Abstract
Mechanotransduction plays a crucial role in physiology. Biological processes including sensing touch and sound waves require yet unidentified cation channels that detect pressure. Mouse piezo1 (mpiezo1) and mpiezo2 induce mechanically activated cationic currents in cells; however, it is unknown if piezos are pore-forming ion channels or modulate ion channels. We show that Drosophila piezo (dpiezo) also induces mechanically activated currents in cells, but through channels with remarkably distinct pore properties including sensitivity to the pore blocker ruthenium red and single channel conductances. mpiezo1 assembles as a ~1.2 million-Dalton homo-oligomer, with no evidence of other proteins in this complex. Finally, purified mpiezo1 reconstituted into asymmetric lipid bilayers and liposomes forms ruthenium red-sensitive ion channels. These data demonstrate that piezos are an evolutionarily conserved ion channel family involved in mechanotransduction.
Introduction
Mechanically-activated (MA) currents have been described in various mammalian cells, including inner ear hair cells1, somatosensory dorsal root ganglion neurons2, vascular smooth muscle cells3, and kidney primary epithelia4. The majority of these MA currents are cationic with Ca2+-permeability, leading to a search for cation channels able to convert mechanical forces into such currents. Few MA channels have been described to date5-7; however, none of the candidates have been shown convincingly to mediate the physiological relevant non-selective cationic MA currents in mammals.
Mouse piezo1 (mpiezo1) was recently identified as a protein required for MA currents in a mammalian cell line. Expressing mpiezo1 or related mpiezo2 in a variety of mammalian cell lines induces large MA cationic currents8. mpiezo1-induced currents are inhibited by GsMTx4, a toxin widely used to study MA channels9. Piezo1 and piezo2 contain over 30 putative transmembrane domains and do not resemble known ion channels or other protein classes. Piezo proteins could be non-conducting subunits of cationic ion channels required for proper expression or for modulating channel properties10-12. Alternatively, piezo proteins may define a novel class of ion channels involved in mechanotransduction.
Mechanosensitivity of dpiezo
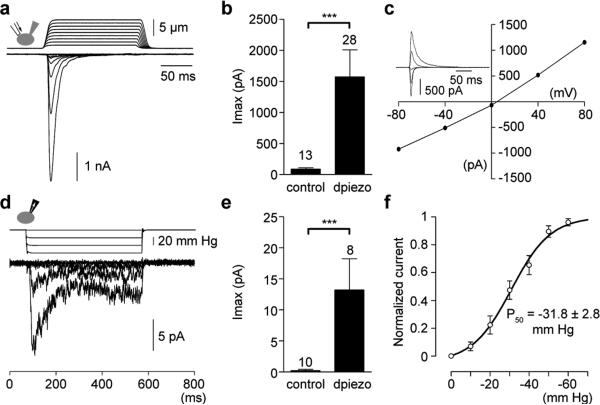
Piezo sequences are present in the genomes of many animal, plant, and other eukaryotic species. Functional analysis of piezos from distant species could demonstrate a conserved role of these proteins in mechanotransduction; furthermore, a comparative analysis of MA currents could elucidate unique pore properties of channels induced by piezos from distinct species. We focused on the apparently single member of D. melanogaster piezo (dpiezo), as this distant invertebrate species is widely used to study mechanotransduction using genetic approaches13-17. dpiezo is 24% identical to mammalian piezos, with sequence conservation throughout the length of the proteins (Supplementary Fig. 1). We cloned the full length dPiezo cDNA into pIRES2-EGFP vector. We recorded MA currents from fluorescent HEK293T cells expressing dPiezo-pIRES2-EGFP by applying force to the cell surface while monitoring transmembrane currents at constant voltage using patch-clamp recordings in the whole-cell configuration2,18,19. dpiezo, but not mock-transfected cells showed large MA currents (Fig. 1a, b). These currents display a time constant of inactivation τ of 6.2 ± 0.3 ms (n = 32 cells) at -80 mV when fitted with mono-exponential function, which is faster than observed for mpiezo1 (~16 ms) and more comparable to mpiezo2 (~7 ms)8. Similar to its mammalian counterparts, dpiezo- MA currents are characterized by a linear current-voltage relationship with a reversal potential around 0 mV, consistent with it mediating a non-selective cationic conductance (Fig. 1c). We further characterized dpiezo-induced currents in HEK293T cells in response to negative pressure pulses applied through the recording pipette in the cell-attached mode, an alternative mechanosensitivity assay. Overexpression of dpiezo induced stretch-activated currents (Fig. 1d-e) with a pressure for half-maximal activation (P50) of -31.8 ± 2.8 mm Hg (Fig. 1f), similar to the P50 calculated for mpiezo1-induced currents (~30 mm Hg)8. Therefore, mechanosensitivity of the piezo family is conserved in invertebrates. Importantly, we demonstrate the physiological relevance of dpiezo in vivo in an accompanying paper20.
Figure 1. Human cells expressing Drosophila piezo show large mechanically activated currents.
MA currents of dpiezo-expressing HE293T cells recorded in the whole-cell (a-c) or cell-attached (d-f) configuration. (a) Representative traces of MA inward currents at -80 mV in dpiezotransfected cells subjected to a series of mechanical steps in 1 μm increments . (b) Average maximal current amplitude of MA inward currents at -80mV. (c) Representative current-voltage relationship of MA currents in dpiezo-transfected cells. (Inset) MA currents evoked at holding potentials ranging from -80 to +80 mV. (d) Representative currents elicited by negative pipette pressure (0 to -60 mmHg, Δ20 mmHg) in dpiezo-transfected cells. (e) Average maximal current amplitude of stretch activated currents at -80mV. (f) Imax normalized current-pressure relationship of stretch activated currents recorded at -80 mV in dpiezo-transfected cells (n = 8 cells) and fitted with a Boltzmann equation. P50 is the average of P50 values determined for individual cells. Bars represent mean ± s.e.m. and the number of cells tested is shown above bars. *** P<0.001, Mann-Whitney test.
