Although recent studies have revealed that heart cells are generated in adult mammals, the frequency and source of new heart cells is unclear. Some studies suggest a high rate of stem cell activity with differentiation of progenitors to cardiomyocytes1. Other studies suggest that new cardiomyocytes are born at a very low rate2,3,4, and that they may be derived from division of pre-existing cardiomyocytes. Thus, the origin of cardiomyocytes in adult mammals remains unknown. Here we combined two different pulse-chase approaches -- genetic fate-mapping with stable isotope labeling and Multi-isotope Imaging Mass Spectrometry (MIMS). We show that genesis of cardiomyocytes occurs at a low rate by division of pre-existing cardiomyocytes during normal aging, a process that increases by four-fold adjacent to areas of myocardial injury. Cell cycle activity during normal aging and after injury led to polyploidy and multinucleation, but also to new diploid, mononucleated cardiomyocytes. These data reveal pre-existing cardiomyocytes as the dominant source of cardiomyocyte replacement in normal mammalian myocardial homeostasis as well as after myocardial injury.
Despite intensive research, fundamental aspects of the mammalian heart’s capacity for self-renewal are actively debated5,6. Estimates of cardiomyocyte turnover range from less than 1% per year2,3,4 to more than 40% per year7. Turnover has been reported to either decrease3 or increase with age7, while the source of new cardiomyocytes has been attributed to both division of existing myocytes8 and to progenitors residing within the heart9 or in exogenous niches such as bone marrow10. Controversy persists regarding the plasticity of the adult heart in part due to methodological challenges associated with studying slowly replenished tissues. Toxicity attributed to radiolabeled thymidine11 and halogenated nucleotide analogues12 limits the duration of labeling and may produce direct biological effects. Tissue autofluorescence can reduce the sensitivity and specificity of immunofluorescent methods of detecting cell cycle activity5,13, including as cell cycle markers or incorporation of halogenated nucleotide analogues. The challenge of measuring cardiomyocyte turnover is further compounded by the faster rate of turnover of cardiac stromal cells relative to cardiomyocytes14.
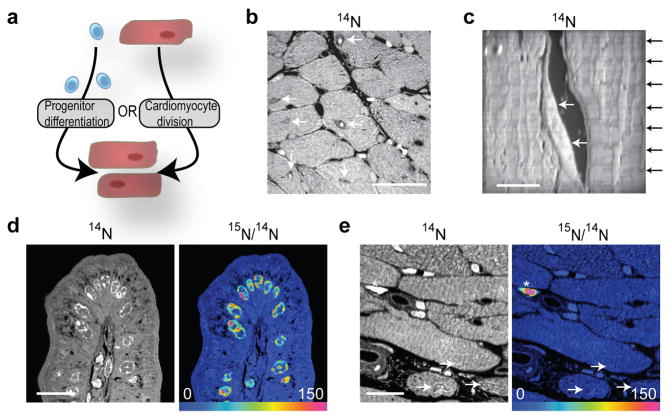
We used Multi-isotope Imaging Mass Spectrometry (MIMS) to study cardiomyocyte turnover and to determine whether new cardiomyocytes are derived from preexisting myocytes or from a progenitor pool (Fig 1a). MIMS uses ion microscopy and mass spectrometry to generate high resolution quantitative mass images and localize stable isotope reporters in domains smaller than one micron cubed15,16,17. MIMS generates 14N quantitative mass images by measuring the atomic composition of the sample surface with a lateral resolution of under 50nm and a depth resolution of a few atomic layers. Cardiomyocyte cell borders and intracellular organelles were easily resolved (Fig 1b). Regions of interest can be analyzed at higher resolution, demonstrating cardiomyocyte-specific subcellular ultrastructure, including sarcomeres (Fig 1c, Supplemental Fig 1a). In all subsequent analyses, cardiomyocyte nuclei were identified by their location within sarcomere-containing cells, distinguishing them from adjacent stromal cells.
Figure 1. Multi-isotope imaging mass spectrometry (MIMS) to study cardiomyocyte turnover.
a) Primary question: are new cardiomyocytes are derived from progenitors or from pre-existing cardiomyocytes?
b) 14N mass image. Subcellular details are evident, including cardiomyocyte nuclei (white arrows). Scale bar = 20μm.
c) MIMS resolves periodic sarcomeres (black arrows) in cardiomyocytes. Non-cardiomyocytes (white arrows) are seen outside cardiomyocyte borders. Scale bar = 5μm.
d) Hue saturation intensity (HSI) 15N:14N image of small intestinal epithelium after 15N-thymidine. The scale ranges from blue, where the ratio is equivalent to natural ratio (0.37%, expressed as 0% above natural ratio), to red, where the ratio is 150% above natural ratio. 15N-labeling is concentrated in nuclei in a pattern resembling chromatin. Scale bar = 15μm.
e) HSI 15N:14N image of heart section (left ventricle). 15N-thymidine administered for 1 wk. Rare 15N-labeled interstitial cells (asterisk). Cardiomyocyte nuclei (white arrows) are unlabeled. Scale bar = 15μm.
