chemotherapies. In mice, the microbiota’s influence on the immune system appears not only important for homeostatic regulation but also for response to cancer therapy. In a second study, Zitvogel and colleagues identified a gut microbe-dependent mechanism by which cyclophosphamide, a chemotherapy used to treat many non-colorectal cancers, injures the small intestine and triggers anti-tumor Th1 and Th17 immune responses (Viaud et al., 2013).
While the studies of Iida et al. and Viaud et al. are of great interest, there are numerous limitations that need to be considered for the translation of such findings to humans. Anatomical, behavioral and dietary differences, which distinguish mice from humans, influence luminal and mucosal intestinal microbial communities, their transcriptomes, and metabolomes. Thus beyond identifying clades that track with chemotherapy responses, subsequent studies need to address how microbial pattern recognition receptor and metabolite signaling in the host, specifically human hosts, can be tuned to both mitigate side effects and optimize tumor responsiveness to chemotherapies. This point requires especial consideration as cancer patients often require antibiotics for bacterial infections while receiving chemotherapy. An over-arching message of the above studies is that we need to develop a well-nuanced understanding of how drug and biologic treatments alter the gut microbiota and immune system function.
Conclusion
Cancer has been called the ‘emperor of all maladies’ (Mukherjee 2010) and in unraveling the role of the microbiota in colorectal carcinogenesis, research efforts are giving this emperor new clothes and laying him bare. With sufficient research support the vast genomic and metabolic potential of the gut microbiota may be realized as the most powerful weapon in the 40-plus year war on cancer. Specific species, microbial consortia, and microbial metabolites generated from ingested foodstuffs are all potential targets for decreasing or increasing cancer risk and perhaps even for diagnosis, treatment stratification, and therapy.
Acknowledgments
We thank current and former members of the Sears and Garrett labs. CLS also thanks Drew Pardoll, Ken Kinzler, Bert Vogelstein and members of their groups for helpful discussions over time; WSG also thanks Aleksandar D. Kostic, Andrew T. Chan, and Curtis Huttenhower for thoughtful discussions. CLS acknowledges the following funding sources: NIH (R01CA151393, R21CA170492, R01CA151325, R01CA179440) and the Merieux Institute. WSG acknowledges the following funding sources: NIH (R01CA154426 and K08AI078942), a Burroughs Wellcome Career in Medical Sciences Award, a Searle Scholars Award, and a Cancer Research Institute Investigator Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol Cancer. 2010;9:249. doi: 10.1186/1476-4598-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, Goedert JJ, Hayes RB, Yang L. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105:1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aries V, Crowther JS, Drasar BS, Hill MJ, Williams RE. Bacteria and the aetiology of cancer of the large bowel. Gut. 1969;10:334–335. doi: 10.1136/gut.10.5.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeke F, Korf H, Overbergh L, Verstuyf A, Thorrez L, Van Lommel L, Waer M, Schuit F, Gysemans C, Mathieu C. The vitamin D analog, TX527, promotes a human CD4+CD25highCD127low regulatory T cell profile and induces a migratory signature specific for homing to sites of inflammation. J Immunol. 2011;186:132–142. doi: 10.4049/jimmunol.1000695. [DOI] [PubMed] [Google Scholar]
- Barrasa JI, Olmo N, Lizarbe MA, Turnay J. Bile acids in the colon, from healthy to cytotoxic molecules. Toxicol in Vitro. 2013;27:964–977. doi: 10.1016/j.tiv.2012.12.020. [DOI] [PubMed] [Google Scholar]
- Boleij A, Tjalsma H. The itinerary of Streptococcus gallolyticus infection in patients with colonic malignant disease. Lancet Infect Dis. 2013;13:719–724. doi: 10.1016/S1473-3099(13)70107-5. [DOI] [PubMed] [Google Scholar]
- Boleij A, Dutilh BE, Kortman GAM, Roelofs R, Laarakkers CM, Engelke UF, Tjalsma H. Bacterial responses to a simulated colon tumor microenvironment. Mol Cell Proteomics. 2012;11:851–862. doi: 10.1074/mcp.M112.019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boleij A, Muytjens CMJ, Bukhari SI, Cayet N, Glaser P, Hermans PWM, Swinkels DW, Bolhuis A, Tjalsma H. Novel clues on the specific association of Streptococcus gallolyticus subsp gallolyticus with colorectal cancer. J Infect Dis. 2011a;203:1101–1109. doi: 10.1093/infdis/jiq169. [DOI] [PubMed] [Google Scholar]
