Abstract
Antarctic surface oceans are well-studied during summer when irradiance levels are high, sea ice is melting and primary productivity is at a maximum. Coincident with this timing, the bacterioplankton respond with significant increases in secondary productivity. Little is known about bacterioplankton in winter when darkness and sea-ice cover inhibit photoautotrophic primary production. We report here an environmental genomic and small subunit ribosomal RNA (SSU rRNA) analysis of winter and summer Antarctic Peninsula coastal seawater bacterioplankton. Intense inter-seasonal differences were reflected through shifts in community composition and functional capacities encoded in winter and summer environmental genomes with significantly higher phylogenetic and functional diversity in winter. In general, inferred metabolisms of summer bacterioplankton were characterized by chemoheterotrophy, photoheterotrophy and aerobic anoxygenic photosynthesis while the winter community included the capacity for bacterial and archaeal chemolithoautotrophy. Chemolithoautotrophic pathways were dominant in winter and were similar to those recently reported in global ‘dark ocean' mesopelagic waters. If chemolithoautotrophy is widespread in the Southern Ocean in winter, this process may be a previously unaccounted carbon sink and may help account for the unexplained anomalies in surface inorganic nitrogen content.
Keywords: Antarctic bacterioplankton, metagenomics, chemolithoautotrophy
Introduction
The Southern Ocean has long been the focus of microbial ecology studies, especially in relation to primary productivity and carbon cycling (Karl, 1993; Fuhrman and Azam, 1980; Ducklow et al., 2001). These waters are a controlling factor in regulating atmospheric CO2 concentration and are characterized by high nitrate concentrations but low primary productivity limited to varying degrees by iron, silica and light (Coale et al., 2004). Seasonal variation of oceanographic, weather and ice conditions in the Southern Ocean is dramatic. The austral summer is characterized by continuous high solar irradiance; water column stratification; and an intense but spatially patchy period of primary productivity that fuels a food web of krill, penguins, fish, seals and whales while supporting heterotrophic microbial production up to 10 mmol C m−2 d−1 (Ducklow et al., 2001). A natural experiment occurs each winter in waters south of the Polar Front when sea ice forms, light fades and a deeply mixed water column results in little net primary productivity from photoautotrophy as phytoplankton disappear. This winter bacterial productivity is likely driven by drawdown of phytoplankton produced dissolved organic carbon (DOC) and at least in the zone near the polar front, relatively high bacterial carbon production (nearly 10% of which can be chemoautotrophic) that can support an active protist and viral community (Manganelli et al., 2009).
High-latitude winter microbial oceanography studies have been limited mostly because of challenging access and the perception that most primary productivity is limited to summer phytoplankton or late winter/early spring sea-ice algae (Kottmeier and Sullivan, 1987). Most studies have focused on variability in microbial abundance (Delille et al., 1996) or activity (Ducklow et al., 2001; Pearce et al., 2007) and showed winter lows followed by increases in abundance that precede increases in carbon production. Cyclic patterns in bacterial community structure were identified off the Antarctic Peninsula, through two annual cycles (Murray et al., 1998) where greater richness in winter and dramatic shifts between winter and summer were found (Murray and Grzymski, 2007). A similar pattern was seen in a comparative pyrotag sequence study conducted at Kerguelen Islands and the Antarctic Peninsula (Ghiglione and Murray, 2012). Archaea, first described in Antarctic waters almost 20 years ago (DeLong et al., 1994), have a distinct seasonal cycle in which Marine Group I crenarchaeota (MGI hereafter) are abundant in late winter surface Antarctic waters (Murray et al., 1998; Church et al., 2003). These findings contrast with the original studies reporting that these organisms were dominant in deeper ocean habitats (Fuhrman et al., 1992; Karner et al., 2001). The MGI patterns in the Arctic also contrast with the Antarctic. In the Arctic, seasonal patterns were not identified, though the MGI were the most dominant archaeal group in surface waters (Galand et al., 2009). Low organic matter levels and deep mixing in winter could be major drivers in shifting bacterioplankton community composition and metabolic potential; this may, as a result, impact the global carbon cycle—a possibility that has largely gone unnoticed due to lack of studies in winter. Similarly, little is known about possible transformations in inorganic nutrient pools due to winter microbial processes. Southern Ocean environmental genomic studies focusing on identifying organisms and metabolic capabilities of the microbial community are limited (Béja et al., 2002; Grzymski et al., 2006), but promise to revise our understanding of the plankton dynamics.
Motivated by observations of strong temporal shifts in the Antarctic bacterioplankton community, we were interested in better defining the community diversity and genome-encoded capabilities at two contrasting times of the annual cycle. Here, we analyzed small subunit ribosomal RNA (SSU rRNA) gene sequences, end sequences from large insert (40 kb) environmental clones and a subset of the fully sequenced environmental clones from winter and summer near-surface seawater samples collected in the coastal waters off Anvers Island, Antarctic Peninsula. In addition, we interpreted our data in light of a companion metaproteomic study conducted during the summer and winter of 2008 at the same location near Anvers Island. This area is experiencing rapid regional warming (Vaughan et al., 2003) and recently was shown to have reduced levels in summer primary productivity due to decreases in winter sea-ice cover (Montes-Hugo et al., 2009).
Materials and methods
Sample collection
Seawater samples were collected from coastal surface waters at Palmer Station, on the west coast of the Antarctic Peninsula. The winter metagenomic library was created from the microorganisms in the 1.6–0.2 μm seawater fraction (50 l) collected on 20 August 2002 in the nearshore surface waters of Arthur Harbor, Anvers Island (−64° 46.433 S, −64° 03.269 W). Samples were collected directly from the station's seawater intake at 6 m depth 16 m from the shore (water column is 7 m). The summer metagenomic library was created from the microorganisms in the 1.6–0.2 μm seawater fraction (100 l) collected on 28 February 2006 about 500 m offshore (−64° 47.009 S, −64° 04.656 W, 10 m depth, near LTER Station B, accessed by Zodiac). Summer sampling was accomplished using a submersible pump system and acid washed silicone tubing. In both cases, a GF/A Whatman filter (Millipore, Billerica, MA, USA) was used to screen larger organisms and the picoplankton were collected onto a 0.2 μm Sterivex (Millipore) filter. The summer sample was processed initially using a Pellicon tangential flow filtration system (50 KD cassettes) for rapid concentration of the<1.6 μm fraction, and then the>0.2 μm fraction was collected on Sterivex filters.
Physiochemical data accompanying bacterioplankton sample from winter 2002 only included temperature and chlorophyll a due to limited sampling capabilities (seawater temperature, −1.73 °C and chlorophyll a concentration, 0.073 μg l−1). Associated data for the summer 2006 sample seawater physical, chemical and biological properties were determined using standard LTER methods (http://pal.lternet.edu/publications/documents/protocols/) and include the following: seawater temperature, 2.2 °C; salinity, 33.38 PSU; nutrient concentrations PO4−2, 1.02 μℳ; Si(OH)4, 64.66 μℳ, and NO2−+NO3−, 14.93 μℳ; dissolved organic carbon: 48.70 μℳ; bacterioplankton cell abundance, 1.02 106 cells ml−1; leucine incorporation rate, 112 pM hr−1; thymidine incorporation rate, 4.6 pℳ h−1; chlorophyll a concentration, 3.0 μg l−1; and primary production of 35.8 mg C m−3 d−1.
Nucleic acid preparation
DNA for the fosmid libraries and SSU rRNA libraries was prepared according to instructions from the Joint Genome Institute (JGI; http://www.jgi.doe.gov/sequencing/protocols/prots_production.html). Briefly, high-molecular-weight DNA was extracted from Sterivex filters using a sucrose buffer, phenol/chloroform extraction following Massana et al. (1997). High-molecular-weight DNA was treated with RNase, verified by gel electrophoresis and sent to the JGI for fosmid library preparation and sequencing.