Pore properties of piezos
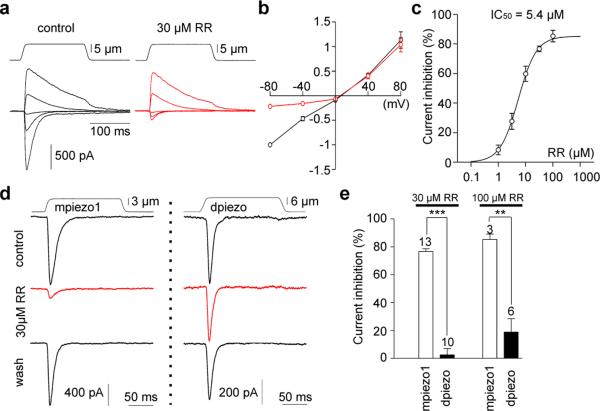
We next compared fundamental permeation properties of mpiezo1- and dpiezo. Ruthenium red (RR), a polycationic pore blocker of TRP channels21,22, blocks mpiezo1- and mpiezo2-induced MA currents8. We found that RR is a voltage-dependent blocker of mpiezo1, with an IC50 value of 5.4 ± 0.9 μM at -80mV (Fig. 2a-c): At a concentration of 30 μM, extracellular RR inhibited inward MA currents without affecting outwards currents. Such voltage-dependence is a characteristic of open channel block. A high concentration of RR (50 μM) included in the pipette solution in the whole cell configuration showed no evidence of block, as large MA currents still displayed a linear current-voltage relationship (Supplementary Fig. 2). These results suggest RR blocks the pore of mpiezo1-induced MA channels from the extracellular side. Remarkably, dpiezo-induced MA currents were insensitive to RR concentrations that potently blocked mPiezo1-induced currents (Fig. 2d-e). Together, these results demonstrate that overexpression of dpiezo or mpiezo1 gives rise to MA channels with distinct channel properties.
Figure 2. Ruthenium red is a channel pore blocker of mpiezo1- but not dpiezo-induced currents.
(a) Representative traces of MA currents in mpiezo1-transfected cells evoked at holding potentials ranging from -80 to +80 mV before (left panel) and during perfusion of 30 μM of RR (right panel, red traces). (b) Average current-voltage relationship of MA currents in mpiezo1-transfected cells (n = 7 cells) before (black symbols) and during (red symbols) perfusion of 30 μM RR. Currents were normalized to the value of control current evoked at -80 mV for each individual cell. (c) Concentration-inhibition curve for RR on MA currents evoked at -80 mV in mpiezo1-transfected cells and fitted with a Boltzmann equation. Each data point is the mean ± s.e.m. of 3-13 observations. (d) Representative traces of piezo-dependent MA currents evoked at -80 mV in the presence of RR. (e) Blocking effect of RR on piezo-dependent MA currents evoked at -80 mV. Bars represent mean ± s.e.m. and the number of cells tested is shown above bars. *** P<0.001; ** P<0.01, unpaired t-test.
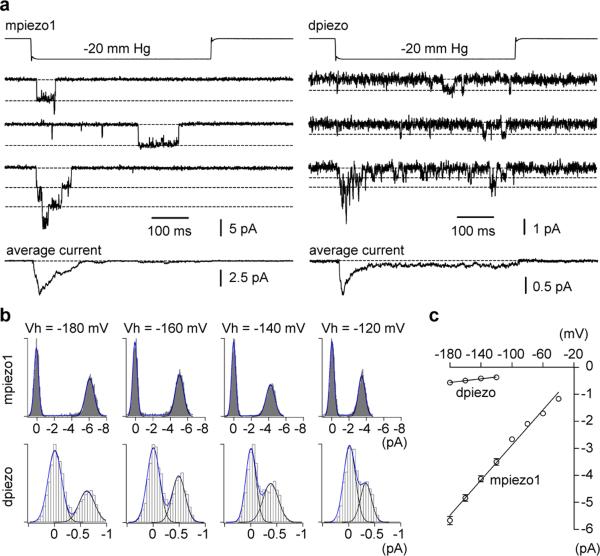
Next, we set out to determine the single channel conductance (γ) of MA channels induced by piezo proteins by using negative-pressure stimulations of membrane patches in cell-attached mode. Figure 3 illustrates the single MA channel properties of mpiezo1 or dpiezo. Openings of stretch-activated channels showed a striking difference in amplitude of single channel currents (Fig. 3a), as determined from the single channel current-voltage relationship for mpiezo1- and dpiezo (Fig. 3b, c). Linear regression of these I-V relationships resulted in slope-conductance values in these recording conditions of 29.9 ± 1.9 and 3.3 ± 0.3 pS for mpiezo1- and dpiezo-induced MA currents, respectively (n = 7 and 5 cells; mean ± s.e.m.). Therefore, dpiezo-dependent channels are 9-fold less conductive than mpiezo1-dependent channels.
Figure 3. mpiezo1- and dpiezo-induced stretch-activated channels have different conductances.
(a) Representative piezo-dependent stretch-activated channel openings elicited at -180 mV. Bottom traces represent average of 40 individual recording traces. (b) All-point histograms of single channel opening events (average of 10 and 20 individual events for mpiezo1 and dpiezo, respectively) at different holding potentials. (c) Average current-voltage relationships of stretch activated single channels in mpiezo1 and dpiezo transfected cells (n = 7 and 5 cells, respectively; mean ± s.e.m.). Single channel amplitude was determined as the amplitude difference in Gaussian fits as shown in b.
mpiezo1 oligomerization
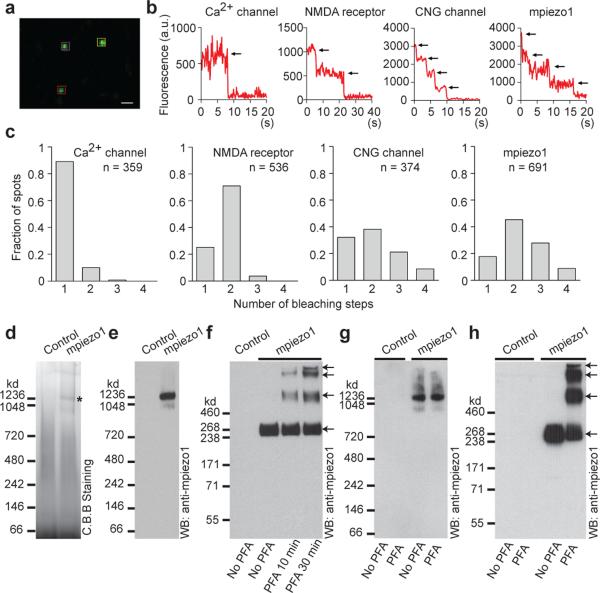
The pore of the majority of ion channels is formed by the assembly of transmembrane domains from distinct subunits (e.g., voltage-gated K+ channels, ligand-gated ion channels) or structurally repetitive domains within a large protein (e.g., voltage gated Na+ and Ca2+ channels). Since piezos lack repetitive transmembrane motifs presumably they oligomerize to form ion channels. To test this hypothesis, we determined the number of subunits in piezo complexes by expressing GFP-mpiezo1 fusion proteins in Xenopus oocytes, imaging individual spots with total internal reflection microscopy (TIRF), and counting discrete photobleaching steps (Fig. 4a-b and ref. 8). N-terminal GFP-mpiezo1 functionality was confirmed by overexpression in HEK293T cells (Supplementary Fig. 3). We used several GFP-fusion constructs of ion channels with known stoichiometry as controls: voltage-gated Ca2+ channel (α1E-GFP; monomer), NMDA receptor (NR1 co-expressed with NR3A-GFP; dimer of dimers), and cyclic nucleotide gated (CNG) channel (XfA4-GFP; tetramer)23. We found that complexes of mpiezo1 frequently exhibited at most four photobleaching steps, consistent with the idea that piezos homo-multimerize. Fluorescent mpiezo1 (or CNG) complexes exhibiting bleaching in fewer than four steps can be explained by non-functional GFP or pre-bleached GFP23 or general bias against noisier multi-step traces during data analysis (see Methods). Histograms of the number of photobleaching steps observed for mpiezo1 complexes were comparable to histograms obtained from tetrameric CNG channels (Fig. 4c). These results suggest that in living cells, piezos assemble as homo-multimers.
