Abstract
Summary: The phylum Actinobacteria harbors many important human pathogens and also provides one of the richest sources of natural products, including numerous antibiotics and other compounds of biotechnological interest. Thus, a reliable phylogeny of this large phylum and the means to accurately identify its different constituent groups are of much interest. Detailed phylogenetic and comparative analyses of >150 actinobacterial genomes reported here form the basis for achieving these objectives. In phylogenetic trees based upon 35 conserved proteins, most of the main groups of Actinobacteria as well as a number of their superageneric clades are resolved. We also describe large numbers of molecular markers consisting of conserved signature indels in protein sequences and whole proteins that are specific for either all Actinobacteria or their different clades (viz., orders, families, genera, and subgenera) at various taxonomic levels. These signatures independently support the existence of different phylogenetic clades, and based upon them, it is now possible to delimit the phylum Actinobacteria (excluding Coriobacteriia) and most of its major groups in clear molecular terms. The species distribution patterns of these markers also provide important information regarding the interrelationships among different main orders of Actinobacteria. The identified molecular markers, in addition to enabling the development of a stable and reliable phylogenetic framework for this phylum, also provide novel and powerful means for the identification of different groups of Actinobacteria in diverse environments. Genetic and biochemical studies on these Actinobacteria-specific markers should lead to the discovery of novel biochemical and/or other properties that are unique to different groups of Actinobacteria.
INTRODUCTION
The phylum Actinobacteria, which is comprised mainly of Gram-positive organisms with a high G+C content (>55 mol% in genomic DNA), constitutes one of the largest phyla within the Bacteria (76, 103, 192, 193, 283, 284). The different genera that are part of this phylum exhibit enormous diversity in terms of their morphology, physiology, and metabolic capabilities (76, 277, 313). The morphologies of actinobacterial species vary from coccoid (e.g., Micrococcus) or rod-coccoid (e.g., Arthrobacter) to fragmenting hyphal forms (e.g., Nocardia) or highly differentiated branched mycelia (e.g., Streptomyces) (8). Spore formation, although common, is not ubiquitous among actinobacteria, and they could range from motile zoospores to specialized propagules (182). The species of this group also exhibit enormous physiological diversity, as evidenced by their production of numerous extracellular enzymes and thousands of metabolic products that they synthesize and excrete (42, 256), many of which are antibiotics (65, 146, 182). The phylum Actinobacteria also constitutes one of the earliest lineages within the prokaryotes (119, 122, 168, 179), and the production of antibiotics by them has been indicated to be an important determining factor in the evolution of both the Archaea and Gram-negative (diderm) bacteria from Gram-positive (monoderm) bacteria (119, 120, 124, 129, 311).
The most extensively studied representatives of this group include soil-dwelling Streptomyces spp., which are the major producers of antibiotics (18, 41, 145, 146, 219, 314), and important human pathogens of the genus Mycobacterium (M. tuberculosis and M. leprae), which are responsible for the largest number of human deaths from bacterial infections (17, 53, 56, 252, 305). However, the genera Streptomyces and Mycobacterium constitute only 2 of the genera within this large phylum that contains >300 genera (77, 343). In addition, there are huge populations of poorly studied actinobacteria that are prevalent in soil, water, deep-sea, or extreme environments, such as arctic ice, chemically contaminated sites, and radioactive environments, or that reside with humans, animals, and plants in a friendly or hostile way (14, 35, 85, 202, 205, 270, 307, 314). In recent years, due to rapid advances in genome-sequencing technologies, increasing progress is being made in studying the diversity and biology of Actinobacteria. The main focuses of these studies have been on bacteria that either produce or have the potential for the discovery of novel useful natural products (e.g., Streptomyces, Salinispora, Saccharopolyspora, Cellulomonas, Verrucosispora, Pseudonocardia, and Micromonospora) (12, 16, 21, 36, 86, 220, 249) or on pathogenic Actinobacteria that cause severe human and animal diseases or agricultural losses (e.g., Mycobacterium, Actinomyces, Renibacterium, Atopobium, Gordonia, Gardnerella, Leifsonia, and Clavibacter) (36, 69, 105, 219, 287). Extensive work has also been carried out on the Bifidobacteriales, which form a major component of the microbial flora in the gastrointestinal tracts of humans and other mammals and are believed to exhibit useful probiotic activities (183, 307, 313, 314, 317). In addition, the exploration of other industrially important species (e.g., Corynebacterium, Rhodococcus, Micrococcus, Cellulomonas, Acidothermus, Thermobifida, and Nocardioides) and environmentally beneficial species (e.g., Arthrobacter, Kocuria, Frankia, Kineococcus, Pseudonocardia, and Rubrobacter) has been greatly facilitated by the development of technology and the urgency for new biosources (9, 14, 85, 150, 159, 189, 194, 196, 202, 216, 296).