- Boleij A, van Gelder MMHJ, Swinkels DW, Tjalsma H. Clinical Importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin Infect Dis. 2011b;53:870–878. doi: 10.1093/cid/cir609. [DOI] [PubMed] [Google Scholar]
- Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A, Pezet D, Bonnet R. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE. 2013;8:e56964. doi: 10.1371/journal.pone.0056964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey EJ, Lindor KD. Chemoprevention of colorectal cancer with ursodeoxycholic acid: cons. Clin Res Hepatol Gastroenterol. 2012;36(Suppl 1):S61–S64. doi: 10.1016/S2210-7401(12)70023-2. [DOI] [PubMed] [Google Scholar]
- Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrède J-P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USa. 2010;107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva M, Jaggers GK, Verstraeten SV, Erlejman AG, Fraga CG, Oteiza PI. Large procyanidins prevent bile-acid-induced oxidant production and membrane-initiated ERK1/2, p38, and Akt activation in Caco-2 cells. Free Radic Biol Med. 2012;52:151–159. doi: 10.1016/j.freeradbiomed.2011.10.436. [DOI] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013 doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejea C, Wick E, Sears CL. Bacterial oncogenesis in the colon. Future Microbiol. 2013;8:445–460. doi: 10.2217/fmb.13.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AS. Causation and disease: the Henle-Koch postulates revisited. 1976. [PMC free article] [PubMed] [Google Scholar]
- Fredericks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev. 1996;9:18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, Punit S, Karlsson M, Bry L, Glickman JN, Gordon JI, Onderdonk AB, Glimcher LH. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell host & microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill CIR, Rowland IR. Diet and cancer: assessing the risk. Br J Nutr. 2002;88(Suppl 1):S73–S87. doi: 10.1079/BJN2002632. [DOI] [PubMed] [Google Scholar]
- Gordon JI, Knowlton N, Relman DA, Rohwer F, Youle M. Superorganisms and Holobionts. Microbe Magazine. Forum online 2013 [Google Scholar]
- Goodwin AC, Destefano Shields CE, Wu S, Huso DL, Wu X, Murray-Stewart TR, Hacker-Prietz A, Rabizadeh S, Woster PM, Sears CL, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci USA. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, Datz C, Feng Y, Fearon ER, Oukka M, Tessarollo L, Coppola V, Yarovinsky F, Cheroutre H, Eckmann L, Trinchieri G, Karin M. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491:254–8. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MJ, Drasar BS, Hawksworth G, Aries V, Crowther JS, Williams RE. Bacteria and aetiology of cancer of large bowel. Lancet. 1971;1:95–100. doi: 10.1016/s0140-6736(71)90837-3. [DOI] [PubMed] [Google Scholar]
- Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobin C. Colorectal cancer: CRC-all about microbial products and barrier function? Nat Rev Gastroenterol Hepatol. 2012;9:694–696. doi: 10.1038/nrgastro.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawalek JC, Hallmark RK, Andrews AW. Effect of lithocholic acid on the mutagenicity of some substituted aromatic amines. J Natl Cancer Inst. 1983;71:293–298. [PubMed] [Google Scholar]
- Kawato Y, Aonuma M, Hirota Y, Kuga H, Sato K. Intracellular roles of SN-38, a metabolite of the camptothecin derivative CPT-11, in the antitumor effect of CPT-11. Cancer Res. 1991;51:4187–4191. [PubMed] [Google Scholar]
- Kim GB, Lee BH. Biochemical and molecular insights into bile salt hydrolase in the gastrointestinal microflora: a review. Asian-Aust J Anim Sci. 2005;18:1505–1512. [Google Scholar]
- Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Meyerson M, Garrett WS. Microbes and Inflammation in Cancer. Cancer Immunology Research. 2013;1:150–157. doi: 10.1158/2326-6066.CIR-13-0101. [DOI] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host and Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier J-C, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- Lax S, Schauer G, Prein K, Kapitan M, Silbert D, Berghold A, Berger A, Trauner M. Expression of the nuclear bile acid receptor/farnesoid X receptor is reduced in human colon carcinoma compared to nonneoplastic mucosa independent from site and may be associated with adverse prognosis. Int J Cancer. 2012;130:2232–2239. doi: 10.1002/ijc.26293. [DOI] [PubMed] [Google Scholar]