Figure 4. mpiezo1 forms tetramers.
(a) Representative image of an acquired sequence showing three selected GFP-mpiezo1 spots in the cell membrane. Levels were adjusted for clarity. Scale bar, 0.8 μm. (b) Representative traces of fluorescence intensities of indicated single GFP-fusion constructs. Black arrows indicate photo-bleaching steps. (c) Histograms of the average number of bleaching steps observed in 10 or more movies from 4 or more oocytes of single fluorescent complexes of indicated constructs. (d,e) Indicated samples purified and separated on native gels and visualized by Coomassie staining (d) or western blotting (e). Asterisk in (d) indicates a strong protein band specifically present in the mpiezo1 sample. (f) Purified mpiezo1-GST proteins treated with or without PFA with the indicated time period, separated on a denaturing gel and detected with the anti-mpiezo1 antibody. Sample purified from cells without transfection served as a negative control. (g, h) mpiezo1-GST-transfected HEK293T cells or untransfected cells treated with or without 0.25% PFA for 10 minutes. The crosslinked mpiezo1-GST proteins were purified and separated on native gel (g) or denaturing gels (h), followed by western blotting. Panels d-h are representatives of at least 3 independent experiments.
We further characterized piezo proteins biochemically by heterologously expressing and purifying mpiezo1 C-terminally fused with a glutathione S-transferase (mpiezo1-GST). Functionality of mpiezo1-GST was confirmed by overexpression in HEK293T cells (Supplementary Fig. 3). We observed a protein band at a position near the 260 kDa protein marker on a Coomassie blue-stained denaturing protein gel (Supplementary Fig. 4a). Western blot with a GST (S. japonicum form) antibody (Supplementary Fig. 4b) or a mpiezo1 specific antibody8 (Fig. 4) confirmed the presence of mpiezo1-GST in the mpiezo1-GST sample. Using native gel electrophoresis and Coomassie blue staining, we detected a prominent protein band at a position near the 1,236 kDa protein marker only in the mpiezo1-GST sample (Fig. 4d). Western blot using mpiezo1 antibody confirmed that this major band contains mpiezo1 (Fig. 4e). These data indicate that the purified mpiezo1-GST protein complex has a molecular weight of about 1.2 million Daltons, four times the predicted molecular weight of a single mpiezo1-GST polypeptide (318 kDa). Next, we asked whether any endogenous proteins are present in this mpiezo1-containing complex. Mass spectrometry of the ~1.2 million Dalton protein complex mainly detected peptides derived from mpiezo1-GST, but not from other endogenous membrane proteins. Although several non-transmembrane proteins were also detected, most of them were also present in the control sample, indicating an absence of specific interacting proteins in the complex (Supplementary Table 1). Moreover, mass spectrometry of the whole purified solution samples prior to gel electrophoresis confirmed that no other ion channel protein was detected (Supplementary Table 2). This argues that mpiezo1 is not tightly associated with any endogenous pore-forming protein.
To further examine whether this piezo complex is indeed a tetramer, we treated the purified mpiezo1-GST protein with the crosslinker paraformaldehyde (PFA) and subjected the samples to denaturing gel electrophoresis and western blotting. PFA-treated samples contained three major additional higher-order piezo containing bands, with longer PFA treatments increasing the prominence of the higher bands (Fig. 4f). The distribution of the bands on the 3-8% gradient gel suggests that the four bands correspond to monomer, dimer, trimer and tetramer of mpiezo1-GST (Fig. 4f). The observation that mpiezo1 is crosslinked by formaldehyde, a crosslinker with a relative short spacer arm (2.3-2.7 Å), suggests that the subunits form a tetramer.
It is possible that mpiezo1 oligomers associate with other proteins; however such an association might not withstand the GST purification step. To probe this, we performed PFA crosslinking experiments on living cells prior to the purification procedure. On a native gel, the mpiezo1-GST complex purified from PFA-treated cells also migrated to the position near the 1236 kDa protein marker, similar to the sample from untreated cells (Fig. 4g). On a denaturing gel, on-cell PFA treatment resulted in four distinct Piezo1-specific bands, similar to results of PFA treatment on the purified complex (Fig. 4h). This suggests that mpiezo1 is not tightly associated with other proteins large enough to discernibly alter its size on denaturing gels, and confirms the results from mass spectrometry. However, our cross-linking studies with paraformaldehyde could miss weak interactors with mpiezo1. Regardless, together with the results obtained from single molecule photobleaching analysis in living cells, our biochemical data suggest that mpiezo1 forms a homomultimeric ion channel, most likely as a homotetramer.
mpiezo1 reconstitution in lipid bilayers
Finally, to assess if piezo proteins were sufficient to recapitulate the channel properties recorded from piezo-overexpressing cells, we reconstituted purified mpiezo1 proteins into lipid bilayers in two distinct configurations: droplet interface lipid bilayers (DIBs) assembled from two monolayers24-26 (Fig. 5a-e, and 5l-q) and proteoliposomes27 (Fig. 5f-i). In the first configuration, mpiezo1 was reconstituted into asymmetric bilayers that mimic the cellular environment: The extracellular facing lipid monolayer is predominantly neutral whereas the intracellular facing leaflet is negatively charged28. In contrast, the lipid composition of the bilayer in the second configuration is uniform.