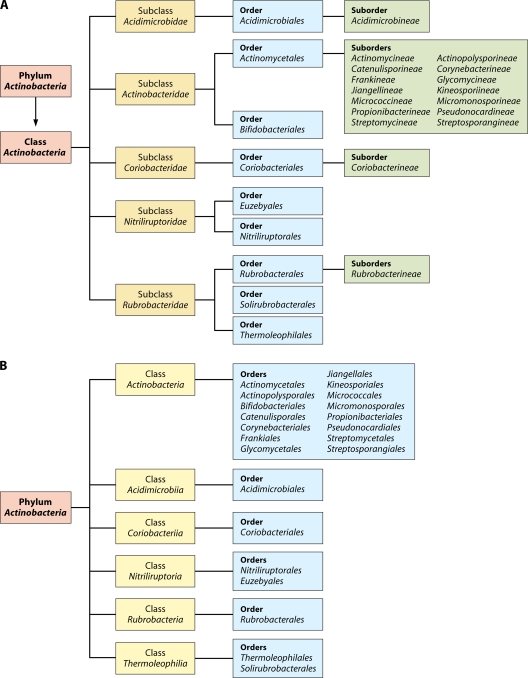
In view of the medical, biotechnological, and ecological importance of the Actinobacteria, an understanding of the evolutionary relationships among members of this large phylum and what unique biochemical or physiological characteristics distinguish species of different clades of Actinobacteria is of great importance and significance (97, 110, 130, 132, 283, 323, 324). Currently, the phylum Actinobacteria is delineated from other bacteria solely on the basis of its branching position in 16S rRNA gene trees. The most recently published taxonomy of Actinobacteria, by Zhi et al. (343), divided this phylum at the highest level into four subclasses, namely, Actinobacteridae, Acidimicrobidae, Coriobacteridae, and Rubrobacteridae, which together encompassed 219 genera in 50 families (104, 280). In an updated version of this taxonomy in the List of Prokaryotic Names with Standing in Nomenclature, maintained by J. P. Euzeby (http://www.bacterio.cict.fr), the phylum Actinobacteria at the highest level is now divided into five subclasses, namely, Actinobacteridae, Acidimicrobidae, Coriobacteridae, Nitriliruptoridae, and Rubrobacteridae. These subclasses are further subdivided into a number of different orders and suborders (Fig. 1A) (343). It is noteworthy that in this taxonomy, 47 of the 57 families within the phylum Actinobacteria are part of a single subclass, Actinobacteridae, whereas the other four subclasses together contained only 10 families.
Fig 1.
Current taxonomic outline for the phylum Actinobacteria based upon the List of Prokaryotic Names with Standing in Nomenclature (http://www.bacterio.cict.fr/classifphyla.html#Actinobacteria) (A) and proposed taxonomy for Actinobacteria in the forthcoming Bergey's Manual of Systematic Bacteriology (191) (B).