An immense advantage of MIMS is the detection of nonradioactive stable isotope tracers. As an integral part of animate and inanimate matter, they do not alter biochemical reactions and are not harmful to the organism18. MIMS localizes stable isotope tracers by simultaneously quantifying multiple masses from each analyzed domain; this enables the generation of a quantitative ratio image of two stable isotopes of the same element15. The incorporation of a tracer tagged with the rare stable isotope of nitrogen (15N) is detectable with high precision by an increase in 15N:14N above the natural ratio (0.37%). Nuclear incorporation of 15N-thymidine is evident in cells having divided during a one-week labeling period, as observed in the small intestinal epithelium, which turns over completely in 3–5 days16 (Fig 1d); in contrast, 15N-thymidine labeled cells are rarely observed in the heart (Fig 1e) after 1 week of labeling. In subsequent studies, small intestine was used as a positive control to confirm label delivery.
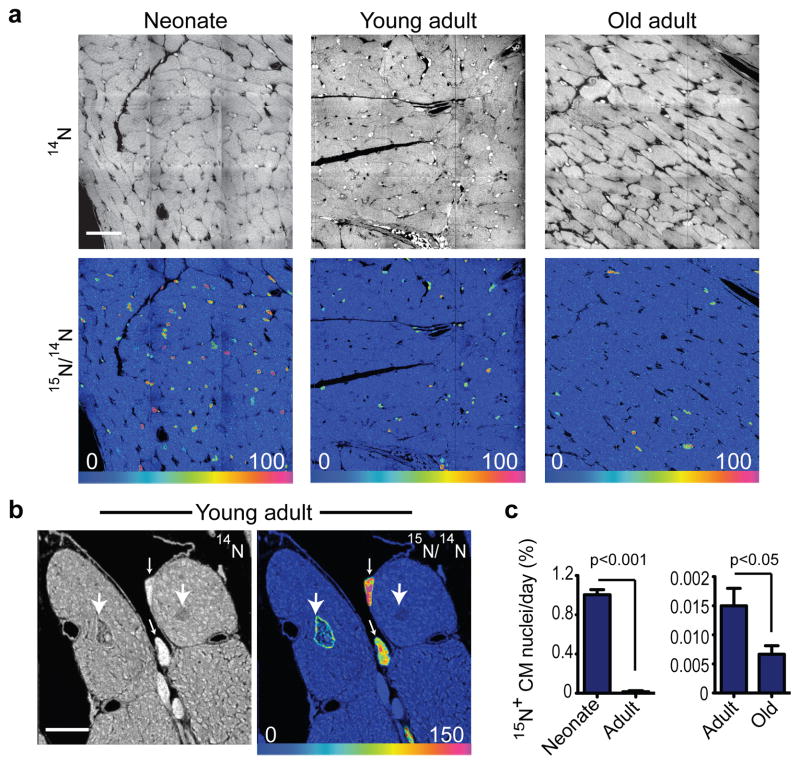
To evaluate for an age-related change in cell cycle activity, we administered 15N-thymidine for 8 weeks to three age groups of C57BL6 mice starting at day-4 (neonate), at 10-weeks (young adult) and at 22-months (old adult) (Supplemental Fig 2). We then performed MIMS analysis (Fig 2a, b, Supplemental Fig 3). In the neonatal group, 56% (±3% s.e.m., n=3 mice) of cardiomyocytes demonstrated 15N nuclear labeling, consistent with the well-accepted occurrence of cardiomyocyte DNA synthesis during post-natal development19. We observed a marked decline in the frequency of 15N-labeled cardiomyocyte nuclei (15N+CM) in the young adult (neonate= 1.00%15N+CM/day ±0.05 s.e.m. vs young adult=0.015% 15N+CM/day ±0.003 s.e.m., n=3 mice/group, p<0.001) (Fig 2a, c; Supplemental Fig 3). We found a further reduction in cardiomyocyte DNA synthesis in old mice (young adult=0.015%15N+CM/day ±0.003 s.e.m. vs. old adult=0.007 %15N+CM/day ±0.002 s.e.m., n=3/group, p<0.05) (Fig 2c). The observed pattern of nuclear 15N-labeling in cardiomyocytes is consistent with the known chromatin distribution pattern in cardiomyocytes20 (Supplemental Fig 1b) and was measured at levels that could not be explained by DNA repair (Supplemental Fig 4). Extrapolating DNA synthesis measured in cardiomyocytes over 8 weeks yields a yearly rate of 5.5% in the young adult and 2.6% in the old mice. Given that cardiomyocytes are known to undergo DNA replication without completing the cell cycle19,21,22, these calculations represent the upper limit of cardiomyocyte generation under normal homeostatic conditions, indicating a low rate of cardiogenesis.