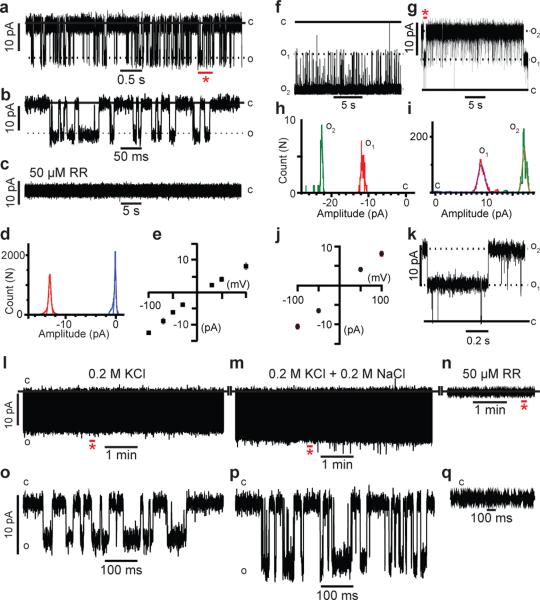
Figure 5. mpiezo1 forms ruthenium red sensitive ion channels.
(a-e) Reconstitution of purified mpiezo1 into asymmetric lipid bilayers. (a) Representative single channel currents at –100 mV. The section of the recordings indicated by the red asterisk is shown in (b) at a 100-fold higher time resolution. (c) After 35 minutes of recording the channel activity shown in (a), injection of 50 μM RR onto the neutral facing compartment blocks mpiezo1 currents. (d) All-event current amplitude histogram of a 6 minute recording; γ = 124 ± 7 pS. The total number of opening events (N) analyzed was 18,424. (e) Single channel current-voltage (IV) relationship, n=6 experiments. (f-k) Reconstitution of purified mpiezo1 into asolectin proteoliposomes. Representative channel currents recorded at –100 mV (f) and +100 mV (g) in presence of 50 μM RR inside the recording pipette. Two open channels are present in the membrane. The segment of the 15 minute recording shown in (g) indicated by the red asterisk is displayed in (k) at a 25-fold higher time resolution. (h,i) All-event current amplitude histograms from a 30 s (h) and 15 min (i) recordings: γ = 110 ± 10 pS (h) and 80 ± 5 pS (i); N was 9,938 events. (j) Single channel I-V relationship, n=8 experiments. (l-q) Representative single channel currents at -100 mV of purified mpiezo1 reconstituted into asymmetric lipid bilayers in symmetric 0.2 M KCl (l), after addition of 0.2 M NaCl (m) and after addition of 50 μM RR (n). Segments indicated by red asterisks in panels l-n are displayed in panels o-q, respectively. C and O denote the closed and open states.
In the DIBs setting, representative segments from a 6 minute recording obtained at –100 mV show brief, discrete channel openings (Fig. 5a, b) blocked by addition of 50 μM RR to the neutral facing compartment (Fig. 5c). In contrast, no effect was observed when RR was introduced into the negative facing compartment (not shown). We detected efficient block of channel activity even at 5 μM RR (not shown). The asymmetric accessibility of RR block of reconstituted channels agrees with the data obtained from mpiezo1-overexpressing HEK293T cells (Fig. 2 and Supplementary Fig. 2), thereby establishing the fidelity of the assays and validating mpiezo1 protein as an authentic ion channel. The piezo currents exhibit ohmic behavior; records displayed at higher resolution (Fig. 5b) clearly demonstrate the occurrence of unitary events with γ values obtained from conductance histograms of 118 ± 15 pS and 80 ± 6 pS (n = 6) in symmetric 0.5 M KCl from the negative and positive branches of I-V plots, respectively (Fig. 5d, e).
A similar pattern of activity was obtained from mpiezo1 reconstituted in asolectin liposomes27 (Fig. 5f-k). A selection of recordings shows the presence of two channels in the membrane which reside predominantly in the open state (Fig.5f, g), as discerned in a higher time resolution display (Fig. 5k) . These recordings were obtained in the presence of 50 μM RR inside the recording pipette, to ensure functional selection of a single population of mpiezo1 channels facing the RR-free compartment. mpiezo1 in asolectin proteoliposomes under these conditions (symmetric 0.2 M KCl) exhibits a γ = 110 ± 10 pS at V = –100 mV and 80 ± 5 pS at V = 100 mV (Fig. 5h-j) (n=8). Finally, reconstitution of control samples purified from nontransfected cells as well as heat-denatured purified mpiezo1-GST into either bilayer systems under otherwise identical conditions failed to reproduce this pattern of channel activity (not shown).
We then tested the ability of the reconstituted mpiezo1 to conduct sodium (Fig. 5 l-q). Initially, single channel currents were recorded from asymmetric bilayers in symmetric 0.2 M KCl; γ = 58 ± 5 pS (Fig. 5 l, o). Subsequent addition of 0.2M NaCl in presence of 0.2M KCl increased the unitary conductance of reconstituted channels to 95 ± 5 pS (Fig. 5 m, p) while retaining sensitivity to RR block (Fig. 5n, q). These results confirm that these channels conduct both sodium and potassium as would be expected from a cationic non-selective channel. This assertion was further substantiated by recording mpiezo1 currents from proteoliposomes under bi-ionic conditions (0.2 M KCl/0.2M NaCl) (Supplementary Fig. 5a-h). A summary of the current–voltage relation for the mpiezo1 channel, extracted from 204,088 events obtained in three experiments, shows that the single channel current is ohmic between –100 and 200 mV with a slope conductance of 102 ± 2 pS (Supplementary Fig. 5i). The current reversed direction at 0.0± 0.3 mV demonstrating that the channel does not select between K+ and Na+, and importantly, displays open channel block by RR (Supplementary Fig. 5j-l).
The difference in γ between overexpressed mpiezo1 in cells and reconstituted mpiezo1 in lipid bilayers may be attributed to many variables, including the distinct lipid environments which are known to strongly influence conductance measurements29-33. Moreover the ionic conditions used in the two systems are different, as divalent cations present in HEK293T cell-attached experiments also affect the conductance values. Indeed, when divalent cations are excluded from the recording pipette, γ of mpiezo1-induced currents in HEK293T cells is 58.0 pS ± 1.5 pS (150 mM NaCl solution, Supplementary Fig. 6), compared to 29.9 ± 1.5 pS in the presence of divalent ions (Fig. 3). The near equivalence of γ values together with the similar pattern of channel activity demonstrates that reconstitution of mpiezo1 into two distinct bilayer systems produces channels with identical functional properties (Supplementary Table 3).
Future reconstitution and recording of dpiezo in lipid bilayers will show whether the difference in conductance between mpiezo1 and dpiezo arises from intrinsic properties. The membrane milieu and lipid composition are known to modulate the activity of the embedded channel proteins in a drastic and deterministic manner [e.g., see29-33]. It is not entirely surprising that the conditions to emulate the cellular environment in the reconstituted system in terms of the mechanical state of the membrane or its lipid composition have thus far been inadequate to retrieve the activation features of MA ion channels. Furthermore, the complexity of protein clusters and dynamic cytoskeletal interacting partners at the cell membrane34 introduce regulatory constraints on channel activity. Further investigation may clarify whether piezo ion channel subunits are intrinsically mechanosensitive or use unknown interacting partners to sense membrane tension.