Recently, another update of the taxonomy of the phylum Actinobacteria based upon 16S rRNA trees was reported (191), which will form the basis of the section on Actinobacteria in the forthcoming Bergey's Manual of Systematic Bacteriology (191). Although the phylogenetic information on which this update is based is not posted on the Bergey's Manual Trust website, in the revised taxonomy, the taxonomic ranks of subclasses and suborders are eliminated, and they are now elevated to the ranks of classes and orders, respectively (Fig. 1B). At the highest level, the phylum Actinobacteria is now divided into six classes, namely, Actinobacteria, Acidimicrobiia, Coriobacteriia, Nitriliruptoria, Rubrobacteria, and Thermoleophilia. The class Actinobacteria now contains a total of 15 orders, including both previously proposed orders Actinomycetales and Bifidobacteriales (343). However, the order Actinomycetales is now restricted to the members of the family Actinomycetaceae, and the other suborders that were previously part of this order are now designated as distinct orders.
Although the taxonomic classification of the phylum Actinobacteria deduced on the basis of 16S rRNA trees represents an important advancement (103, 191, 283, 343), the compact clustering of different actinobacterial orders in the rRNA trees makes it difficult to determine reliably the interrelationships or branching order of the higher taxonomic clades within this phylum. This is especially true for its largest class, Actinobacteria, which accounts for >80% of all known actinobacterial families/genera (97, 103). Additionally, in the current classification scheme, all taxa higher than the rank of genus are distinguished primarily on the basis of taxon-specific 16S rRNA signature nucleotides (343). However, these signature nucleotides are based on published 16S rRNA sequences of type strains, and they change when new sequences are added to the databases (283, 343). There is also not much information available regarding the specificity of these signatures or their predictive ability to identify species belonging to these taxa. Although other phenotypic characteristics, such as morphological, physiological, and chemotaxonomic features, are useful for preliminary classifications and identifications of many spore-forming Actinobacteria, their levels of congruence are low (76, 103). Thus, in order to develop a reliable and stable understanding of this phylum, novel and more definitive characteristics need to be identified to define and distinguish the phylum Actinobacteria and its different lineages in clearer terms.
The rapidly increasing numbers of genome sequences provide an important resource to study Actinobacteria from different perspectives (211). This review focuses on the use of available genome sequences to discover novel molecular characteristics that are specific for the phylum Actinobacteria and its various lineages and their applications to develop a reliable evolutionary framework for the members of this phylum. However, before focusing on these aspects, a brief overview of some general features of the sequenced actinobacterial genomes is provided.
OVERVIEW OF GENOMIC CHARACTERISTICS OF ACTINOBACTERIA
Genomic characteristics of limited numbers of Actinobacteria have been described by various authors (17, 167, 314, 339) (see Table 1 for other references). In the latest comprehensive review on this subject by Ventura et al. (314), the features of 20 actinobacterial genomes that were available in 2007 were summarized. However, since the publication of that review, the number of sequenced actinobacterial genomes has increased more than 8 times (157 complete and 474 in progress), providing an abundant resource for such studies. The enormous phenotypic diversity of the Actinobacteria is well reflected in their genotypes. Some features of the completed actinobacterial genomes are summarized in Table 1. The sequenced genomes varied in size from 0.93 Mb (Tropheryma whipplei) to 12 Mb (Streptomyces bingchenggensis), and their GC contents varied from 41.5% (Gardnerella vaginalis ATCC 14019) to 74.2% (Kineococcus radiotolerans SRS30216) (9, 20, 30, 196). Interestingly, of these genomes, species of at least 4 genera have linear chromosomes, including Streptomyces, Rhodococcus, Gordonibacter, and Kineococcus (9, 44, 165, 196, 257, 261), These linear chromosomes are characterized by a central replication origin (oriC) and terminal inverted repeats (9, 47, 196, 257, 314). The mechanism for chromosome linearization was proposed previously to arise from recombination with linear plasmids that have evolved by the integration of bacteriophages (44, 321). Based upon the current taxonomy of the Actinobacteria (Fig. 1) and a phylogenetic tree for the sequenced species of this phylum (Fig. 2), the 4 genera containing the linear chromosomes belong to 4 distinct suborders, and they are distantly related (103, 283, 343). Thus, the chromosome linearization characteristic has evolved more than once during the evolution of the Actinobacteria (165).
Table 1.