SSU rRNA gene libraries and analysis
Bacterial and archaeal SSU rRNA gene clone libraries were prepared following a JGI protocol (http://my.jgi.doe.gov/general/protocols/SOP_16S18S_rRNA_PCR_Library_ Creation.pdf) from the same high-molecular-weight DNAs that the metagenomic libraries were prepared. Clone sequences were assembled, aligned against the Greengenes database (DeSantis et al., 2006) and chimeras were identified using Mallard (Ashelford et al., 2006) and removed from the data set. Sequences in the larger of the two libraries (summer, 656) were resampled to the size of the smaller library (winter, 411) using a daisychopper (Gilbert et al., 2009). Diversity statistics for the two bacterial libraries and single winter archaeal library were generated using Mothur (Schloss et al., 2009). SSU rRNA gene sequences were deposited in GenBank and have accession numbers GU234182-GU235864.
Phylogenetic analysis
Neighboring ribosomal RNA gene sequences were identified and aligned using the nearest alignment space termination alignment tool (DeSantis et al., 2006). The evolutionary history of nearly full-length SSU rRNA gene sequences was inferred using neighbor-joining (complete deletion for the bacterial analysis and pairwise deletion for the archaeal analysis) in MEGA4 (Tamura et al., 2007). The bootstrap consensus trees inferred from 1000 replicates are shown. Evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al., 2004) and are in units of the number of base substitutions per site. There were a total of 1075 positions in the final bacterial data set and 1474 positions in the archaeal data set.
Amino acid alignments for ribulose bisphosphate carboxylase/oxygenase (RuBisCO) sequences identified in the metagenome and neighboring sequences identified with BLASTP were created in ClustalX (Larkin et al., 2007). The evolutionary history was inferred using the Minimum Evolution method and a bootstrap consensus tree inferred from 1000 replicate trees generated using a neighbor-joining algorithm is shown. The analysis was based on 474 aligned positions. The analysis was conducted in MEGA4.
Sequence annotation
Fosmid end sequences and fosmids selected for complete sequencing (288 in total) were assembled, annotated and managed through the Integrated Microbial Genomes-Metagenome (IMG-M) database (Markowitz et al., 2008) at the Joint Genome Institute (JGI). The fosmid end-sequence component of this Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession IDs ADIF00000000 and ADKQ00000000. The versions described in this paper are the first versions, ADIF01000000 and ADKQ01000000. The IMG-M Taxon Object IDs for these two data sets are 2008193000 and 2008193001, respectively. In addition, IMG-M Taxon Object IDs for the fully sequenced fosmids are 2040502004, 2040502005, 2077657013 and 2077657020. The fosmid sequences selected for complete sequencing were used only in genome alignments and 3-hydroxypropionate/4-hydroxybutyrate pathway reconstruction, as their inclusion in comparative genomic analysis would have been highly biased. Custom analyses are detailed below.
MEGAN analysis
Winter Environmental Genome (WEG) and Summer Environmental Genome (SEG) end-sequences were analyzed and normalized using the software MEGAN according to the recommendations of the authors (Huson et al., 2007) after BlastX analyses run against the July 2011 non redundant and taxonomy database from the National Center for Biotechnology Information. The least common ancestor assignment algorithm had parameters: min support=5; min score=35; top percent=10 and win score=0.
Genome alignments
Nucleotide sequences from winter (including sequences from fully sequenced fosmids) were BLASTn searched against the whole-genome sequences from Candidatus Ruthia magnifica and Nitrosopumilus maritimus. Matches with e values less than 10−10 were plotted on the circle graph using the software CIRCOS (Krzywinski et al., 2009).
COG analysis
To account for the different sizes of the winter and summer end-sequence data sets, clusters of orthologous groups (COG) distributions were randomly re-sampled 100 times to estimate variance and a Student's two tailed t-test was used to test for significance for each category. Specific COG distributions also were quantified from the boot strapped data. Here, counts with s.d. from the bootstraps are reported. COG data for the end-sequence data sets were also compared using STAMP (Parks and Beiko, 2010).
Metagenome size estimation
Metagenome size estimation for the two end-sequence libraries was calculated using the GAAS software package according to the recommendations of the authors (Angly et al., 2009).
Carbon fixation estimates
Estimates of potential chemolithoautotrophic carbon fixation of 0.05 Gt of carbon were based on autrotrophic half reactions for ammonia and nitrite oxidation and on an assumption that 85% of the electrons went to cellular energetics and 15% went to growth. The final reaction normalized to one mole of NH4+:
NH4++1.8000 NO2−+2.3300 O2+0.3300 CO2+0.0825 HCO3−=2.7500 NO3−+0.8500 H2O+0.0825 C5H7O2N+1.8100 H+
The calculations further assume a Southern Ocean area of 2013 m2, a depth of 100 m and a NH4+ utilization rate (for autotrophy) of 0.1 mmol m−3.
Results and discussion
Phylogenetic differences between winter and summer bacterioplankton SSU rRNA clone libraries
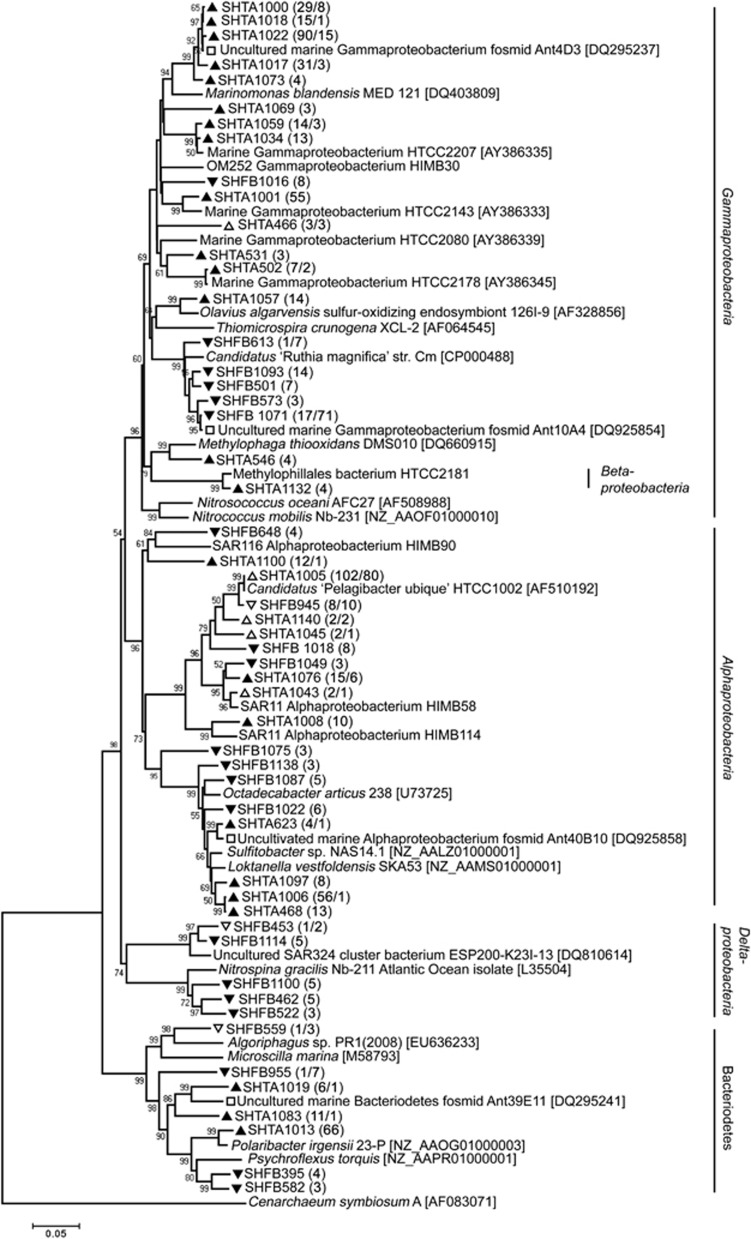
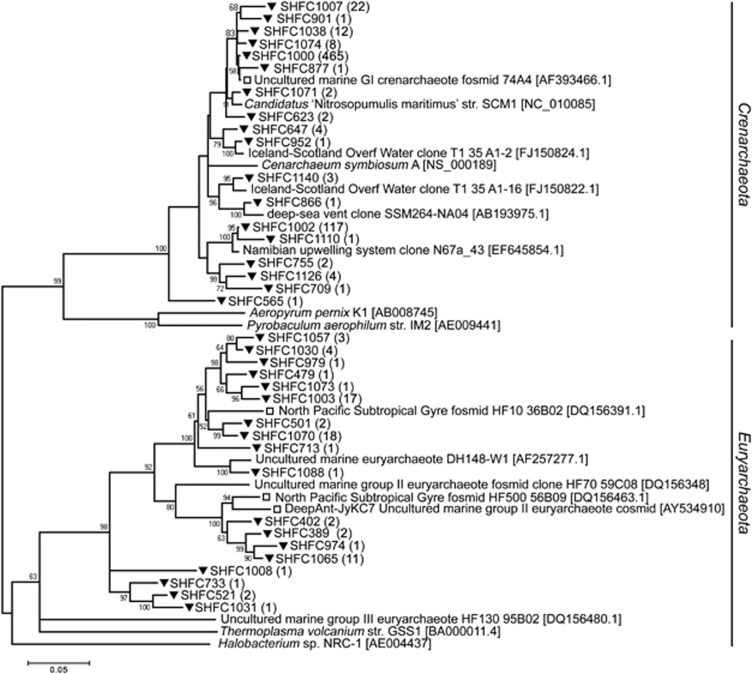