Conclusion
Here, we provide compelling evidence to support the hypothesis that piezo proteins are indeed ion channels. First, overexpression of dpiezo or mpiezo1 in a human cell line gives rise to MA channels with distinct biophysical and pore-related properties. Second, isolated piezo complexes do not contain detectable amounts of other channel-like proteins. Finally, purified mpiezo1 protein reconstituted into proteoliposomes and planar lipid bilayers in the absence of any other cellular components gives rise to RR-sensitive cationic ion channel activity. The mouse piezo1 complex is estimated to weigh ~1.2 million Daltons with 120-160 transmembrane segments, being, to our knowledge, the largest plasma membrane ion channel complex identified to date.
METHODS SUMMARY
Electrophysiology
Mechanical stimulation was achieved as previously described8.
Subunit counting
The preparations were imaged on an inverted Nikon Ti-E fluorescence TIRF microscope (Nikon Corporation, Melville NY) and imaged with a high numerical aperture objective (Nikon 100x PlanApo, NA1.49). eGFP-fusion proteins were excited with a 488nm Coherent laser (Coherent, Inc., Santa Clara, CA) and images were collected with an Andor iXon DU-897 EMCCD camera.
mpiezo1-GST purification
Cells were collected and lysed 24h after transfection, followed by an affinity purification. Initially, purification was conducted from whole cell lysates. Thereafter, purification was performed using the membrane fraction as starting material, resulting in significantly enhanced frequency of retrieval of channel activity after reconstitution. Untransfected cells were subjected to the same purification procedure to serve as a negative control. Purified samples were kept at 4°C till further analysis.
Native Gel electrophoresis
The purified mpiezo1-GST proteins or negative control samples were subjected to 3-12% NativePAGE Novex Bis-Tris gel for native (non-denaturing) electrophoresis according to the User Manuel (Invitrogen). After electrophoresis, the native gel was then either visualized by a fast coomassie G-250 staining or transferred to a PVDF membrane for western blotting.
Reconstitution in lipid bilayers and proteoliposomes
Purified mpiezo1 was reconstituted into proteoliposomes by detergent dilution. Excised patches from giant asolectin proteoliposomes were used for channel recordings. Asymmetric lipid bilayers were formed using the droplet interface strategy; one monolayer was composed of 1,2-diphytanoyl-sn-glycero-3 phosphocholine (DPhPC), and the other of 90% DPhPC and 10 % of the negatively charged lipid, 1,2-dioleoyl-sn-glycero-3-phosphatidic acid (DOPA) (mole/mole) (Avanti Polar Lipids, Alabaster, AL).
Supplementary Material
Aknowledgments
We thank M.H. Ulbrich for providing Ca2+ channel-, NMDA receptor- and CNG channel- GFP-fusion constructs used as controls for photo-bleaching experiments. This research was supported by grants from the National Institutes of Dental and Craniofacial Research, Neurological Disorders, General Medical Sciences, and by The Genomics Institute of the Novartis Research Foundation. B.X. and J.G. are postdoctoral fellowship recipients from the American Heart Association and the NIH, respectively.
METHODS
Cloning of Drosophila Piezo full-length cDNA
Drosophila Piezo gene was cloned from adult drosophila poly A+ RNAs (Clonetech) by RT-PCR. Primers for RT-PCR were designed based on the annotated sequence of CG8486. Two fragments of 2kb and 6.5kb were amplified and cloned sequentially into pIRES2-EGFP expression vector. Each cloning step was sequence verified. Full-length drosophila Piezo gene is 8355bp in length. The protein sequence of dpiezo is shown in Supplementary Figure 1.
Cell culture and transient transfection
Human Embryonic Kidney 293T (HEK293T), NIH-3T3, F11 and Hela cells were grown in Dulbecco's Modified Eagle Medium containing 4.5 mg.ml-1 glucose, 10% fetal bovine serum, 50 units.ml-1 penicillin and 50 μg.ml-1 streptomycin. Cells were plated onto poly-lysine coated 12-mm round glass coverslips placed in 24-well plates and transfected using lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. 500 to 1000 ng.ml-1 of plasmid DNA was transfected and cells were recorded 12-48 hours later.
Electrophysiology
Patch-clamp experiments were performed in standard whole-cell or cell attached recordings using an Axopatch 200B amplifier (Axon Instruments). Patch pipettes had resistance of 2–3 MΩ when filled with an internal solution consisting of (in mM) 133 CsCl, 10 HEPES, 5 EGTA, 1 CaCl2, 1 MgCl2, 4 MgATP, and 0.4 Na2GTP (pH adjusted to 7.3 with CsOH). The extracellular solution consisted of (in mM) 130 NaCl, 3 KCl, 1 MgCl2, 10 HEPES, 2.5 CaCl2, 10 glucose (pH adjusted to 7.3 with NaOH). For cell attached recordings, pipette were filled with a solution consisting of (in mM) 130 NaCl, 5 KCl, 10 HEPES, 1 CaCl2, 1 MgCl2, 10 TEA-Cl (pH 7.3 with NaOH), excepted for Supplementary Fig. 6 where the internal solution was (in mM) 150 NaCl, 10 HEPES (pH adjusted to 7.3 with NaOH). External solution used to zero the membrane potential consisted of (in mM) 140 KCl, 10 HEPES, 1 MgCl2, 10 glucose (pH 7.3 with KOH). All experiments were done at room temperature. Currents were sampled at 50 or 20 kHz and filtered at 5 or 2 kHz. Voltages were not corrected for a liquid junction potential. Leak currents before mechanical stimulations were subtracted off-line from the current traces. 10 mM ruthenium red stock solution was prepared in water.
Mechanical Stimulation
For whole-cell recordings mechanical stimulation was achieved using a fire-polished glass pipette (tip diameter 3–4 μm) positioned at an angle of 80° to the cell being recorded. Downward movement of the probe toward the cell was driven by a Clampex controlled piezo-electric crystal microstage (E625 LVPZT Controller/Amplifier; Physik Instrumente). The probe was typically positioned ~2 μm from the cell body. The probe had a velocity of 1 μm/ms during the ramp segment of the command for forward motion and the stimulus was applied for 150 ms. To assess the mechanical sensitivity of a cell, a series of mechanical steps in 1 μm increments were applied every 10 to 20 s, which allowed full recovery of mechanosensitive currents. Inward MA currents were recorded at a holding potential of -80 mV. For I-V relationship recordings, voltage steps were applied 0.7 s before the mechanical stimulation from a holding potential of -60 mV.