Characteristics of sequenced actinobacterial genomesc
| Actinobacterial genome | Size (Mb)e | % GC content | No. of proteins | GOT (°C)a | Habitatb | Source or referenced |
|---|---|---|---|---|---|---|
| Acidimicrobium ferrooxidans DSM 10331 | 2.16 | 68.3 | 1,964 | Ther | S | DOEJGI |
| Acidothermus cellulolyticus 11B | 2.4 | 66.9 | 2,157 | 58 | A | 14 |
| Actinosynnema mirum DSM 43827 | 8.25 | 73.7 | 6,916 | Meso | T | DOEJGI |
| Amycolatopsis mediterranei U32 | 10* | — | 9,228 | Meso | — | 341 |
| Amycolicicoccus subflavus DQS3-9A1 | 4.83* | — | 4,557 | — | — | COE, Beijing University |
| Arcanobacterium haemolyticum DSM 20595 | 2 | — | 1,731 | Meso | H | 336 |
| Arthrobacter arilaitensis Re117 | 3.96* | — | 3,376 | — | — | 203 |
| Arthrobacter aurescens TC1 | 5.23 | 62.4 | 4,041 | 30 | T | 202 |
| Arthrobacter chlorophenolicus A6 | 4.99 | 66 | 3,885 | Meso | T | DOEJGI |
| Arthrobacter phenanthrenivorans Sphe3 | 4.58* | 65.7 | 3,843 | 30 | T | DOEJGI |
| Arthrobacter sp. strain FB24 | 5.08 | 65.4 | 4,146 | Meso | T | DOEJGI |
| Atopobium parvulum DSM 20469 | 1.54 | 45.7 | 1,353 | Meso | H | 60 |
| Beutenbergia cavernae DSM 12333 | 4.7 | 73.1 | 4,197 | Meso | T | DOEJGI |
| Bifidobacterium adolescentis ATCC 15703 | 2.1 | 59.2 | 1,631 | 37 | H | Gifu University, Japan |
| Bifidobacterium animalis subsp. lactis AD011 | 1.9 | 60.5 | 1,528 | 39 | M | 164 |
| Bifidobacterium animalis subsp. lactis BB-12 | 1.9* | 60.5 | — | Meso | M | 102 |
| Bifidobacterium animalis subsp. lactis Bl04 | 1.9 | 60.5 | 1,567 | 39 | M | 15 |
| Bifidobacterium animalis subsp. lactis DSM 10140 | 1.9 | 60.5 | 1,566 | 39 | M | 15 |
| Bifidobacterium animalis subsp. lactis V9 | 1.9* | 60.5 | — | Meso | M | 292 |
| Bifidobacterium bifidum PRL2010 | 2.2* | — | 1,706 | Meso | — | 306 |
| Bifidobacterium bifidum S17 | 2.2 | — | 1,783 | — | — | 344 |
| Bifidobacterium breve ACS-071-V-Sch8b | 2.3* | — | — | — | — | JCVI |
| Bifidobacterium dentium Bd1 | 2.6* | 58.5 | 2,129 | Meso | — | 318 |
| Bifidobacterium longum DJO10A | 2.41 | 60.2 | 1,990 | 37-41 | H | 183 |
| Bifidobacterium longum NCC2705 | 2.26 | 60.1 | 1,727 | 37-41 | H | 254 |
| Bifidobacterium longum subsp. infantis 157F | 2.41 | 59.9 | 1,991 | Meso | H | 93 |
| Bifidobacterium longum subsp. infantis ATCC 15697 | 2.8 | 59.9 | 2,416 | 37-41 | H | 262 |
| Bifidobacterium longum subsp. longum BBMN68 | 2.3* | — | 1,806 | — | — | 141 |