The SSU rRNA gene sequence analysis indicated that planktonic bacterial richness was significantly greater in winter than summer (301 and 183 operational taxonomic units, respectively; Supplementary Figure S1; Supplementary Table S1). This phylogenetic diversity is represented in neighbor-joining trees of bacteria (Figure 1) and archaea (Figure 2). Coverage of diversity was nearly saturating in the summer library (Supplementary Figure S1b), and only 10% of the operational taxonomic unit clusters (distance of 0.03; Supplementary Table S1) overlapped with the winter library. The Sorensen similarity coefficient was 0.72 when community structure based on abundance was taken into consideration, and only 0.18 when just richness was considered (Supplementary Table S1), a result that supports earlier SSU clone library analysis (Murray and Grzymski, 2007). These results are also consistent with a survey of V6 pyrotags from samples collected in January, July and August, 2002 (10 798 tags per sample; the August sample is the same sample analyzed in the present study) in which the similarity (based on abundance) between summer (January) and winter (both July and August) was 0.41 (Ghiglione and Murray, 2012). Though the samples used for SSU rRNA gene and fosmid library sequencing were collected in different years, multiple surveys over the annual cycle (Murray et al., 1998; Murray and Grzymski, 2007) strongly support a reproducible seasonal pattern over the annual cycle.
Figure 1.
Phylogenetic relationships between austral summer and winter SSU rRNA gene sequences. Inferred neighbor-joining tree for Antarctic bacterial SSU rRNA gene sequences and selected, related marine bacterial and environmental genome-derived SSU rRNA gene sequences. Sequences were clustered at a distance of 0.03, and clusters with at least three representatives are shown in the consensus tree (1000 bootstraps). Symbols designate the following groups: filled triangles=summer dominated; filled inverted triangles=winter dominated; open triangles=relatively equal proportions in summer and winter; open squares=previously reported Antarctic environmental genome sequences.
Figure 2.
Inferred neighbor-joining tree for Antarctic winter SSU rRNA gene sequences. As in Figure 1, but in this tree all clusters, including those with a membership of 1, are shown. There were no summer archaeal sequences—archaeal SSU rRNA was not recovered following PCR amplification.
Results from the SSU rRNA gene comparative analysis suggested that SAR11 was the only phylogenetic lineage that did not vary seasonally. This lineage represented ∼20% of sequences in both libraries spread over nine, 0.03 distance clusters. A lack of seasonal patterns in relative abundances in SAR11, and specifically Ca. Pelagibacter ubique, was reported in pyrotag surveys in Kerguelen Islands and the Antarctica Peninsula (Ghiglione and Murray, 2012), and in a study between winter and summer bacterioplankton in the Arctic Ocean. However, studies such as the English Channel (Gilbert et al., 2011) and Sargasso Sea (Carlson et al., 2009), where ecotypes in the SAR11 cluster were more thoroughly studied over the annual cycle, showed ecotype variation associated with peaks in winter or following deep-mixing events—deeper than the surface waters studied here.
A significant component of the winter bacterioplankton (19.7% of the winter library compared with 2.7% of the summer library) were Gammaproteobacteria falling into five closely related 0.03 distance bins that were affiliated with the GSO-EOSA-1 complex (Walsh et al., 2009). This complex encompasses SUP05 (which includes sulfur-oxidizing chemolithotrophic symbionts such as Ca. Ruthia magnifica) and ARCTIC96BD-19 clusters (Figure 1 and Supplementary Figure S1). Members of these clusters have been reported in an increasing number of studies in marine habitats including the Arctic Ocean (ARCTIC96BD-19; Bano and Hollibaugh, 2002), the Antarctic Peninsula (Ant10A4, Murray and Grzymski, 2007), seamount hydrothermal plume waters (SUP05; Sunamura et al., 2004), Chilean oxygen minimum zone (Stevens and Ulloa, 2008), Canadian fjords (Walsh et al., 2009) and mesopelagic ocean (Swan et al., 2011). These clusters were previously thought to be favored in hypoxic waters (for example, Walsh et al., 2009) but are now recognized to be ubiquitous in the ‘dark oxygenated ocean' (Swan et al., 2011). Our results suggest they are widely distributed in Antarctic surface ocean winter waters. Length heterogeneity-PCR surveys conducted in the Scotia Sea (June, 2008) and Weddell Sea (March–April, 2009) also suggested sequence fragments matching Ant10A4 were dominant and ubiquitous representing ∼20% of the signal detected (Murray et al., 2011).
Another hallmark of the winter bacterioplankton community was the presence of archaea (Figure 2 and Supplementary Figure S1). There were very low levels of archaea in summer surface waters, as previously reported (Murray et al., 1998); levels of PCR-amplified DNA were not detectable and thus insufficient for SSU rRNA gene library preparation from the February 2006 sample. The winter archaeal SSU rRNA gene library was dominated (84%, Supplementary Figure S1) with sequences closely related (>99% sequence identity) to genomic clone 74A4 (Béja et al., 2002) that also is related to Ca. N. maritimus, a known chemolithoautotrophic ammonia oxidizer (Francis et al., 2005; Könneke et al., 2005). Proteins that showed high matches to Ca. N. maritimus also dominated the winter metaproteome (30% of identified proteins) but were not detected in the summer (Williams et al., 2012).
Two other winter-only SSU rRNA gene clusters that also are suspected to have chemolithoauthotrophic or mixotrophic lifestyles are the Deltaproteobacteria-affiliated nitrite-oxidizing genus, Nitrospina (three clusters; Watson and Waterbury, 1971) and the Deltaproteobacteria-affiliated SAR324 cluster (two clusters; Swan et al., 2011). Nitrospina were detected in numerous SSU rRNA gene-targeted and genomic studies including waters 200 m and deeper at the HOT Station (DeLong et al., 2006), in 55 m winter waters in the Arctic Ocean (Alonso-Sáez et al., 2010) and in Monterey Bay below 80 m (Suzuki et al., 2004). The distribution of Nitrospina was correlated with planktonic amoA-containing Crenarchaeota in Monterey Bay (Mincer et al., 2007) as they did in this study. The ARCTIC96BD-19 cluster (in which the Ant10A4-affiliated sequences are the dominant member in Antarctic winter waters) and SAR324 organisms may be mixotrophic, based on evidence from single-cell genome sequencing (Swan et al., 2011).