For cell-attached recordings, membrane patches were stimulated with brief negative pressure pulses through the recording electrode using a Clampex controlled pressure clamp HSPC-1 device (ALA-scientific). Unless otherwise stated, stretch-activated channels were recorded at a holding potential of -80 mV with pressure steps from 0 to -60 mm Hg (-10 mm Hg increments). Current-pressure relationships were fitted with a Boltzmann equation of the form: I(P) = [1 + exp (-(P – P50)/s)]-1, where I is the peak of stretch-activated current at a given pressure, P is the applied patch pressure (in mm Hg), P50 is the pressure value that evoked a current value which is 50% of Imax, and s reflects the current sensitivity to pressure.
Single channel amplitude characterization was performed on patches that showed strong stretch activated current activity at -80 mV using increasing steps of negative pressure up to -60 mmHg. Similar activity was never present in control transfected cells. Negative pressure steps were then reduced to low to moderate level (-5 to -20 mm Hg) allowing detection of single channel openings.
Subunit counting
Oocyte injection
All construct plasmids were linearized at C-terminus with NheI, HindIII or NotI and DNA transcribed with T7 mMessage mMachine Kit (Ambion) and poly(A)-tailing Kit (Ambion) and cleaned with LiCl precipitation. 50 nl of 0.2 μg/μl mRNA was injected into Xenopus oocytes (Nasco).
Acquisition
12-24 hours after injection, oocytes were osmotically shocked in stripping buffer (in mM: 220 N-Methyl Glucamine aspartate, 10 HEPES, 1 MgCl2) and mechanically de-vitellinated. MatTek dishes (MatTek Corporation, Ashland, MA) were prepared by sonication in 1M KOH to remove background fluorescence and further sonicated in MilliQ dH2O. Oocytes were placed onto MatTek dishes into SOS buffer (in mM: 100 NaCl, 2 KCl, 1.8 CaCl2-H2O, 1MgCl2-6H2O, 5 HEPES, 2.5 Na Pyruvate and 50 μg/ml gentamicin, pH 7.0). The preparations were imaged on an inverted Nikon Ti-E fluorescence TIRF microscope (Nikon Corporation, Melville NY) and imaged with a high numerical aperture objective (Nikon 100x PlanApo, NA1.49) with an additional 1.5x Optovar magnification. eGFP-fusion proteins were excited with a 488nm Coherent laser (Coherent, Inc., Santa Clara, CA) and images were collected with an Andor iXon DU-897 EMCCD camera (Belfast, Northern Ireland). Sixty second movies were collected at 100ms exposures, for a frame rate of 10Hz.
Analysis
Using Nikon Elements software, movies were duplicated and processed with a rolling average of 2. A second duplicate was filtered with a low-pass kernel of 7, to remove background. The low-pass images were subtracted from the averaged images, to produce the movies used for analysis. Non-overlapping 4×4 pixel regions of interest were drawn around randomly selected spots that were clearly separated from neighboring bright pixels. The spots were required to fit entirely within the 4x4 pixel regions. Pixel size was 0.11 μm. The average intensity of each region was plotted over the length of the movies. Traces were discarded, a) if the intensity increased after an initial decrease, b) if the fluorescent spot moved out of the region, or c) if the fluorescent signal showed a continuous decay instead of step-wise bleaching. Finally, the number of bleaching steps was counted for each spot.
mpiezo1-GST purification
The mpiezo1-GST construct was subcloned by inserting a GST encoding sequence from Schistosoma japonicum into the mpiezo1 construct8 at the 3’ end of mpiezo1 cDNA sequence using the AscI and SacII restriction enzyme sites. The resulting mpiezo1-GST fusion protein has 2773 amino acids.
After incubationwith cell lysates overnight at 4 degrees, the glutathione beads were washed four times in a buffer containing 25 mM NaPIPES, 140 mM NaCl, 0.6% CHAPS, 0.14% PC, 2.5 mM DTT, and a cocktail of protease inhibitors and eluted with 100 mM glutathione in a buffer containing 25 mM NaPIPES, 50 mM Tris, 0.6% CHAPS, 0.14% PC, 2.5 mM DTT, and a cocktail of protease inhibitors. The eluant was dialyzed against a buffer containing 25 mM NaPIPES, 0.6% CHAPS, 0.14% PC, 2.5 mM DTT, and a cocktail of protease inhibitors. The purified samples were kept at 4°C. Samples purified according to this protocol were used for all the biochemical work and the initial reconstitution experiments. However, since retrieval of channel activity from the reconstituted mpiezo1-GST fluctuated from preparation to preparation, we adopted an alternative purification protocol involving the membrane fraction as the starting material. Specifically, 24 hours after transfection, cells were collected and homogenized in a buffer containing 25 mM NaPIPES, 50 mM NaCl, 2.5 mM DTT, and a cocktail of protease inhibitors. The cell suspension was forced to go through a 25½ G needle for 20 times and centrifuged at 1000 g for 15 min at 4°C. The supernatant was collected and centrifuged at 167,000 g for 30 min at 4°C. The resulting membrane fraction was washed three times (using a buffer containing 25 mM NaPIPES, 150 mM NaCl, 2.5 mM DTT, and a cocktail of protease inhibitors), and used as the starting material for mpiezo1-GST purification using the same procedure described above. Purification from the membrane fraction greatly reduced the content of endogenous GST proteins and significantly enhanced the frequency of retrieval of mpiezo1 channel activity after reconstitution (Fig 5, Supplementary Fig. 5 and Supplementary Table 3).
NativePAGE Novex Bis-Tris Gel
The purified mpiezo1-GST proteins and control samples were subjected to 3-12% NativePAGE Novex Bis-Tris gel for native (non-denaturing) electrophoresis according to the User Manual (Invitrogen). In brief, samples were mixed with NativePAGE Sample Buffer and NativePAGE 5% G-250 Sample Additive and then subjected to electrophoresis at 150 V for 2 hours. The use of G-250 charge-shift in NativePAGE gels results in protein resolution based upon protein size and therefore allows accurate size estimation of native protein complexes 35. However, the native protein conformation may give an expected size estimation error of ~15%. After electrophoresis, the native gel was then either visualized by a fast Coomassie G-250 staining or transferred to a PVDF membrane for western blotting with an antibody specifically against piezo1 proteins.
Paraformaldehyde (PFA) cross-linking
The purified mpiezo1-GST proteins were treated with or without 0.1% PFA at room temperature for different periods of time and then mixed with NuPAGE LDS Sample Buffer and NuPAGE Reducing Agent (Invitrogen), followed by heating at 70°C for 10 min to denature the protein. The treated samples were subjected to 3-8% NuPAGE Tris-Acetate gel electrophoresis under denaturing conditions. For live cell cross-linking, 0.25% concentrations of PFA was added to the cell culture medium and kept at room temperature for 10 min, followed by adding 125 mM glycine to stop the PFA cross-linking reaction. Treated cells were collected and subjected to sequential steps of protein purification, 3-8% NuPAGE Tris-Acetate gel electrophoresis under denaturing conditions or 3-12% NativePAGE Novex Bis-Tris gel for native (non-denaturing) electrophoresis, and western blotting with the anti-piezo1 antibody.