| Bifidobacterium longum subsp. longum F8 | 2.4* | — | — | Meso | H | MetaHIT |
| Bifidobacterium longum subsp. longum JCM 1217 | 2.4* | — | 1,924 | Meso | — | 93 |
| Bifidobacterium longum subsp. longum JDM301 | 2.5* | — | 1,958 | 37-41 | H | 326 |
| Brachybacterium faecium DSM 4810 | 3.6 | 72.0 | 3,068 | — | T | 180 |
| Catenulispora acidiphila DSM 44928 | 10.47 | 69.8 | 8,913 | Meso | T | 59 |
| Cellulomonas fimi ATCC 484 | 4.3* | — | 3,761 | Meso | T | DOEJGI |
| Cellulomonas flavigena DSM 20109 | 4.1* | 74.1 | 3,678 | Meso | T | 2 |
| Clavibacter michiganensis NCPPB 382 | 3.4 | 72.5 | 2,984 | 25-28 | M | 105 |
| Clavibacter michiganensis subsp. sepedonicus | 3.44 | 72.4 | 2,941 | 25-28 | M | 19 |
| Conexibacter woesei DSM 14684 | 6.4* | 72.7 | 5,914 | Meso | T | 234 |
| Coriobacterium glomerans PW2 | 2.1* | 60 | 1,768 | Meso | H | DOEJGI |
| Corynebacterium aurimucosum ATCC 700975 | 2.83 | 60.6 | 2,531 | Meso | H | 304 |
| Corynebacterium diphtheriae NCTC 13129 | 2.49 | 53.5 | 2,272 | 37 | M | 39 |
| Corynebacterium efficiens YS-314 | 3.1* | 63.1 | 2,938 | 30-45 | M | 213 |
| Corynebacterium glutamicum ATCC 13032 | 3.3 | 53.8 | 2,993 | 30-40 | M | 159 |
| Corynebacterium glutamicum R | 3.35 | 54.1 | 3,052 | 30-40 | M | 339 |
| Corynebacterium jeikeium K411 | 2.51* | 61.4 | 2,104 | Meso | M | 297 |
| Corynebacterium kroppenstedtii DSM 44385 | 2.4 | 57.5 | 2,018 | Meso | H | 298 |
| Corynebacterium pseudotuberculosis 1002 | 2.3* | — | — | Meso | — | 267 |
| Corynebacterium pseudotuberculosis C231 | 2.3* | — | — | Meso | — | 267 |
| Corynebacterium pseudotuberculosis FRC41 | 2.3* | — | 2,110 | — | — | 267 |
| Corynebacterium pseudotuberculosis I19 | 2.3* | — | — | — | — | 267 |
| Corynebacterium ulcerans 809 | 2.5* | — | — | — | — | Bielefeld University |
| Corynebacterium ulcerans BR-AD22 | 2.6* | — | — | — | — | Bielefeld University |
| Corynebacterium urealyticum DSM 7109 | 2.4 | 64.2 | 2,024 | Meso | H | 299 |
| Cryptobacterium curtum DSM 15641 | 1.6 | 50.9 | 1,357 | — | — | 195 |
| Eggerthella lenta DSM 2243 | 3.63 | 64.2 | 3,070 | Meso | H | 251 |
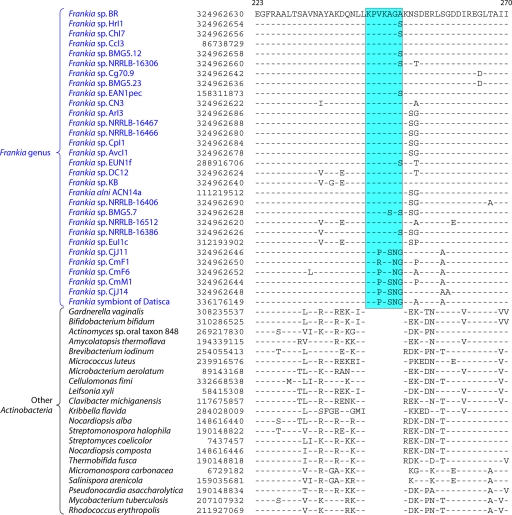
| Frankia alni ACN14a | 7.5 | 72.8 | 6,711 | Meso | H | 216 |
| Frankia sp. strain CcI3 | 5.4 | 70.1 | 4,499 | Meso | M | 216 |
| Frankia sp. strain EAN1pec | 9 | 71.2 | 7,191 | Meso | M | DOEJGI |