Figure 2. Cardiomyocyte DNA synthesis decreases with age.
a) 15N-thymidine administered for 8 wks to mice of different ages: Neonate: starting post-natal day 4, Young adult: starting at 2 mos, Old adult: starting at 22 mos. Top: 14N mass images show histologic details. Bottom: 15N:14N HSI images show 15N-labeled nuclei. Mosaics constructed of 9 tiles, 60μm each. Scale bar = 30μm.
b) High magnification analysis demonstrates cardiomyocyte from the young adult with nuclear 15N-labeling (large arrow), two labeled non-cardiomyocytes (small arrows), and an adjacent unlabeled cardiomyocyte nucleus (medium arrow). Scale bar = 10μm.
c) Age-related decline in cardiomyocyte DNA synthesis. Left: neonate compared to young adult. Right: Scale reduced to compare young adult to old adult (n=3 mice per group). Mean% ± S.E.M.
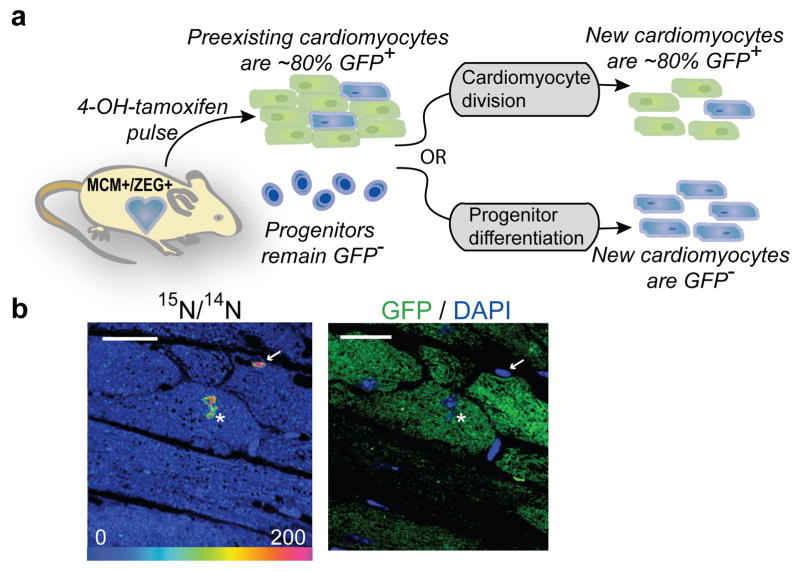
To test whether cell cycle activity occurred in preexisting cardiomyocytes or was dependent on a progenitor pool, we performed 15N-thymidine labeling of double-transgenic MerCreMer/ZEG mice, previously developed for genetic lineage mapping (Fig 3a)23,24. MerCreMer/ZEG cardiomyocytes irreversibly express green fluorescent protein (GFP) after treatment with 4OH-tamoxifen, allowing pulse labeling of existing cardiomyocytes with a reproducible efficiency of approximately 80%. Although some have reported rare GFP expression by non-cardiomyocytes with this approach25, we did not detect GFP expression in interstitial cells isolated from MerCreMer/ZEG hearts nor did we detect GFP expression by Sca1 or ckit-expressing progenitors in histological sections (Supplemental Fig 5). Thus, during a chase period, cardiomyocytes generated from progenitors should be GFP−, whereas cardiomyocytes arising from preexisting cardiomyocytes should express GFP at a frequency similar to the background rate induced by 4OH-tamoxifen. We administered 4OH-tamoxifen for two weeks to 8 wk-old mice (n=4); during a subsequent 10-week chase, mice received 15N-thymidine via osmotic minipump (20μg/h). With MIMS analysis, we identified 35 15N-labeled cardiomyocyte nuclei (of 4190 analyzed) over 10 weeks, yielding a projected yearly rate of cardiomyocyte DNA replication of 4.4%. Extrapolating from prior reports of high stem cell-dependent cardiomyocyte turnover7, we had anticipated detecting >320 cardiomyocytes entering the cell cycle; these results exclude such a high rate of turnover (expected 15N+ cardiomyocytes=321; observed=35; Fisher’s exact<0.0001). Immunofluorescent staining for GFP was performed on adjacent sections and an observer unaware of MIMS analysis results assessed GFP status. Of 15N+ cardiomyocytes, 77% expressed GFP, a frequency essentially identical to that of surrounding 15N− cardiomyocytes (15N+ CM, 77% vs 15N− cardiomyocytes, 84%; Fisher’s exact=n.s.) (Table 1). If new cardiomyocytes were derived from progenitors, 15N+ cardiomyocytes would have been GFP− (expected=0/35; observed=27/35, Fisher’s exact<0.0001). These data reveal that 15N-labeled cardiomyocytes resulted from DNA synthesis by preexisting cardiomyocytes and exclude a substantial contribution from stem cells to cardiomyocyte replacement in the uninjured heart.