Western blotting
After electrophoresis, either the native or denaturing PAGE gels were transferred to PVDF membranes. Transferring protein from native gel to PVDF membranes was conducted according to instructions for NativePAGE Novex Bis-Tris Gel System. Transferred PVDF membranes were blocked with 5% milk in TBS buffer with 0.1% Tween-20 (TBST buffer) at room temperature for 1 hour, and then incubated with the anti-piezo1 antibody (1:200) at 4°C overnight. The membranes were washed with TBST buffer and incubated with peroxidaseconjugated anti-rabbit IgG secondary antibody (1:10000) at room temperature for 1 hour. Proteins were detected with the ECL plus detection kit (GE Healthcare).
Mass spectrometry
After electrophoresis, purified samples were separated on the 3-12% NativePAGE Novex Bis-Tris gel and visualized by fast Coomassie G-250 staining. The gel band containing the mpiezo1-GST complex or the corresponding blank band from the control sample near the 1236 kDa molecular marker was excised and subjected to the Scripps Center for Mass Spectrometry for analysis. In brief, the gel bands were destained, reduced with 10 mM DTT, alkylated with 55 mM iodoacetamide, and digested with Trypsin overnight before analysis using the nano-LC-MS/MS. The nano-LC-MS/MS data obtained on a LTQ ion trap mass spectrometer was searched using the mpiezo1-GST protein sequence and NCBInr Homo sapiens database. In separate sets of experiments, the purified mpiezo1-GST and control solution samples prior to gel electrophoresis were subjected to mass spectrometry (Supplementary table 2).
Reconstitution into proteoliposomes or droplet interface bilayers (DIBs), single channel recordings and analysis
Purified mpiezo1-GST protein was reconstituted into asolectin (Soybean polar lipid extract, Avanti) liposomes (10 mg/ml) by incubating the mixture (lipid/protein mass ratios between 2000:1 and 1000:1 - this corresponds to a molar lipid/protein ratio of ~800,000-400,000 to 1) on ice for 5 min followed by 20x dilution in 200 mM KCl, 5 mM MOPS pH 7.0 and incubated with rotation at room temperature for 20 minutes. Biobeads were added to mixture and incubated with rotation for 1 hour. Thereafter, biobeads were removed by filtration and a new batch of beads was added. After 30 min incubation, the biobeads were filtered, and the sample was centrifuged at 60,000 rpm for 60 min at 8 °C. The proteoliposome pellet was resuspended in 40 μl of the same buffer and used to place two 25 μl drops on a cover slide. The samples were dried under vacuum for >16 hours at 4 °C. Samples were hydrated with 25 μl of the same buffer and allowed to sit for 2 hours before starting recordings. Thereafter, 2-3 μl of proteoliposomes were withdrawn from the edge of the spots on the cover slide and transferred to the recording chamber. After 5 min, the chamber was slowly filled with recording solution. Multi-GΩ seals were made to proteoliposomes immobilized at the bottom of the recording chamber. At that time, the proteoliposome patch was excised and brought through the air-water interface. Excised patches were used36. Pipette and bath solution contained (in mM) 200 KCl, 5 MOPS titrated to pH 7.0 with KOH. Capillaries of borosilicate glass from Sigma were pulled to yield resistances of 1–2 MΩ when immersed in recording solution.
Droplet interface lipid bilayers (DIBs) were formed between two lipid monolayer-encased aqueous nanoliter droplets submerged in hexadecane 24. Liposomes were composed of 1,2-diphytanoyl-sn-glycero-3 phosphocholine (DPhPC) or 90% (mole/mole) DPhPC and 10 % of the negatively charged lipid, 1,2-dioleoyl-sn-glycero-3-phosphatidic acid (DOPA) (Avanti Polar Lipids, Alabaster, AL). mpiezo1 was diluted directly into the liposome suspension to yield a final concentration of 2 to 5 ng/ml. The electrode carrying the droplet with mpiezo1 and desired buffer-lipid mix (in mM, 500 KCl, 10 HEPES, pH 7.4, 0.5 lipid solution of DPhPC) was connected to the grounded end of the amplifier head-stage (Axopatch 200B). The second electrode, in a droplet containing the same buffer and 10% DOPA: 90% DPhPC, was connected to the working end of the head-stage. Where indicated, RR or 0.2M NaCl was injected using a nano-injector (WPI, Inc).
For proteoliposome patches, records were acquired at a sampling frequency of 40 kHz and filtered online to 5 kHz with a 3-pole Bessel filter before digitization; for DIBs, data acquisition was at 10 kHz and filtered at 2 kHz. For analysis and presentation, records were filtered to 1 kHz with a low-pass Gaussian filter. Transitions were detected by the half-threshold method implemented in Clampfit (proteoliposomes) and by the segmental k-means method (SKM) implemented in QuB (DIBs). Transitions ≤ 0.5 ms were excluded from the pool for analysis to correct for detection of false and missed events. Data were analyzed using Clampfit v.9.2 software (Axon Instruments), QuB, Excel 2007 (Microsoft), and IGOR Pro (Wavemetrics). γ was calculated from Gaussian fits to currents histograms. All statistical values represent means ± s.e.m., unless otherwise indicated. n and N denote number of experiments and number of events, respectively. All experiments were done at room temperature.
Footnotes
Author Informations Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Author Contributions B.C. performed and analyzed electrophysiological experiments. B.X. performed and analyzed biochemical experiments. J.S.S. and R.S. performed the reconstitution experiments and together with M.M. analyzed the single channel data. J.G. and K.S. performed and analyzed photo-bleaching experiments. S.K. cloned dPiezo gene. M.S. initiated biochemical experiments. J.M. generated GFP-mPiezo1 and the mRNA used for oocyte injection. A.D. provided technical help for oocyte experiments. A.P., B.C., B.X., J.G., J.S.S., R.S., and M.M. wrote the manuscript.