The SSU rRNA data showed that the largest bacterial group in summer (21% compared with 8% of the winter library) was related to uncultivated gammaproteobacterium Ant4D3 (Grzymski et al., 2006), a group with the closest cultivated relatives (Gammaproteobacteria HTCC 2143 and HTCC 2180) with only 91% sequence identity across the SSU rRNA gene. Ant4D3 cluster-associated cells were abundant (10% of total cells, and ½ of the Gammaproteobacteria) and shown to readily incorporate amino acids in combined fluorescent in situ hybridization-microautoradiography studies in Antarctic Peninsula waters (Straza et al., 2010). The other highly represented Gammaproteobacteria in the summer library were affiliated with the Oligotrophic Marine Gammaproteobacteria (Cho and Giovannoni, 2004) including HTCC 2207 and HTCC 2143, which both harbor biosynthetic capacity for carotenoids and proteorhodopsin (Oh et al., 2010). Sequences affiliated with the psychrophilic, proteorhodopsin-containing Polaribacter genus (11% of the summer library) were only detected in summer and potentially could be seeded from melting sea ice. Clusters affiliated with the globally distributed, and metabolically versatile (demethylation of DMSP, aerobic anoxygenic photosynthesis, CO metabolism, sulfur oxidation, and so on) Roseobacter clade, were dominant in the summer SSU rRNA library, compared with the winter library with the most abundant representatives falling into the Sulfitobacter (>6% of the library; SHTA1006 and SHTA468) and Loktanella, genera. These organisms often are found in surface waters, and numerous interactions with phytoplankton have been reported (Moran et al., 2007), supporting their quantitative dominance in the summer bacterioplankton. End-sequences (discussed below) matching Polaribacter-like functional genes are almost exclusively associated with summer, and no winter end-sequences matched Sulfitobacter.
Winter and summer end-sequence library comparisons
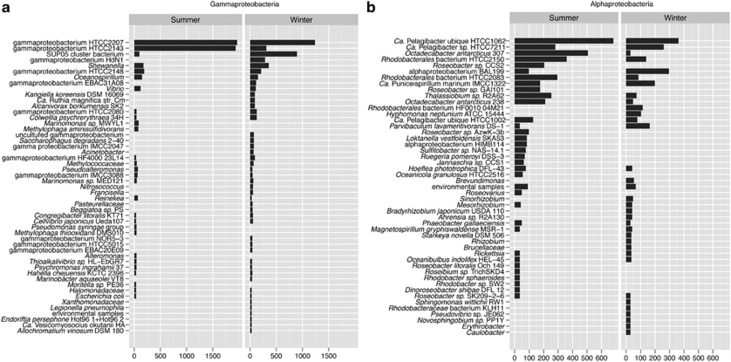
We generated a total of 23.3 Mb of end-sequence data from fosmid libraries prepared from summer (February, SEG) and winter surface waters (August; WEG; Supplementary Table S2). BLASTn and MEGAN were used to characterize the closest phylogenetic match for each end-sequence from the fosmid libraries (Supplementary Figure S2). There are more than three times as many genera occurring predominantly in winter (135) compared with 40 that occur predominantly in summer, according to phylogenetic assignment of end-sequences. Forty-three genera were shared between both libraries. Differences between winter and summer representation of Gammaproteobacteria genera were evident (Figure 3a). End sequences associated with 21 genera were unique to winter compared with only six genera that were unique to summer. End sequences assigned to the SUP05 cluster bacterium (Walsh et al., 2009), a putative sulfur oxidizer, are in the second most abundant lineage following gammaproteobacterium HTCC 2207 in WEG, with only very minor read assignment in the SEG. WEG-only patterns were also prevalent with the other sulfur-oxidizing genera assigned (Ca. Vesicomyosocius okutanii, Ca. R. magnifica, Endoriftia persephone). Conversely, sequences assigned to marine gammaproteobacterium HTCC 2143 were over 6 × more represented in SEG than WEG and were the second most abundant summer Gammaproteobacteria genera with assigned reads. As mentioned above, this isolate has the most closely related SSU rRNA gene sequence to the abundant, Ant4D3 group of bacterioplankton in summer. Thus, unlike the conclusion drawn in Manganelli et al. (2009) for south Drake Passage—that Gammaproteobacteria do not dominate in winter—our results of SSU rRNA and end-sequence analyses suggest that Gammaproteobacteria are abundant year-round in this coastal ecosystem, but the abundances of various genera shift based presumably on metabolic potential of the organisms.
Figure 3.
End-sequence reads assigned to specific genera with bacterial genome sequence representation based on the MEGAN analysis. (a) Reads assigned to Gammaproteobacteria, categorized by genera, from winter and summer end-sequence libraries. These are the genera based on normalized hits—only those genera that had >20 hits are shown for ease of data presentation. (b) Reads assigned to the Alphaproteobacteria, categorized by genera and labeled as in (a). Only those with>20 hits per genera are shown. Full results for both proteobacterial classes are found in Supplementrary Figure S6.
Similarly, given their common association with phytoplankton blooms and high coastal abundances (Moran et al., 2007), it was reported that Alphaproteobacteria dominate the summer community (Manganelli et al., 2009). Our analyses of end-sequence data reveal that Alphaproteobacteria are equally abundant in both libraries, though the membership changes (Figure 3b and Supplementary Figure S2). End sequences with high sequence identity to Ca. P. ubique HTCC1062 were almost twice as abundant in the SEG as the WEG, while the strain HTCC 7211 was equally abundant in the libraries. End sequences affiliated with Octadecabacter antarcticus ranked second among Alphaproteobacteria (Figure 3b) and were almost exclusively found in the summer (12% of the ends assigned in all of the Alphaproteobacteria). O. antarcticus, a gas vacuolate bacterium found in Antarctic sea ice and seawater, has a neighboring species in the Arctic (Staley and Gosink, 1999) that is prevalent in Arctic sea ice (Brinkmeyer et al., 2003); the dominance of O. antarcticus-related sequences in summer surface waters, like Polaribacter, suggested that it also could have been seeded from the melting sea ice.
Though the end sequences are distributed among more than 17 genera in the Roseobacter clade and are found in winter and summer, they were more abundant in the summer library (Figure 3)—a result that was also found in the SSU rRNA gene sequence comparisons. The organisms in this clade are ecologically important in coastal waters, have versatile metabolic capabilities as mentioned previously and have close affiliations with phytoplankton (Moran et al., 2007).
The MEGAN analysis also pointed at significant shifts in the end-sequence bins of Bacteroidetes-affiliated organisms. In particular, Polaribacter-related sequences were dominant in summer, while sequences were spread across numerous genera in winter. Polaribacter sp. also have gas vacuoles bacteria and are consistently found as members of sea-ice communities in both the Antarctic and Arctic similar to the Octadecabacter spp. (Staley and Gosink, 1999; Brinkmeyer et al., 2003). The shifts in Bacteroidetes membership and dominance of Polaribacter spp. in SEG were consistent with observations of the SSU rRNA gene analyses (Supplementary Figure S1a). Polaribacter-related proteins were also exclusive to the summer metaproteome (Williams et al., 2012).
We detected seven orthologs of form I RuBisCO (six in the WEG and one in the SEG) that were phylogenetically related to chemolithoautotrophic sulfur and hydrogen oxidizers (Supplementary Figure S3). These sequences are similar (average of 92% identity at the amino acid level) to the form IA RuBisCO sequences reported from mesopelagic Gammaproteobacteria (Swan et al., 2011). There is strong support that these organisms are highly related; sequence identity for the SSU rRNA of Ant10A4 and SCGC AA007-O20 is 99% sequence identity for cbbL is 83% at the nucleotide level and 96% identical at the amino acid level. BLAST was used to compare the AAA007-O20 AprA (adenylyl sulfate reductase, alpha subunit) with the WEG and SEG data sets; five hits (two in the SEG, three in the WEG) were with amino acid identities ∼80%, suggesting the potential for the same capacity in Antarctic Gammaproteobacteria. In addition, the Ant10A4 fosmid sequence is highly syntenous with Monterey Bay BAC clone EBAC080-L31E09.65, which is part of the same ARCTIC96BD-19 group. We were unable to link (via contiguous DNA) the Ant10A4 SSU rRNA gene and the fosmids with the RuBisCO genes.