Figure 3. New cardiomyocytes are derived from pre-existing cardiomyocytes during aging.
a) Experimental strategy. MerCreMer+/ZEG+ mice (n=4) treated for 2 wks with 4OH-tamoxifen to induce cardiomyocyte-specific GFP expression. 15N-thymidine administered continuously during 10 wk chase, then cycling cells identified by 15N-labeling. New cardiomyocytes (15N+) derived from preexisting cardiomyocytes should express GFP at a rate similar to surrounding quiescent (15N−) cardiomyocytes. New cardiomyocytes (15N+) derived from progenitors should be GFP−.
b) Left: 15N:14N HSI image demonstrating 15N-thymidine labeled cardiomyocyte nucleus (white asterisk) and 15N-labeled non-cardiomyocyte (white arrow). Right: Immunofluorescent image showing 15N-labeled cardiomyocyte is GFP+. Scale bar = 15μm.
Table 1. 15N-labeled cardiomyocytes (15N+ CM) during normal aging.
Includes new mononucleated/diploid fraction, demonstrated a similar rate of GFP expression compared to background 15N− cardiomyocytes.
| Total counted | GFP + | % GFP + | |
|---|---|---|---|
| 15N− CM | 1111 | 933 | 84% |
| 15N+ CM | 35 | 27 | 77% |
| Polyploid CM | 12 | 10 | 83% |
| Diploid CM | 22 | 17 | 77% |
| Multinucleated CM | 18 | 14 | 78% |
| Mononucleated CM | 17 | 14 | 82% |
| Diploid/Mono CM | 6 | 5 | 83% |
Cardiomyocytes can undergo DNA replication without completing the cell cycle. Although multinucleation and polyploidization occur during early post-natal development and in response to myocardial stress2,19, we considered the possibility these processes could account for 15N+ cardiomyocytes in the uninjured adult mouse. We performed fluorescent in situ hybridization in adjacent sections to assess the ploidy state of each 15N+ cardiomyocyte and surrounding 15N− cardiomyocytes, and an observer unaware of the results of MIMS analysis identified fluorescently-labeled chromosomes (Supplemental Fig 6). Although we found 15N+ cardiomyocytes that were polyploid (4n or greater), we observed a higher frequency of diploid nuclei in the 15N+ pool compared to surrounding 15N− cardiomyocytes (15N+ diploid:polyploid = 22:12 vs 15N− diploid:polyploid = 9:56, Fisher’s exact p<0.0001), consistent with ongoing cell division. We then assessed each cell for multinucleation using serial 0.5μm sections adjacent to the section used for MIMS analysis (Supplemental Fig 6). We observed that 49% of 15N+ cardiomyocytes were mononucleated, in contrast to a frequency of 24% for surrounding 15N− cardiomyocytes (Fisher’s exact p<0.01), also consistent with cell division. The majority of cardiomyocyte DNA synthesis occurred in polyploid and/or multinucleated cardiomyocytes as might be expected with a physiologic hypertrophic response and thus unlikely to indicate cardiomyocyte division; however, 17% (6 of 35 15N+ CM) were diploid and mononucleated, consistent with newly generated cardiomyocytes (Supplemental Fig 7). The mononucleated, diploid, 15N+ cardiomyocytes were also predominantly GFP+ (5 of 6 = 83% vs. 82% background frequency, p=n.s.), suggesting that they arose from preexisting cardiomyocytes at a slow annual rate of 0.76% (n=6 of 4190 over 10 weeks).