Full methods and associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Corey DP, Hudspeth AJ. Response latency of vertebrate hair cells. Biophys J. 1979;26:499–506. doi: 10.1016/S0006-3495(79)85267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarter GC, Reichling DB, Levine JD. Mechanical transduction by rat dorsal root ganglion neurons in vitro. Neurosci Lett. 1999;273:179–182. doi: 10.1016/s0304-3940(99)00665-5. [DOI] [PubMed] [Google Scholar]
- 3.Davis MJ, Donovitz JA, Hood JD. Stretch-activated single-channel and whole cell currents in vascular smooth muscle cells. Am J Physiol. 1992;262:C1083–1088. doi: 10.1152/ajpcell.1992.262.4.C1083. [DOI] [PubMed] [Google Scholar]
- 4.Praetorius HA, Spring KR. Bending the MDCK cell primary cilium increases intracellular calcium. J Membr Biol. 2001;184:71–79. doi: 10.1007/s00232-001-0075-4. [DOI] [PubMed] [Google Scholar]
- 5.Chalfie M. Neurosensory mechanotransduction. Nat Rev Mol Cell Biol. 2009;10:44–52. doi: 10.1038/nrm2595. [DOI] [PubMed] [Google Scholar]
- 6.Delmas P, Hao J, Rodat-Despoix L. Molecular mechanisms of mechanotransduction in mammalian sensory neurons. Nat Rev Neurosci. 2011;12:139–153. doi: 10.1038/nrn2993. [DOI] [PubMed] [Google Scholar]
- 7.Hamill OP, Martinac B. Molecular basis of mechanotransduction in living cells. Physiol Rev. 2001;81:685–740. doi: 10.1152/physrev.2001.81.2.685. [DOI] [PubMed] [Google Scholar]
- 8.Coste B, et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bae C, Sachs F, Gottlieb PA. The mechanosensitive ion channel Piezo1 is inhibited by the peptide GsMTx4. Biochemistry. 2011;50:6295–6300. doi: 10.1021/bi200770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reed-Geaghan EG, Maricich SM. Peripheral somatosensation: a touch of genetics. Curr Opin Genet Dev. 2011;21:240–248. doi: 10.1016/j.gde.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bichet D, Haass FA, Jan LY. Merging functional studies with structures of inward-rectifier K(+) channels. Nat Rev Neurosci. 2003;4:957–967. doi: 10.1038/nrn1244. [DOI] [PubMed] [Google Scholar]
- 12.Nilius B. Pressing and squeezing with Piezos. EMBO Rep. 2010;11:902–903. doi: 10.1038/embor.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong Z, et al. Two interdependent TRPV channel subunits, inactive and Nanchung, mediate hearing in Drosophila. J Neurosci. 2004;24:9059–9066. doi: 10.1523/JNEUROSCI.1645-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, et al. A TRPV family ion channel required for hearing in Drosophila. Nature. 2003;424:81–84. doi: 10.1038/nature01733. [DOI] [PubMed] [Google Scholar]
- 15.Tracey WD, Jr., Wilson RI, Laurent G, Benzer S. painless, a Drosophila gene essential for nociception. Cell. 2003;113:261–273. doi: 10.1016/s0092-8674(03)00272-1. [DOI] [PubMed] [Google Scholar]
- 16.Walker RG, Willingham AT, Zuker CS. A Drosophila mechanosensory transduction channel. Science. 2000;287:2229–2234. doi: 10.1126/science.287.5461.2229. [DOI] [PubMed] [Google Scholar]
- 17.Zhong L, Hwang RY, Tracey WD. Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr Biol. 2010;20:429–434. doi: 10.1016/j.cub.2009.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coste B, Crest M, Delmas P. Pharmacological Dissection and Distribution of NaN/Nav1.9, T-type Ca2+ Currents, and Mechanically Activated Cation Currents in Different Populations of DRG Neurons. J Gen Physiol. 2007;129:57–77. doi: 10.1085/jgp.200609665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drew LJ, Wood JN, Cesare P. Distinct mechanosensitive properties of capsaicin-sensitive and -insensitive sensory neurons. J Neurosci. 2002;22:RC228. doi: 10.1523/JNEUROSCI.22-12-j0001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho H, et al. A novel mechanosensitive channel identified in sensory neurons. Eur J Neurosci. 2006;23:2543–2550. doi: 10.1111/j.1460-9568.2006.04802.x. [DOI] [PubMed] [Google Scholar]
- 21.Heinemann SH, Terlau H, Stuhmer W, Imoto K, Numa S. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 1992;356:441–443. doi: 10.1038/356441a0. [DOI] [PubMed] [Google Scholar]
- 22.Voets T, et al. TRPM6 forms the Mg2+ influx channel involved in intestinal and renal Mg2+ absorption. J Biol Chem. 2004;279:19–25. doi: 10.1074/jbc.M311201200. [DOI] [PubMed] [Google Scholar]
- 23.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayley H, et al. Droplet interface bilayers. Mol Biosyst. 2008;4:1191–1208. doi: 10.1039/b808893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montal M. Asymmetric lipid bilayers. Reponse to multivalent ions. Biochimica et Biophysica Acta. 1973;298:750–754. doi: 10.1016/0005-2736(73)90092-8. [DOI] [PubMed] [Google Scholar]
- 26.Montal M, Mueller P. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc Natl Acad Sci U S A. 1972;69:3561–3566. doi: 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos JS, Grigoriev SM, Montal M. Molecular template for a voltage sensor in a novel K+ channel. III. Functional reconstitution of a sensorless pore module from a prokaryotic Kv channel. J Gen Physiol. 2008;132:651–666. doi: 10.1085/jgp.200810077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- 29.Ermakov YA, Kamaraju K, Sengupta K, Sukharev S. Gadolinium ions block mechanosensitive channels by altering the packing and lateral pressure of anionic lipids. Biophys J. 2010;98:1018–1027. doi: 10.1016/j.bpj.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gambale F, Montal M. Characterization of the channel properties of tetanus toxin in planar lipid bilayers. Biophys J. 1988;53:771–783. doi: 10.1016/S0006-3495(88)83157-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliver D, et al. Functional conversion between A-type and delayed rectifier K+ channels by membrane lipids. Science. 2004;304:265–270. doi: 10.1126/science.1094113. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt D, MacKinnon R. Voltage-dependent K+ channel gating and voltage sensor toxin sensitivity depend on the mechanical state of the lipid membrane. Proc Natl Acad Sci U S A. 2008;105:19276–19281. doi: 10.1073/pnas.0810187105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao X, MacKinnon R. Functional analysis of Kv1.2 and paddle chimera Kv channels in planar lipid bilayers. J Mol Biol. 2008;382:24–33. doi: 10.1016/j.jmb.2008.06.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartman NC, Groves JT. Signaling clusters in the cell membrane. Current opinion in cell biology. 2011;23:370–376. doi: 10.1016/j.ceb.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schagger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 36.Gambale F, Montal M. Voltage-gated sodium channels expressed in the human cerebellar medulloblastoma cell line TE671. Brain Res Mol Brain Res. 1990;7:123–129. doi: 10.1016/0169-328x(90)90089-v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.