| Frankia symbiont of Datisca glomerata | 5.32* | 70.1 | — | Meso | — | DOEJGI |
| Gardnerella vaginalis 409-05 | 1.6 | 42.0 | 1,261 | — | — | JCVI |
| Gardnerella vaginalis ATCC 14019 | 1.7* | 41.5 | 1,365 | — | — | 337 |
| Gardnerella vaginalis HMP9231 | 1.7* | — | — | — | — | JCVI |
| Geodermatophilus obscurus DSM 43160 | 5.3* | 74.0 | 4,810 | Meso | T | 154 |
| Gordonia bronchialis DSM 43247 | 5.28 | 67.1 | 4,616 | Meso | H | 153 |
| Gordonibacter pamelaeae 7-10-1-b | 3.6* | — | — | — | — | Sanger Institute |
| Intrasporangium calvum DSM 43043 | 4* | — | 3,563 | Meso | — | 66 |
| Isoptericola variabilis 225 | 3.3 | — | 2,881 | — | — | DOEJGI |
| Jonesia denitrificans DSM 20603 | 2.75 | 58.4 | 2,511 | — | — | 233 |
| Kineococcus radiotolerans SRS30216 | 4.99 | 74.2 | 4,480 | 32 | M | DOEJGI |
| Kocuria rhizophila DC2201 | 2.7 | 71.2 | 2,357 | Meso | M | 296 |
| Kribbella flavida DSM 17836 | 7.6* | 70.6 | 6,943 | Meso | T | 235 |
| Kytococcus sedentarius DSM 20547 | 2.8 | 71.6 | 2,554 | Meso | — | 268 |
| Leifsonia xyli subsp. xyli strain CTCB07 | 2.58 | 67.7 | 2,030 | 20-25 | H | 205 |
| Microbacterium testaceum StLB037 | 4 | — | 3,676 | — | — | 207 |
| Micrococcus luteus NCTC 2665 | 2.5 | 72.9 | 2,236 | Meso | M | DOEJGI |
| Micromonospora aurantiaca ATCC 27029 | 7* | 72.9 | 6,222 | Meso | M | DOEJGI |
| Micromonospora sp. L5 | 7* | 72.9 | 6,150 | Meso | — | DOEJGI |
| Mobiluncus curtisii ATCC 43063 | 2.1* | 55.6 | 1,909 | — | — | Baylor College |
| Mycobacterium abscessus ATCC 19977 | 5.09 | 64.1 | 4,920 | 37 | M | 242 |
| Mycobacterium avium 104 | 5.5 | 69 | 5,120 | 37 | H | TIGR |
| Mycobacterium avium subsp. paratuberculosis K-10 | 4.8 | 69.3 | 4,350 | 37 | M | 186 |
| Mycobacterium bovis AF2122/97 | 4.35 | 65.6 | 3,920 | 37 | H | 101 |
| Mycobacterium bovis BCG strain Pasteur 1173P2 | 4.4 | 65.6 | 3,052 | Meso | H | 32 |
| Mycobacterium bovis BCG strain Tokyo 172 | 4.4 | 65.6 | 3,947 | Meso | H | 260 |
| Mycobacterium gilvum PYR-GCK | 5.96 | 67.7 | 5,241 | Meso | DOEJGI | |
| Mycobacterium leprae Br4923 | 3.3 | 57.8 | 1,604 | 37 | H | 204 |
| Mycobacterium leprae TN | 3.27 | 57.8 | 1,605 | 37 | H | 56 |
| Mycobacterium marinum M | 6.62 | 65.7 | 5,423 | 32 | M | 287 |
| Mycobacterium smegmatis strain MC2 155 | 7 | 67.4 | 6,716 | 37 | H | TIGR |
| Mycobacterium sp. JDM601 | 4.6* | — | 4,346 | — | — | Shanghai JT University |