We found three other cbbL genes in the WEG that were related to Betaproteobacteria and Gammaproteobacteria ammonium oxidizers. Two form IV RuBisCO-like genes (one in summer and the other in winter), which are regarded as non-carboxylating or oxygenating RuBisCOs (Li et al., 2005), were detected that are highly related to an ortholog in Octadecabacter antarctica 307, and several other Roseobacter clade organisms such as Roseobacter sp. MED1913. Their function is currently unknown in these organisms—the enzyme may have enolization capabilities (Li et al., 2005)
Evidence for further metabolic complexity in the WEG (both oxidative and reductive enzymes) included genes important in sulfur metabolism such as sulfate permeases, dissimilatory sulfite reductase, disulfide isomerases and PAPS reductase (Supplementary Table S3). Nitrate reductase, nitrite reductase, nitrous oxide reductase and nitroreductase were also proportionally more abundant in the WEG.
Recruitment of winter chemolithoautotroph environmental genome sequences
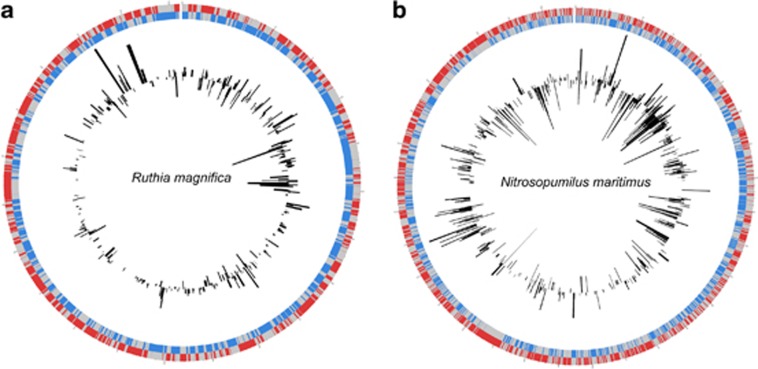
To further investigate the coverage of two organisms prevalent in the WEG, we used recruitment (Rusch et al., 2007) and BLASTn analyses of whole fosmids and fosmid end sequences against genomes from Ca. R. magnifica and Ca. N. maritimus.
More than half (539/976) of the open reading frames of the Ca. R. magnifica genome were recovered from end sequences and fosmid assemblies from the WEG at an average read depth of three (Figure 4a). Genes with a high level of amino acid identity (>55%) include adenylyl-sulfate reductase; the Sox A, Y and B genes; Dsr C, E, F and H genes and carbonic anhydrase. In fact, the entire 15-gene Sox cluster was identified in our data set when searched against Sox loci from a broader group of sulfur-oxidizing bacteria and archaea (Ca. Vesicomyosocius okutanii, Sulfolobus solfataricus, Rhodopseudomonas palustris, Ca. Ruthia magnifica); more than 100 (>0.5%) of the winter end sequences had top hits to one of these Sox genes (e-value<e−10) compared with<0.1% for the SEG. These results support the suggestion that a common metabolic strategy including carbon fixation, sulfur oxidation and possibly mixotrophic growth exists for the SUP05 (Walsh et al., 2009) and ARCTIC96BD-19 clusters (Swan et al., 2011). However, as the free-living organisms in this group have evaded cultivation to date, functional predictions have been limited. The Antarctic bacterioplankton metaproteome survey conducted in 2008 at the same location found Ca. R. magnifica-associated proteins in summer but three times as many in winter (likely reflecting the relative differences in abundance), including detection of AprA during both seasons (Williams et al., submitted).
Figure 4.
Genome alignments of winter end-sequences and whole fosmids to two important chemolithoautotrophic organisms. (a) Circle plot of the chromosome from Ca. R. magnifica. Outer gray bars are the leading strand coding regions; inner gray bars are the lagging strand coding regions. Bar graphs (positive leading strand, negative lagging strand) correspond to matches from the WEG with the height corresponding to read depth (average=3). (b) As in (a), but for the genome of Ca. N. maritimus.
Nearly half (881/1796) of the Ca. N. maritimus genome was recovered from end sequences and fosmid assemblies from the winter library, with an average read depth of three (Figure 4). In particular, sequence matches had a high level of identity to the Ca. N. maritimus genome—more than 100 genes had full-length matches with >86% nucleotide identity (average length 1220, bp). Many genes were affiliated with the 3-hydroxypropionate/4-hydroxybutyrate (3-HP/4-HB) cycle (Figure 5), and the ammonia oxidation and associated nitrogen cycling genes were identified in the WEG (Supplementary Table 5) through homology matches to Ca. N. maritimus (Könneke et al., 2005) and Cenarchaeum symbiosum (Hallam et al., 2006a and 2006b), suggesting that capabilities existed for nitrification, urease utilization and potentially nitrite reduction and nitrous oxide production in the winter MGI.
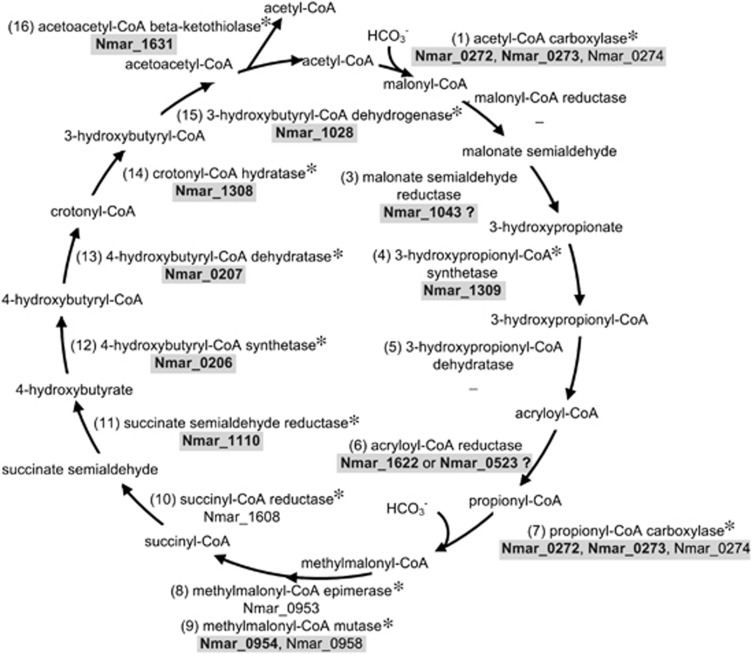
Figure 5.
Carbon flow for 3-hydroxypropionate/4-hydroxybutyrate cycle inferred for the Ca. N. maritimus-like MGI found in Antarctic Peninsula coastal waters. For each enzyme, genes detected in the metagenome are indicated with an asterisk, and proteins detected in the metaproteome are shown in bold with gray background for their respective N. maritimus homologs. Questionable identities for a given enzyme are shown with a question mark. The two carboxylation reactions are at steps (1) and (7).
The results of a companion metaproteome study (Williams et al., 2012) augment these findings particularly with regard to the MGI, due to their strong dominance in winter waters. In addition to identifying most of the genes associated with the 3-HP/4-HB pathway, peptide matches to the majority of the proteins in the 3-HP/4-HB pathway were detected in the winter metaproteome data (highlighted in Figure 5) based on homologs of proteins demonstrated to be involved in this pathway in Metallosphaera sedula or inferred to be involved in this pathway in Ca. N. maritimus (Walker et al., 2010; Williams et al., 2012).
These organisms have a half-saturation constant (133 nℳ) and substrate threshold (<10 nℳ) for ammonium that make them highly suited to the physico-chemical conditions that pervade high-latitude surface waters in winter—low organic carbon, low heterotrophic bacterioplankton and low autotrophic phytoplankton concentrations. The high affinity for ammonium by these organisms (Martens-Habbena et al., 2009) and their consistent presence in Antarctic winter surface waters (Murray et al., 1998) suggest a more dynamic carbon and nitrogen cycle in winter than was previously thought to exist; this is congruent with a similar reinterpretation of the mesopelagic ocean (Swan et al., 2011).