- Lee DK, Park SY, Baik SK, Kwon SO, Chung JM, Oh ES, Kim HS. Deoxycholic acid-induced signal transduction in HT-29 cells: role of NF-kappa B and interleukin-8. Korean J Gastroenterol. 2004;43:176–185. [PubMed] [Google Scholar]
- Lepercq P, Gérard P, Béguet F, Raibaud P, Grill J-P, Relano P, Cayuela C, Juste C. Epimerization of chenodeoxycholic acid to ursodeoxycholic acid by Clostridium baratii isolated from human feces. FEMS Microbiol Lett. 2004;235:65–72. doi: 10.1016/j.femsle.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Liu W, Chen Y, Golan MA, Annunziata ML, Du J, Dougherty U, Kong J, Musch M, Huang Y, Pekow J, et al. Intestinal epithelial vitamin D receptor signaling inhibits experimental colitis. J Clin Invest. 2013;123:3983–3996. doi: 10.1172/JCI65842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Wu S, Xia Y, Sun J. The Vitamin D Receptor, Inflammatory Bowel Diseases, and Colon Cancer. Curr Colorectal Cancer Rep. 2012;8:57–65. doi: 10.1007/s11888-011-0114-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks ODK, Short AJ, Donnenberg MS, Bader S, Harrison DJ. Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS ONE. 2009;4:e5517. doi: 10.1371/journal.pone.0005517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee EA, Richardson CJ, Hughes R, Cummings JH. Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am J Clin Nutr. 2000;72:1488–1494. doi: 10.1093/ajcn/72.6.1488. [DOI] [PubMed] [Google Scholar]
- McCoy AN, Araujo-Perez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium Is Associated with Colorectal Adenomas. PLoS ONE. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S. Scribner. 2010. The Emperor of All Maladies: A Biography of Cancer. [Google Scholar]
- Mühlbauer M, Allard B, Bosserhoff AK, Kiessling S, Herfarth H, Rogler G, Schölmerich J, Jobin C, Hellerbrand C. Differential effects of deoxycholic acid and taurodeoxycholic acid on NF-kappa B signal transduction and IL-8 gene expression in colonic epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2004;286:G1000–G1008. doi: 10.1152/ajpgi.00338.2003. [DOI] [PubMed] [Google Scholar]
- Zitomersky NL, Coyne MJ, Comstock LE. Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect Immun. 2011;79:2012–20. doi: 10.1128/IAI.01348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- Nowrouzian FL, Oswald E. Escherichia coli strains with the capacity for long-term persistence in the bowel microbiota carry the potentially genotoxic pks island. Microb Pathog. 2012;53:180–182. doi: 10.1016/j.micpath.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Ota H, Katsuyama T, Nakajima S, El-Zimaity H, Kim JG, Graham DY, Genta RM. Intestinal metaplasia with adherent Helicobacter pylori: a hybrid epithelium with both gastric and intestinal features. Human Path. 1998;29:846–50. doi: 10.1016/s0046-8177(98)90455-5. [DOI] [PubMed] [Google Scholar]
- Payne CM, Weber C, Crowley-Skillicorn C, Dvorak K, Bernstein H, Bernstein C, Holubec H, Dvorakova B, Garewal H. Deoxycholate induces mitochondrial oxidative stress and activates NF-kappaB through multiple mechanisms in HCT-116 colon epithelial cells. Carcinogenesis. 2007;28:215–222. doi: 10.1093/carcin/bgl139. [DOI] [PubMed] [Google Scholar]
- Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- Reddy BS. Types and amount of dietary fat and colon cancer risk: Prevention by omega-3 fatty acid-rich diets. Environ Health Prev Med. 2002;7:95–102. doi: 10.1265/ehpm.2002.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BS. Role of bile metabolites in colon carcinogenesis. Animal models. Cancer. 1975;36:2401–2406. doi: 10.1002/1097-0142(197512)36:6<2401::aid-cncr2820360619>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Wallace BD, Venkatesh MK, Mani S, Redinbo MR. Molecular insights into microbial β-glucuronidase inhibition to abrogate CPT-11 toxicity. Mol Pharmacol. 2013;84:208–217. doi: 10.1124/mol.113.085852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host and Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitta KS, Zhang C, Lee KK, Fujimoto K, Redinbo MR, Boelsterli UA. Bacterial β-glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen or diclofenac: mode of action and pharmacokinetics. Xenobiotica. 2013 doi: 10.3109/00498254.2013.811314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. 2009;22:349–69. doi: 10.1128/CMR.00053-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis. 2011;203:306–311. doi: 10.1093/jinfdis/jiq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serfaty L. Chemoprevention of colorectal cancer with ursodeoxycholic acid: pro. Clin Res Hepatol Gastroenterol. 2012;36(Suppl 1):S53–S60. doi: 10.1016/S2210-7401(12)70022-0. [DOI] [PubMed] [Google Scholar]