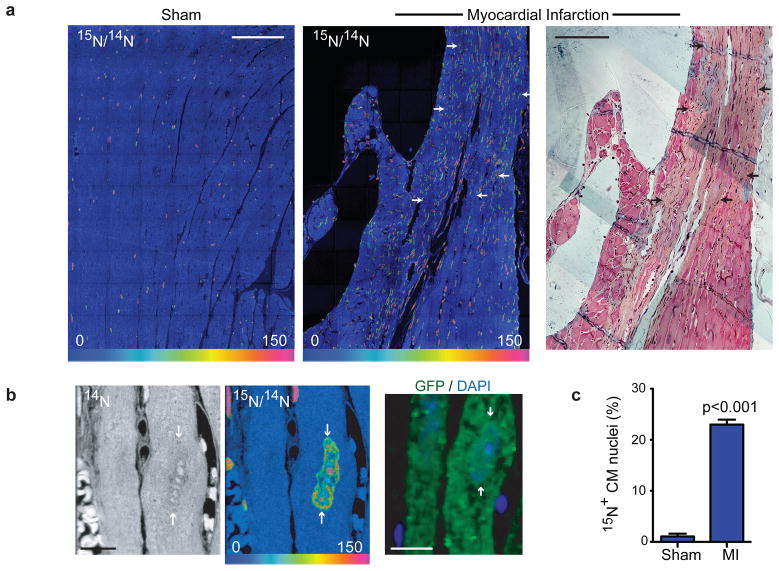
We next used MIMS and genetic fate mapping to study myocardial injury. Cardiomyocyte GFP labeling was induced in MerCreMer/ZEG mice with 4OH-tamoxifen. Mice then underwent experimental myocardial infarction or sham surgery followed by continuous labeling with 15N-thymidine for 8wks. The frequency of 15N-labeled cardiomyocytes in sham-operated mice was similar to prior experiments in unoperated mice (yearly projected rates: sham=6.8%; unoperated=4.4%), but increased significantly adjacent to infarcted myocardium (total 15N+ cardiomyocyte nuclei: MI=23.0% vs sham=1.1%, Fig 4a–b, Supplemental Fig 8). We examined GFP expression, nucleation and ploidy status of 15N-labeled cardiomyocytes and surrounding unlabeled cardiomyocytes. We found a significant dilution of the GFP+ cardiomyocyte pool at the border region as previously shown23,24 (67% vs. 79%, p<0.05, Table 2, Supplemental Fig 9); however, 15N+ myocytes demonstrated a similar frequency of GFP expression compared to unlabeled myocytes (71% vs. 67%, Fisher’s exact=n.s.), suggesting that DNA synthesis was primarily occurring in pre-existing cardiomyocytes. Of 15N-labeled cardiomyocytes, approximately 14% were mononucleated and diploid consistent with division of pre-existing cardiomyocytes (Supplemental Fig 6, 7). We observed higher DNA content (>2N) in the remaining cardiomyocytes as expected with compensatory hypertrophy after injury. Thus, in the 8wks after myocardial infarction, approximately 3.2% of the cardiomyocytes adjacent to the infarct had unambiguously undergone division (total 15N+ × mononucleated diploid fraction = 23% × 0.14 = 3.2%). The low rate of cardiomyocyte cell cycle completion is further supported by the absence of detectable Aurora B Kinase, a transiently expressed cytokinesis marker, which was detected in rapidly proliferating small intestinal cells but not in cardiomyocytes (Supplemental Fig 10). We also considered the possibility that a subset of 15N+ myocytes that were multinucleated and/or polyploid resulted from division followed by additional rounds of DNA synthesis without division. However, quantitative analysis of the 15N+ population did not identify a subpopulation that had accumulated additional 15N-label as would be expected in such a scenario (Supplemental Fig 11). Together, these data suggest that adult cardiomyocytes retain some capacity to reenter the cell cycle, but that the majority of DNA synthesis after injury occurs in preexisting cardiomyocytes without completion of cell division.
Figure 4. Myocardial injury stimulates division of pre-existing cardiomyocytes.
a) Myocardial infarction (MI) leads to extensive DNA synthesis within and adjacent to scar (arrows). MerCreMer+/ZEG+ mice were treated for 2 wks with 4OH-tamoxifen to induce cardiomyocyte-specific GFP expression before MI or sham surgery, then 15N-thymidine administered continuously for 8wks. Mosaics of 70 60×60μm MIMS tiles. Trichrome stained adjacent section (far right) shows scar. Scale bars = 90μm.
b) 15N-thymidine labeled cardiomyocyte nucleus (white arrows) from MI border region. Immunofluorescent staining demonstrates that the cardiomyocyte is GFP+. Scale bars = 10 μm.
c) Mean % 15N+ cardiomyocyte nuclei after MI (n=4) in the scar border region compared to sham operated mice (n=3). Mean% ± S.E.M.
Table 2. GFP analysis of 15N-labeled cardiomyocytes (15N+ CM).
Pooled analysis of 4 mice in MI group and 3 mice in sham group. MI led to dilution of GFP+ cardiomyocytes adjacent to scar. New cardiomyocytes generated after MI (the 15N+/diploid/mononucleated pool) were predominantly GFP+ consistent with division of preexisting cardiomyocytes.