Oligotrophic and copiotrophic signatures in winter and summer COG distributions
COG category distributions revealed broader functional and genome content differences between WEG and SEG (Supplementary Table S4 and Supplementary Figure S4). There were more post-translational modification, protein turnover and chaperone genes (COG category O) found in winter (3.43% vs 2.61% P<0.001), consistent with observations that protein damage under stressful conditions may require frequent protein re-folding (Lauro et al., 2009; Williams et al., 2011). Stress in winter is likely a combination of lower temperature (1–2 °C decrease in winter) and oligotrophic conditions (especially with respect to carbon). The WEG had more cell envelope biogenesis (COG M), coenzyme metabolism (COG H), energy production and conversion (COG C) genes than in summer (Supplementary Table S4). This is consistent with a more metabolically diverse population. There were also significantly more translational proteins (COG category J) in the winter library (5.56% vs 4.62% P<0.001)—consistent with a smaller average genome size for organisms in winter versus summer as the frequency of these conserved proteins is correlated with genome size (Grzymski et al., 2008). This was supported using average genome size estimation re-sampling algorithms (Angly et al., 2009). We calculated the average genome size in winter to be 1.6 Mb (SE=890 Kb) while the average genome size in summer was 1.9 Mb (SE=1.4 Mb). Three of the COG class results were confirmed using the analytical package STAMP (COGs O, H, and J; Parks and Beiko, 2010), which also highlighted specific COG categories with significant differences between the two libraries.
Although SSU rRNA and end-sequence libraries from winter and summer have high proportions of the oligotrophic SAR11, the SEG has more traits found in copiotrophic organisms (Lauro et al., 2009) and in organisms that perform light-harvesting reactions than the WEG. Pathways such as biosynthesis of porphyrin rings—found in bacteriochlorophyll and carotenoid biosynthesis, hydrolytic enzymes (for example, hydrolases and numerous peptidases), DNA repair associated genes (Supplementary Figure S4) and genes involved in iron metabolism (for example, heme utilization) that reflect access to high energy reduced carbon and light—were all prominent in summer (Supplementary Table S3). The presence of pufL and pufM photosynthetic reaction center subunits homologous to Roseovarius sp. 217 provided further evidence for anoxygenic photosynthesis in summer.
The SEG contained numerous phage-encoded genes (for example, phage terminases and site-specific recombinases; Supplementary Table S3, Supplementary Figure S4) implying a more active role of phage in summer when bacterioplankton growth rates and biomass were greater, a finding also reported for the Arctic (Payet and Suttle, 2008) but contradicted observations of winter viral counts from the polar front region (Manganelli et al., 2009). There was 2.2 × the frequency of transcriptional regulators (P<0.001), 2.4 × the frequency of FOG:EAL domain proteins (P<0.001) and a higher frequency of signal transduction proteins (2.0% vs 1.5% P<0.001; also supported by STAMP results, Supplementary Figure S4) in summer compared with winter, findings that are consistent with copiotrophic cells that undergo more cellular responses to environmental stimuli (Lauro et al., 2009).
Genome structure differences between winter and summer
Smaller genomes also tended to have low GC content (student's t-test on marine microbial genomes P<10−6; Firmicutes were an exception, but they are poorly represented in the Antarctic waters studied here). Closer inspection of environmental genome data sets in which the average GC content of WEG and SEG were similar (43.4% and 41.25%, respectively) revealed that the GC distribution was very different, providing additional evidence that bacterioplankton genomes and hence community structure were distinct in each season (Supplementary Figure S5). The GC distribution from summer was bi-modal while there was a much stronger low GC influence in the winter data set attributable to genomes from Archaea, SAR11 and Ruthia-related organisms. The SAR11 cluster of Alphaproteobacteria is the only abundant cluster during both seasons (20% of the total composition each; Figure 3). This is consistent with the metaproteomic data regarding SAR11, with proteins that showed the best matches to the SAR11 clade abundant in both seasons (Williams et al., 2012). The GC distribution of the libraries also revealed significant differences in structure between Antarctic environmental genomes and those from lower latitudes. We found common distribution pattern averaging, ∼34% GC for four Global Ocean Survey surface water data sets (Rusch et al., 2007) that differed from the Hawaii Ocean Time Series surface and deep water libraries (50% and 60%, respectively) (DeLong et al., 2006). The Global Ocean Survey and Hawaii Ocean Time Series patterns were distinct from the Antarctic libraries (Supplementary Figure S5; Kolmogorov–Smirnov test P<10−225). This finding appears to underscore fundamental differences between winter and summer Antarctic environmental genomes and those from mid-latitude surface and deep oceanic bacterioplankton.
A new conceptual model of Antarctic Peninsula surface ocean ecology
Through synthesis of the findings presented here, we have developed a new conceptual model of Antarctic Peninsula surface ocean ecology (Figure 6). This model reflects complexity of the community and versatile metabolic strategies of winter bacterioplankton. We divided the assemblage into three archaeal and bacterial groups: photoheterotrophs, chemolithoautotrophs and chemoheterotrophs. Given very low light levels during winter, proteorhodopsin-based photoheterotrophy would not be expected to be fundamental to winter microbial ecology. Nevertheless, these organisms, especially SAR11, and genes encoding proteorhodopsin (Supplementary Table S3) represented a consistent component in both summer and winter, as in the Arctic (Cottrell and Kirchman, 2009). The role of proteorhodopsin in winter is currently unknown, though expression of this protein was also detected in the metaproteome in the winter library (Williams et al., 2012).
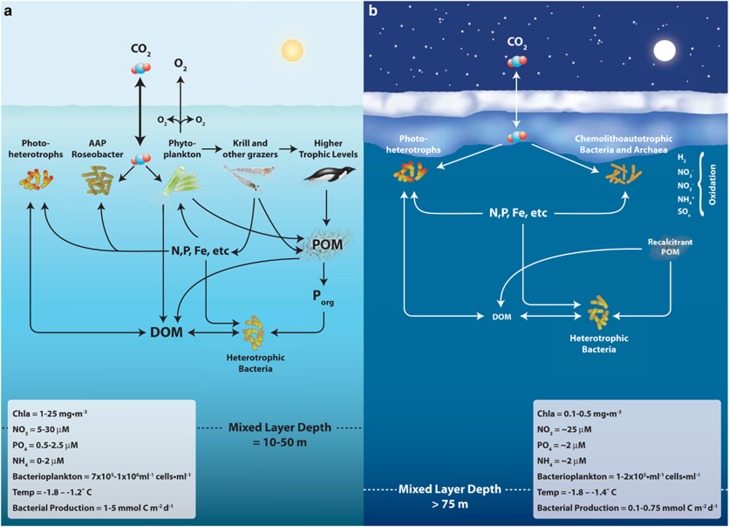
Figure 6.
Schematic comparison of summer and winter surface water food webs from the Antarctic Peninsula region with emphasis on bacterioplankton-driven processes. (a) Summer bacterioplankton-driven processes based on analysis of SSU rRNA gene sequences, end-sequence gene content and metaproteome data. Trophic levels of phytoplankton, krill and higher levels are inserted for reference as they are a major component of surface water summer activity. The major groups of bacterioplankton are photoheterotrophs, anoxygenic photosynthetic Roseobacter and heterotrophic bacteria. Basic chemical, physical and biological properties also are shown and represent the range of data in summer (Materials and methods). (b) As in (a), but for winter bacterioplankton-driven processes. The dominant groups shift significantly and are comprised of chemolithoautotrophic bacteria and archaea, organisms that have photoheterotrophic capabilities and heterotrophic bacteria. Chemical, physical and biological properties are shown and represent the range of data in winter (Materials and methods).