| Mycobacterium sp. strain JLS | 6 | 68.4 | 5,739 | Meso | M | DOEJGI |
| Mycobacterium sp. strain KMS | 6.22 | 68.2 | 5,460 | Meso | M | DOEJGI |
| Mycobacterium sp. strain MCS | 5.92 | 68.4 | 5,391 | Meso | — | DOEJGI |
| Mycobacterium sp. strain Spyr1 | 5.73* | — | 5,130 | Meso | T | DOEJGI |
| Mycobacterium tuberculosis CDC1551 | 4.4 | 65.6 | 4,189 | 37 | H | 88 |
| Mycobacterium tuberculosis F11 | 4.4 | 65.6 | 3,941 | 37 | H | The Broad Institute |
| Mycobacterium tuberculosis H37Ra | 4.4 | 65.6 | 4,034 | 37 | H | 342 |
| Mycobacterium tuberculosis H37Rv | 4.4 | 65.6 | 3,989 | 37 | H | 55 |
| Mycobacterium tuberculosis KZN 1435 | 4.4 | 65.6 | 4,059 | 37 | H | The Broad Institute |
| Mycobacterium tuberculosis KZN 4207 | 4.4* | 65.4 | — | 37 | H | The Broad Institute |
| Mycobacterium ulcerans Agy99 | 5.77 | 65.4 | 4,160 | 32 | H | 288 |
| Mycobacterium vanbaalenii PYR-1 | 6.5 | 67.8 | 5,979 | 24-37 | — | DOEJGI |
| Nakamurella multipartita DSM 44233 | 6.06 | 70.9 | 5,240 | Meso | T | 302 |
| Nocardia farcinica IFM 10152 | 6.29 | 70.7 | 5,683 | 37 | M | 151 |
| Nocardioides sp. JS614 | 5.31 | 71.4 | 4,645 | 30 | T | DOEJGI |
| Nocardiopsis dassonvillei subsp. dassonvillei DSM 43111 | 6.58* | 72.7 | 4,798 | — | M | 291 |
| Olsenella uli DSM 7084 | 2.1* | — | 1,739 | 37 | — | 108 |
| Propionibacterium acnes 266 | 2.5* | 60 | — | — | — | G.-A. University |
| Propionibacterium acnes KPA171202 | 2.56 | 60 | 2,297 | 37 | H | 34 |
| Propionibacterium acnes SK137 | 2.5 | 60.1 | 2,352 | Meso | — | JCVI |
| Propionibacterium freudenreichii subsp. shermanii CIRM-BIA1 | 2.6 | — | 2,375 | Meso | M | 79 |
| Pseudonocardia dioxanivorans CB1190 | 7.3 | — | 6,495 | 30 | — | DOEJGI |
| Renibacterium salmoninarum ATCC 33209 | 3.2 | 56.3 | 3,507 | 15 | H | 328 |
| Rhodococcus equi 103S | 5* | — | 4,512 | Meso | M | 184 |
| Rhodococcus erythropolis PR4 | 6.88 | 62.3 | 6,030 | 20 | — | 261 |
| Rhodococcus jostii RHA1 | 9.67 | 67 | 7,211 | 30 | T | 196 |
| Rhodococcus opacus B4 | 7.9 | 67.9 | 7,246 | — | T | 209 |
| Rothia dentocariosa ATCC 17931 | 2.5 | — | 2,217 | Meso | H | Baylor College |
| Rothia mucilaginosa | 2.5* | 59.6 | 1,904 | Meso | H | Osaka Dental University |
| Rubrobacter xylanophilus DSM 9941 | 3.23 | 70.5 | 3,140 | 60 | S | DOEJGI |
| Saccharomonospora viridis DSM 43017 | 4.3 | 67.3 | 3,828 | 37 | H | 228 |
| Saccharopolyspora erythraea NRRL 2338 | 8.2 | 71.1 | 7,197 | 28 | T | 222 |