| Total | GFP+ | % GFP+ | |
|---|---|---|---|
| Sham | 3411 | 2693 | 79% |
| 15N+ CM | 23 | 19 | 83% |
| MI | 7063 | 4766 | 67% |
| 15N+ CM | 205 | 146 | 71% |
| PolyploidCM | 126 | 97 | 77% |
| DiploidCM | 65 | 47 | 72% |
| Multinucleated CM | 59 | 42 | 71% |
| Mononucleated CM | 41 | 29 | 71% |
| Diploid/MonoCM | 16 | 11 | 69% |
If dilution of the GFP+ cardiomyocyte pool cannot be attributed to division and differentiation of endogenous progenitors, do these data exclude a role for progenitors in the adult mammalian heart? These data could be explained by preferential loss of GFP+ cardiomyocytes after injury, a process that we have previously considered but for which we have not found supporting evidence23. Such an explanation excludes a role for endogenous progenitors in cardiac repair and would be consistent with data emerging from lower vertebrates8,26 and the neonatal mouse27 in which preexisting cardiomyocytes are the cellular source for cardiomyocyte repletion. A second possibility to explain the dilution of the GFP+ cardiomyocyte pool is that injury stimulates progenitor differentiation without division; inevitably, this would lead to exhaustion of the progenitor pool, which if true could explain the limited regenerative potential of the adult mammalian heart.
In summary, this study demonstrates birth of cardiomyocytes from preexisting cardiomyocytes at a projected rate of approximately 0.76%/year (15N+ annual rate × mononucleated diploid fraction = 4.4% × 0.17) in the young adult mouse under normal homeostatic conditions, a rate that declines with age but increases by approximately four-fold after myocardial injury in the border region. This study shows that cardiac progenitors do not play a significant role in myocardial homeostasis in mammals and suggests that their role after injury is also limited.
Online Methods
Mice
All experiments were conducted in accordance with the Guide for the Use and Care of Laboratory Animals and approved by the Harvard Medical School Standing Committee on Animals. C57Bl/6 male mice were obtained from Charles River.
We generated double transgenic Mer-CreMer-ZEG male mice by crossbreeding cardiomyocyte-specific MerCreMer mice and ZEG mice (Jackson Laboratory). β-galactosidase-GFP is under the control of a cytomegalovirus (CMV) enhancer/chicken actin promoter (Actb); the background strain was C57BL/6J (N7). The background strain of the MerCreMer mice was C57Bl/6SV129. Genotyping was performed by PCR on tail DNA using the following primers: MerCreMer forward: 5′-GTCTGAC TAGGTGTCCTTCT-3′; MerCreMer backward: 5′-CGTCCTCCTGCTGGTA TAG-3′; ZEG forward: 5′-AAGTTCATCTGCACCACCG-3′; ZEG backward: 5′-TCCTTGAAGAAGATGGTGCG-3′; ZEG control forward: 5′-CTAGGCCA CAGAATTGAAAGATCT-3′; and ZEG control backward: 5′-GTAGGTG GAAATTCTAGCATCATCC-3′. To induce Cre recombination and GFP labeling in cardiomyocytes, we injected 4-OH-tamoxifen (provided by a generous gift from Laboratoires Besins), dissolved in peanut oil (Sigma) at a concentration of 5 mg/ml, intraperitoneally into 8-week-old MerCreMer-ZEG mice daily at a dosage of 0.5 mg/day for 14 days.
Experimental myocardial infarction
Mice were subjected to experimental myocardial infarction as described. Surgeries were performed by a single operator with more than 20 years of experience in the performance of coronary ligation in rodents. In brief, the left coronary artery was permanently ligated approximately 2 mm below the left atrial appendage. For sham operations, the thoracic cavity was opened and the heart exposed, but no intramyocardial sutures were placed. 15N-thymidine (Cambridge Isotopes) was administered at a rate of 20μg/hr via osmotic minipump (Alzet), implanted subcutaneously at the time of experimental myocardial infarction after a single intraperitoneal bolus dose of 500μg.
MIMS data acquisition
The factory prototype of the NanoSIMS50 as well as a standard NanoSIMS 50 and a large radius NanoSIMS 50L (Cameca, Gennevilliers, France) was used for MIMS analysis as previously described15. A focused beam of Cs+ ions was used to sputter a few atomic layers and generate secondary ions from the left ventricular free wall of heart section samples. The Cs+ primary ions were scanned over a raster pattern of either 256 × 256 pixels or 512 × 512 pixels. At each pixel location, the secondary ion intensities for 12C−, 13C−, 12C14N− and 12C15N−were recorded in parallel from the same sputtered volume. The detection of nitrogen requires the use of cluster ions 12C14N− and 12C15N− for 14N and 15N, respectively, due to the low ionization efficiency of nitrogen as N−.
MIMS data analysis
From a single field image acquisition, we first extracted four image files: the four original quantitative mass images (QMIs; 12C, 13C, 12C14N, 12C15N) and the two ratio images (13C/12C and 12C15N/12C14N), derived from the pixel-by-pixel division of the 13C QMI by the 12C QMI and of the 12C15N QMI by the 12C14N QMI, respectively. We then used a hue saturation intensity transformation (HSI) of the ratio image to map 15N-labeled regions. The hue corresponds to the ratio value, and the intensity at a given hue is an index of statistical reliability.