Summer bacterioplankton are divided into three functional groups in our model: (1) photoheterotrophs, dominated by proteorhodopsin-containing SAR11, SAR92 (for example, HTCC 2207 and HTCC 2143) and Flavobacteria related to Polaribacter irgensii; (2) aerobic, anoxygenic photosynthetic Rhodobacterales and some Gammaproteobacteria; and (3) a diverse group of putative heterotrophic bacteria that includes members of the Roseobacter clade, Gammaproteobacteria related to Shewanella, Vibrio and Oceanospirillum genera and an abundant cluster of Gammaproteobacteria related to the genome fragment Ant4D3, which is also numerically important in this region and actively incorporates amino acids (Straza et al., 2010). Given the complexities and diversity of genes from metabolic pathways but the lack of experimental data on these largely, uncultivated organisms, it is likely that organisms can fall into multiple functional groups.
We found a winter signal consistent with bacterial and archaeal chemolithoautotrophy in the GC specific BLAST results, SSU rRNA gene libraries and genome recruitment to Ca. N. maritimus and Ca. R. magnifica—recovering about 50% of the genome of each. The environmental genome also contained genes for multiple means of carbon fixation and sulfur and nitrogen transformations—not all of which can be annotated at present. For example, a small group of chemolithoautotrophic bacterial nitrite oxidizer SSU rRNA genes related to Nitrospina sp. and exclusive to winter was detected, but our ability to identify Nitrospina-affiliated end-sequences was limited due to lack of related sequenced genomes. A diverse community of photoheterotrophic and chemoheterotrophic bacteria was also present in the surface waters that included SAR11, Roseobacter and the Gammaproteobacteria related to Ant4D3, in addition to a large number of rare organisms.
Winter chemolithoautotrophy also could be an important biogeochemical process in high-latitude environments as is now becoming recognized in the mesopelagic zone (Herndl et al., 2005; Swan et al., 2011), and was recently suggested in Drake Passage (Manganelli et al., 2009). The SSU rRNA analysis of known or putative chemolithoautotrophs suggests that between 18 and 37% of the community has the potential for chemolithoautotrophy compared with only 1% in summer. If we make the broad assumption that standing stocks of ammonia-oxidizing archaea and nitrite oxidizers are ∼0.5 × 105 cells ml−1 each (cf. Murray et al., 1998) and NH4+ utilization for autotrophy is ∼0.1 mMol m−3, CO2 fixation in the Southern Ocean (South of 60°S) could exceed 0.05 Gt of carbon in the upper 100 m of the winter water column. This estimate is nearly identical to bacterial chemolithoautotrophic productivity estimated by Manganelli et al. (2009) using carbon production measurements (and an assumption that between 10 and 20% come from chemolithoautotrophs). Export production estimates in the Southern Ocean from temperature and net photosynthesis models are estimated at only 0.6–1.3 Gt C per year (Laws et al., 2000).
Resolving the biogeochemical impacts of even moderate winter bacterial and archaeal primary production is vital to better parameterizing ocean-scale models of carbon and nitrogen. Winter nitrification extending across the Southern Ocean and from surface to mesopelagic waters (Swan et al., 2011) likely will have an impact on the ratio of available N:P in surface waters. These observations could explain the excess N (N*) seen in the Southern Ocean (Deutsch et al., 2007) and help rectify discrepancies in surface ocean N budgets given measurements of nitrogen fixation and denitrification.
Conclusion
We found that the most noteworthy change in the bacterioplankton community in nearshore surface waters of the Antarctic Peninsula was the presence of chemolithoautotrophic organisms in winter and their virtual absence in summer when incident solar irradiance is at a maximum and primary productivity is high. This distinction significantly differentiates the central metabolisms between the two seasons as metabolism shifts from a (CH2O)n/O2 redox couple to one where CO2 fixation is coupled to oxidation of NH4+ to NO2− followed by subsequent oxidation of NO2− to NO3− is energetically favorable. Seasonal differences are likely to be even greater in Southern Ocean waters at higher latitudes than our study site at 64° South. The winter bacterioplankton community, once thought to be merely a subset of the summer population in starvation/survival mode (c.f. Kottmeier and Sullivan, 1987; Karl, 1993), was more phylogenetically and functionally diverse than the community that thrives during summer (Figures 1 and 4, and Supplementary Figure 1). Our results contribute to a new realization of the shifts that occur in the microbial community each year as the physical/chemical environment changes. This new understanding requires further study, especially given rapid reductions in sea-ice cover and climate occurring at high latitudes (Stammerjohn et al., 2008). The boom or bust interpretation of high-latitude microbial ecology is in need of revision.
Acknowledgments
We extend deep appreciation to Palmer Station personnel in the US Antarctic Program for collecting the samples used for creation of the winter fosmid library. The study would not have been possible without the JGI community sequencing program supported by the Department of Energy where library construction, DNA sequencing and automated annotation pipeline were performed. In particular, we thank Kerrie Barry, Susannah Tringe, Jim Bristow and Edward Rubin of the JGI. CSR was supported by an NSF Postdoctoral Fellowship in Biological Informatics (DBI-0532893). The research was supported by National Science Foundation awards: ANT 0632389 (to AEM and JJG), and ANT 0632278 and 0217282 (to HWD), all from the Antarctic Organisms and Ecosystems Program. We thank K Myers for assistance in the field. The Australian Research Council supported research of the Australian contingent.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alonso-Sáez L, Galand PE, Casamayor EO, Pedrós-Alió C, Bertilsson S. High bicarbonate assimilation in the dark by Arctic bacteria. ISME J. 2010;4:1581–1590. doi: 10.1038/ismej.2010.69. [DOI] [PubMed] [Google Scholar]
- Angly FE, Willner D, Prieto-Davó A, Edwards R, Schmieder R, Vega-Thurber R, et al. The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes. PLoS Comp Biol. 2009;5:e1000593. doi: 10.1371/journal.pcbi.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol. 2006;72:5734–5741. doi: 10.1128/AEM.00556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bano N, Hollibaugh T. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl Environ Microbiol. 2002;68:505–518. doi: 10.1128/AEM.68.2.505-518.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmeyer R, Knittel K, Ju J, Weyland H, Amann R, Helmke E. Diversity and structure of bacterial communities in Arctic versus Antarctic pack ice. Appl Env Microiol. 2003;69:6610–6619. doi: 10.1128/AEM.69.11.6610-6619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béja O, Koonin EV, Aravind L, Taylor LT, Seitz H, Stein JL, et al. Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Appl Environ Microbiol. 2002;68:335–345. doi: 10.1128/AEM.68.1.335-345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 2009;3:283–295. doi: 10.1038/ismej.2008.117. [DOI] [PubMed] [Google Scholar]
- Cho J-C, Giovannoni SJ. Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl Environ Microbiol. 2004;70:432–440. doi: 10.1128/AEM.70.1.432-440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church MJ, Delong EF, Ducklow HW, Karner MB, Preston CM, Karl DM. Abundance and distribution of planktonic Archaea and Bacteria in the waters west of the Antarctic Peninsula. Limnol Oceanogr. 2003;48:1893–1902. [Google Scholar]
- Coale KH, Johnson KS, Chavez FP, Buesseler KO, Barber RT, Brzezinski MA, et al. Southern ocean iron enrichment experiment: carbon cycling in high- and low-Si waters. Science. 2004;304:408–414. doi: 10.1126/science.1089778. [DOI] [PubMed] [Google Scholar]
- Cottrell MT, Kirchman DL. Photoheterotrophic microbes in the Arctic Ocean in summer and winter. Appl Environ Microbiol. 2009;75:4958–4966. doi: 10.1128/AEM.00117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delille D, Mallard L, Rosiers C. Inter-annual variability in marine coastal Antarctic bacterioplankton. Polar Biol. 1996;16:19–25. [Google Scholar]