- Shibuya N, Nakadaira H, Ohta T, Nakamura K, Hori Y, Yamamoto M, Saitoh Y, Ogoshi K. Co-mutagenicity of glyco- and tauro-deoxycholic acids in the Ames test. Mutat Res. 1997;395:1–7. doi: 10.1016/s1383-5718(97)00132-0. [DOI] [PubMed] [Google Scholar]
- Shimada K, Bricknell KS, Finegold SM. Deconjugation of bile acids by intestinal bacteria: review of literature and additional studies. J Infect Dis. 1969;119:273–281. doi: 10.1093/infdis/119.3.273. [DOI] [PubMed] [Google Scholar]
- Singh SV, Singh K. Cancer chemoprevention with dietary isothiocyanates mature for clinical translational research. Carcinogenesis. 2012;33:1833–1842. doi: 10.1093/carcin/bgs216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler AP, Miller RD, Laughlin KV, Carp NZ, Klurfeld DM, Mullin JM. Increased tight junctional permeability is associated with the development of colon cancer. Carcinogenesis. 1999;20:1425–1431. doi: 10.1093/carcin/20.8.1425. [DOI] [PubMed] [Google Scholar]
- Sousa T, Paterson R, Moore V, Carlsson A, Abrahamsson B, Basit AW. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm. 2008;363:1–25. doi: 10.1016/j.ijpharm.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, Allen-Vercoe E. Invasive potential of gut mucosa-derived fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011 doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- Summerton J, Goeting N, Trotter GA, Taylor I. Effect of deoxycholic acid on the tumour incidence, distribution, and receptor status of colorectal cancer in the rat model. Digestion. 1985;31:77–81. doi: 10.1159/000199183. [DOI] [PubMed] [Google Scholar]
- Swidsinski A, Khilkin M, Kerjaschki D, Schreiber S, Ortner M, Weber J, Lochs H. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115:281–286. doi: 10.1016/s0016-5085(98)70194-5. [DOI] [PubMed] [Google Scholar]
- Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- Toprak NU, Yagci A, Gulluoglu BM, Akin ML, Demirkalem P, Celenk T, Soyletir G. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12:782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, Berger A, Bruneval P, Fridman W-H, Pagès F, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, Venkatesh M, Jobin C, Yeh LA, Mani S, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke MM. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008;68:9909–9917. doi: 10.1158/0008-5472.CAN-08-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yang Y, Moore DR, Nimmo SL, Lightfoot SA, Huycke MM. 4-hydroxy-2-nonenal mediates genotoxicity and bystander effects caused by Enterococcus faecalis-infected macrophages. Gastroenterology. 2012;142:543–551.e547. doi: 10.1053/j.gastro.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K, Allen-Vercoe E, Holt RA. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1:16. doi: 10.1186/2049-2618-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick E, Rabizadeh S, Albesiano E, Wu X, Wu S, Chan J, Rhee K-J, Ortega G, Huso DL, Pardoll D, Housseau F, Sears CL. Stat3 activation in murine colitis induced by enterotoxigenic Bacteroides fragilis. Inflammatory Bowel Diseases. 2014 doi: 10.1097/MIB.0000000000000019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks J, Beilinson H, Golovkina TV. Dual role of commensal bacteria in viral infections. Immunol Rev. 2013;255:222–229. doi: 10.1111/imr.12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters MD, Schlinke TL, Joyce WA, Glore SR, Huycke MM. Prospective case-cohort study of intestinal colonization with enterococci that produce extracellular superoxide and the risk for colorectal adenomas or cancer. Am J Gastroenterol. 1998;93:2491–2500. doi: 10.1111/j.1572-0241.1998.00710.x. [DOI] [PubMed] [Google Scholar]
- Wu S, Rhee K-J, Albesiano E, Rabizadeh S, Wu X, Yen H-R, Huso DL, Brancati FL, Wick E, McAllister F, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi M, Morikawa T, Kuchiba A, Imamura Y, Qian ZR, Nishihara R, Liao X, Waldron L, Hoshida Y, Huttenhower C, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang X, Huycke T, Moore DR, Lightfoot SA, Huycke MM. Colon Macrophages Polarized by Commensal Bacteria Cause Colitis and Cancer through the Bystander Effect. Transl Oncol. 2013;6:596–606. doi: 10.1593/tlo.13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;207:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]