15N-thymidine labeling
For the neonatal cohort in the aging experiment, labeling was starting at post-natal day 4 with subcutaneous injections of 50μg/g 15N-thymidine (Cambridge Isotopes) every 12h and continued through post-natal week 4. Starting at age 4 wks – and in all other experiments using adult mice – osmotic minipumps (Alzet) were implanted subcutaneously, delivering 15N-thymidine (Cambridge Isotopes) at a rate of 20 μg/hour.
Multinucleation Analysis
Serial adjacent sections (0.5μm) were stained to identify cardiomyocyte borders. A given cardiomyocyte was tracked in the vertical axis by locating it in serial sections. Uninjured hearts were stained using a modified PAS protocol with standard solutions (Electron Microscopy Services), but with longer incubation times optimized for LR white embedding. Slides were incubated in xylene at 37C, 1 hour, rehydrated through graded alcohols, incubated in periodic acid for two hours, and Schiff’s reagent for two nights. Sections were counterstained in hematoxylin and Scott’s Bluing for 1 hour each. Injured hearts were stained using a modified Trichrome staining protocol with standard solutions (Fisher Scientific), but with longer incubation times. Slides were incubated in xylene at 37C, 1 hour, rehydrated through graded alcohols, incubated in bouin’s fluid at 56C, 1 hour, rinsed in tap water, incubated in weigert’s iron hematoxylin stain 1 hour, rinsed in tap water, incubated in scarlet-acid fuchsin solution, 1 hour, rinsed in DI water, incubated in phophotungistic-phophomolybdic acid solution, 30 minutes, incubated in aniline blue stain solution, 30 minutes, and incubated in 1% acetic acid, 20 minutes.
Fluorescent in situ hybridization
Sections were incubated in proteinase K (50ug/ml) at 60C for 15min. After a PBS/MgCl2 (45mM) wash, slides were post-fixed in 4% paraformaldehyde (PBS/MgCl2), dehydrated through graded alcohols. Biotinylated-labeled chromosome Y paint (Star-FISH, Cambio) in hybridization mix was applied to sections, and sealed under glass with rubber cement (note some samples were analyzed with chromosome-18 paint due to product discontinuation of Y-paint). Samples were heated to 90C for 15 min. After an overnight incubation at 37C, slides were washed three times with 50% formamide/2x standard saline citrate at 45C, three washes with 2x standard saline citrate at room temperature, and two washes with 4x standard saline citrate/0.1% Tween at room temperature. Samples were blocked 10 minutes with 4x standard saline citrate/0.1% Tween/ 0.05% milk and incubated for 2 hr in streptavidin-conjugated Alexa Fluor 488 (Invitrogen) prior to washing and mounting. An observer unaware of the MIMS images or 15N-thymidine labeling status of the nuclei assigned ploidy status.
Immunofluorescent staining
Sections were incubated in glycine/tris (50mM glycine/0.05M Tris) at room temperature for 5min. After a brief wash with Tris, sections were incubated with chicken anti-GFP (Abcam), rabbit anti-ckit (Abcam), rat anti-sca1 (Abcam) with fresh 0.1%BSA in TBST (TBS/0.1%Tween) overnight at 4C. After a brief wash with TBS, sections were incubated with anti-chicken Alexa Fluor 488 (Invitrogen) prior to TBS wash and mounting. An observer unaware of the MIMS images or 15N-thymidine labeling status of the nuclei assigned GFP status.
Fluorescence Image analysis
We used a custom-written script in IP Lab version 4.0 (Scanalytics) imaging software for serial image acquisition. Tissue sections were auto-imaged using an Olympus IX-70 microscope with a digital charge-coupled device camera (CoolSNAP EZ, Roper Scientific), an automated stage with a piezoelectric objective positioner (Polytec PI, Auburn MA) to compensate for deviations in the z-axis. Images were compressed and stitched into a mosaic using stitching software (Canon Photostitch). Multichannel images were merged in ImageJ prior to stitching.
Statistical analysis
Statistical testing was performed using Prism 3.0 (Graphpad). Results are presented as mean ± s.e.m. and were compared using T-tests (significance was assigned for p<0.05). Data comparing event rates were tested with a Fisher-Exact test.
Supplementary Material
Acknowledgments
The authors thank J. Gannon for surgical assistance, J. Lammerding and P. Isserman with assistance with microscopy and IP Lab software. We thank the PICT-IBiSA Imaging Facility in the Institut Curie. We thank Catherine MacGillivray for histology. S.E.S is funded by the National Institutes of Health (F32 HL108570). M.L.S. is funded by the American Heart Association (AHA FTF), Future Leaders in Cardiovascular Medicine, Watkins Cardiovascular Leadership Award and the NIH (K08 DK090147). C.P.L. is funded by the National Institutes of Health (EB001974, AG034641) and the Ellison Medical Foundation (AG-SS-2215-08). R.T.L is funded by the National Institutes of Health (AG032977, AG040019).