- DeLong EF, Wu KY, Prézelin BB, Jovine RV. High abundance of Archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N-U, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch C, Sarmiento JL, Sigman DM, Gruber N, Dunne JP. Spatial coupling of nitrogen inputs and losses in the ocean. Nature. 2007;445:163–167. doi: 10.1038/nature05392. [DOI] [PubMed] [Google Scholar]
- Ducklow H, Carlson C, Church M, Kirchman D, Smith D, Steward G. The seasonal development of the bacterioplankton bloom in the Ross Sea, Antarctica, 1994–1997. Deep Sea Res II. 2001;48:4199–4221. [Google Scholar]
- Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Azam F. Bacterioplankton secondary production estimates for coastal waters of British Columbia, Antarctica, and California. Appl Environ Microbiol. 1980;39:1085–1095. doi: 10.1128/aem.39.6.1085-1095.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, McCallum K, Davis A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C. Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J. 2009;3:860–869. doi: 10.1038/ismej.2009.23. [DOI] [PubMed] [Google Scholar]
- Ghiglione JF, Murray AE. Pronounced summer to winter differences and higher wintertime richness in coastal Antarctic marine bacterioplankton. Environ Microbiol. 2012;14:617–629. doi: 10.1111/j.1462-2920.2011.02601.x. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T, et al. The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol. 2009;11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B, et al. Defining seasonal marine microbial community dynamics. ISME J. 2011;6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzymski JJ, Carter BJ, DeLong EF, Feldman RA, Ghadiri A, Murray AE. Comparative genomics of DNA fragments from six Antarctic marine planktonic bacteria. Appl Environ Microbiol. 2006;72:1532–1541. doi: 10.1128/AEM.72.2.1532-1541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzymski JJ, Murray AE, Campbell BJ, Kaplarevic M, Gao GR, Lee C, et al. Metagenome analysis of an extreme microbial symbiosis reveals eurythermal adaptation and metabolic flexibility. Proc Natl Acad Sci USA. 2008;105:17516–17521. doi: 10.1073/pnas.0802782105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y-ichi, Sugahara J, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci USA. 2006a;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006b;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A, et al. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl D.1993Microbial processes in the Southern OceanIn: Friedmann E (ed). Antarctic Microbiology John Wiley & Sons, Inc.: New York; 634 [Google Scholar]
- Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- Kottmeier S, Sullivan C. Late winter primary productivity and bacterial production in sea ice and seawater west of the Antarctic Peninsula. Mar Ecol Prog Ser. 1987;36:287–298. [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl D. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Larkin M, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, et al. The genomic basis of trophic strategy in marine bacteria. Proc Natl Acad Sci USA. 2009;106:15527–15533. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laws EA, Falkowski PG, Smith WO, Ducklow H, McCarthy JJ. Temperature effects on export production in the open ocean. Global Biogeochem Cycles. 2000;14:1231. [Google Scholar]
- Li H, Sawaya MR, Tabita FR, Eisenberg D. Crystal structure of a RuBisCO-like protein from the green sulfur bacterium Chlorobium tepidum. Structure. 2005;13:779–789. doi: 10.1016/j.str.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Manganelli M, Malfatti F, Samo TJ, Mitchell BG, Wang H, Azam F. Major role of microbes in carbon fluxes during austral winter in the Southern Drake Passage. PloS one. 2009;4:e6941. doi: 10.1371/journal.pone.0006941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Szeto E, Palaniappan K, Grechkin Y, Chu K, Chen I-MA, et al. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 2008;36:D528–D533. doi: 10.1093/nar/gkm846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens-Habbena Willm, Berube PM, Hidetoshi Urakawa, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying archaea and bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- Massana R, Murray aE, Preston CM, DeLong EF. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007;9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- Montes-Hugo M, Doney SC, Ducklow HW, Fraser W, Martinson D, Stammerjohn SE, et al. Recent changes in phytoplankton communities associated with rapid regional climate change along the western Antarctic Peninsula. Science. 2009;323:1470–1473. doi: 10.1126/science.1164533. [DOI] [PubMed] [Google Scholar]
- Moran MA, Belas R, Schell MA, González JM, Sun F, Sun S, et al. Ecological genomics of marine Roseobacters. Appl Environ Microbiol. 2007;73:4559–4569. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AE, Grzymski JJ. Diversity and genomics of Antarctic marine micro-organisms. Phil Trans Roy Soc B. 2007;362:2259–2271. doi: 10.1098/rstb.2006.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AE, Preston CM, Massana R, Taylor LT, Blakis A, Wu K, et al. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AE, Peng V, Tyler C, Wagh P. Marine bacterioplankton biomass, activity and community structure in the vicinity of Antarctic icebergs. Deep Sea Res II. 2011;58:1407–1421. [Google Scholar]
- Oh H-M, Kang I, Ferriera S, Giovannoni SJ, Cho J-C. Genome sequence of the oligotrophic marine Gammaproteobacterium HTCC2143, isolated from the Oregon coast. J Bact. 2010;192:4530–4531. doi: 10.1128/JB.00683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26:715–721. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- Payet J, Suttle C. Physical and biological correlates of virus dynamics in the southern Beaufort Sea and Amundsen Gulf. J Mar Sys. 2008;74:933–945. [Google Scholar]
- Pearce I, Davidson AT, Bell EM, Wright S. Seasonal changes in the concentration and metabolic activity of bacteria and viruses at an Antarctic coastal site. Aquatic Micro Ecol. 2007;47:11–23. [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JT, Gosink JJ. Poles apart: biodiversity and biogeography of sea-ice bacteria. Ann Rev Microbiol. 1999;53:189–215. doi: 10.1146/annurev.micro.53.1.189. [DOI] [PubMed] [Google Scholar]
- Stammerjohn SE, Martinson DG, Smith RC, Yuan X, Rind D. Trends in Antarctic annual sea ice retreat and advance and their relation to El Niño–Southern Oscillation and Southern Annular Mode variability. J Geophys Res. 2008;113:1–20. [Google Scholar]
- Stevens H, Ulloa O. Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ Microbiol. 2008;10:1244–1259. doi: 10.1111/j.1462-2920.2007.01539.x. [DOI] [PubMed] [Google Scholar]
- Straza TR, Ducklow HW, Murray AE, Kirchman DL. Abundance and single-cell activity of bacterial groups in Antarctic coastal waters. Limnol Oceanogr. 2010;55:2526–2536. [Google Scholar]
- Sunamura M, Higashi Y, Miyako C, Ishibashi J-I, Maruyama A. Two bacteria phylotypes are predominant in the Suiyo Seamount hydrothermal plume. Appl Environ Microbiol. 2004;70:1190–1198. doi: 10.1128/AEM.70.2.1190-1198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MT, Preston CM, Béjà O, de la Torre JR, Steward GF, DeLong EF. Phylogenetic screening of ribosomal RNA gene-containing clones in bacterial artificial chromosome (BAC) libraries from different depths in Monterey Bay. Microb Ecol. 2004;48:473–488. doi: 10.1007/s00248-004-0213-5. [DOI] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba a, Woyke T, Lamy D, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science. 2011;333:1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan DG, Marshall GJ, Connolley WM, Parkinson C, Mulvaney R, Hodgson DA, et al. Recent rapid regional climate warming on the Antarctic peninsula. Climatic Change. 2003;60:243–274. [Google Scholar]
- Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh Da, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG, et al. Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science. 2009;326:578–582. doi: 10.1126/science.1175309. [DOI] [PubMed] [Google Scholar]
- Williams TJ, Lauro FM, Ertan H, Burg DW, Poljak A, Raftery MJ, et al. Defining the response of a microorganism to temperatures that span its complete growth temperature range (−2 °C to 28 °C) using multiplex quantitative proteomics. Environ Microbiol. 2011;13:2186–2203. doi: 10.1111/j.1462-2920.2011.02467.x. [DOI] [PubMed] [Google Scholar]
- Williams TJ, Long E, Evans F, DeMaere MZ, Lauro FM, Raftery MJ, et al. A metaproteomic assessment of winter and summer bacterioplankton from Antarctica Peninsula coastal surface waters ISME Je-pub ahead of print 12 April 2012; doi: 10.1038/ismej.2012.28 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.