Footnotes
Author Contributions:
S.E.S., M.L.S. and R.T.L. designed the experiments. S.E.S., M.L.S., and V.K.Y. performed the experiments. L.C., C.L.P., V.K.Y., M.L.S., and S.E.S. performed histology. M.W. and S.E.S. operated the prototype nanoSIMS instrument. T-D.W. and J-L.G-K. operated nanoSIMS instrument at Curie. S.E.S. and M.L.S. analyzed the data with input from C.P.L. and R.T.L. C.P.L. supervised all MIMS analyses and provided critical feedback at all junctures. S.E.S. and M.L.S. made the figures. S.E.S., M.L.S., and R.T.L. wrote the manuscript. All authors approved the manuscript.
References
- 1.Hosoda T, et al. Clonality of mouse and human cardiomyogenesis in vivo. Proc Natl Acad Sci U S A. 2009;106:17169–17174. doi: 10.1073/pnas.0903089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soonpaa MH, Field LJ. Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am J Physiol Heart Circ Physiol. 1997;272:H220–226. doi: 10.1152/ajpheart.1997.272.1.H220. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh S, Ponten A, Fleischmann BK, Jovinge S. Cardiomyocyte cell cycle control and growth estimation in vivo--an analysis based on cardiomyocyte nuclei. Cardiovasc Res. 2010;86:365–373. doi: 10.1093/cvr/cvq005. [DOI] [PubMed] [Google Scholar]
- 5.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosoda T, Rota M, Kajstura J, Leri A, Anversa P. Role of stem cells in cardiovascular biology. J Thromb Haemost. 2011;9(Suppl 1):151–161. doi: 10.1111/j.1538-7836.2011.04363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kajstura J, et al. Myocyte turnover in the aging human heart. Circ Res. 2010;107:1374–1386. doi: 10.1161/CIRCRESAHA.110.231498. [DOI] [PubMed] [Google Scholar]
- 8.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beltrami AP, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 10.Orlic D, et al. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410:701–705. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- 11.Hu VW, Black GE, Torres-Duarte A, Abramson FP. 3H-thymidine is a defective tool with which to measure rates of DNA synthesis. Faseb J. 2002;16:1456–1457. doi: 10.1096/fj.02-0142fje. [DOI] [PubMed] [Google Scholar]
- 12.Wilson A, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 13.Laflamme MA, Murry CE. Regenerating the heart. Nat Biotechnol. 2005;23:845–856. doi: 10.1038/nbt1117. [DOI] [PubMed] [Google Scholar]
- 14.Bergmann O, et al. Identification of cardiomyocyte nuclei and assessment of ploidy for the analysis of cell turnover. Exp Cell Res. 2010;317:188–194. doi: 10.1016/j.yexcr.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 15.Lechene C, et al. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. J Biol. 2006;5:1–30. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinhauser ML, et al. Multi-isotope imaging mass spectrometry quantifies stem cell division and metabolism. Nature. 2012;481:516–519. doi: 10.1038/nature10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang DS, et al. Multi-isotope imaging mass spectrometry reveals slow protein turnover in hair-cell stereocilia. Nature. 2012;481:520–524. doi: 10.1038/nature10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein PD, Klein ER. Stable isotopes: origins and safety. J Clin Pharmacol. 1986;26:378–382. doi: 10.1002/j.1552-4604.1986.tb03544.x. [DOI] [PubMed] [Google Scholar]
- 19.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28:1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 20.Nikolova V, et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J Clin Invest. 2004;113:357–369. doi: 10.1172/JCI19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adler CP, Friedburg H. Myocardial DNA content, ploidy level and cell number in geriatric hearts: post-mortem examinations of human myocardium in old age. J Mol Cell Cardiol. 1986;18:39–53. doi: 10.1016/s0022-2828(86)80981-6. [DOI] [PubMed] [Google Scholar]
- 22.Katzberg AA, Farmer BB, Harris RA. The predominance of binucleation in isolated rat heart myocytes. Am J Anat. 1977;149:489–499. doi: 10.1002/aja.1001490406. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh PC, et al. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med. 2007;13:970–974. doi: 10.1038/nm1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loffredo FS, Steinhauser ML, Gannon J, Lee RT. Bone marrow-derived cell therapy stimulates endogenous cardiomyocyte progenitors and promotes cardiac repair. Cell Stem Cell. 2011;8:389–398. doi: 10.1016/j.stem.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong F, et al. Myocardial CXCR4 expression is required for mesenchymal stem cell mediated repair following acute myocardial infarction. Circulation. 126:314–324. doi: 10.1161/CIRCULATIONAHA.111.082453. [DOI] [PubMed] [Google Scholar]
- 26.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.