| Salinispora arenicola CNS-205 | 5.8 | 69.5 | 4,917 | Meso | A | 229 |
| Salinispora tropica CNB-440 | 5.2 | 69.5 | 4,536 | 28 | A | 309 |
| Sanguibacter keddieii DSM 10542 | 4.3* | 71.9 | 3,710 | Meso | H | 155 |
| Segniliparus rotundus DSM 44985 | 3.2* | 68 | 3,006 | Meso | — | 266 |
| Slackia heliotrinireducens DSM 20476 | 3.17 | 60.2 | 2,765 | Meso | M | DOEJGI |
| Stackebrandtia nassauensis DSM 44728 | 6.8* | 68.1 | 6,379 | — | — | DOEJGI |
| Streptomyces avermitilis MA-4680 | 9.09 | 70.7 | 7,580 | 26 | M | 148 |
| Streptomyces bingchenggensis BCW-1 | 12 | — | — | — | — | 322 |
| Streptomyces coelicolor A3(2) | 9.09 | 72 | 7,769 | 25-35 | M | 18 |
| Streptomyces flavogriseus ATCC 33331 | 7.62* | 71.0 | — | Meso | T | DOEJGI |
| Streptomyces griseus subsp. griseus NBRC 13350 | 8.5 | 72.2 | 7,136 | 25-35 | M | 144 |
| Streptomyces scabiei 87.22 | 10* | — | 8,746 | Meso | T | 23 |
| Streptosporangium roseum DSM 43021 | 10.03* | 70.9 | 8,945 | Meso | T | 214 |
| Thermobifida fusca YX | 3.6 | 67.5 | 3,110 | 50-55 | M | 194 |
| Thermobispora bispora DSM 43833 | 4.2 | 70 | 3,546 | Thermo | T | 188 |
| Thermomonospora curvata DSM 43183 | 5.6* | 71.6 | 4,890 | 45-55 | S | 45 |
| Tropheryma whipplei strain Twist | 0.93 | 46.3 | 808 | 37 | H | 239 |
| Tropheryma whipplei TW08/27 | 0.93 | 46.3 | 783 | 37 | H | 20 |
| Tsukamurella paurometabola DSM 20162 | 4.5* | 68.4 | 4,157 | Meso | T | DOEJGI |
| Verrucosispora maris AB-18-032 | 6.75* | — | 5,956 | — | — | CSBL, Korea University |
| Xylanimonas cellulosilytica DSM 15894 | 3.79* | 72.5 | 3,337 | — | S | 89 |
GOT, growth-optimal temperature; Meso, mesophilic; Ther, thermophilic.
A, aquatic; T, terrestrial; H, host associated; M, multiple; S, specialized.
The information in the table was collected from the NCBI website (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). —, information not available.
Abbreviations: DOEJGI, U.S. Department of Energy Joint Genome Institute; COE, College of Engineering; JCVI, J. Craig Venter Institute; TIGR, The Institute for Genomic Research; Shanghai JT University, Jiao Tong University School of Medicine, Shanghai, China; G.-A. University, Georg-August University.
An asterisk indicates that the genome size is estimated; otherwise, the genome size was calculated based on existing sequences.
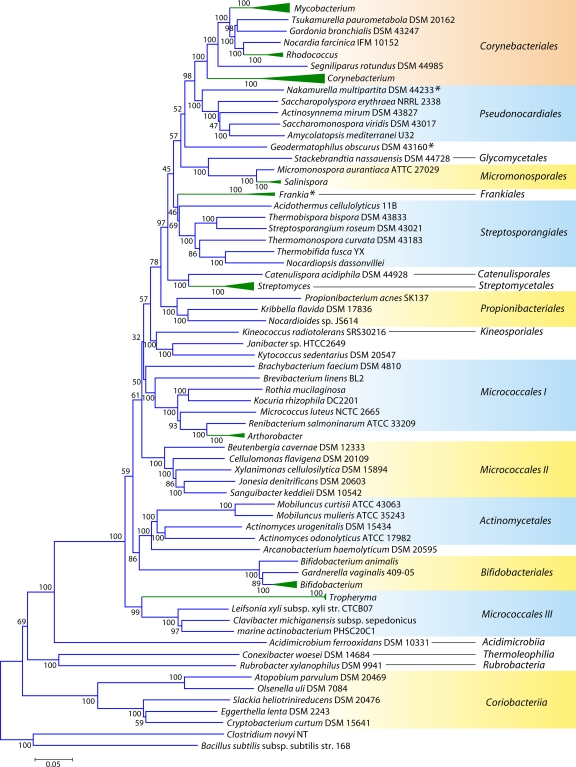
Fig 2.
Phylogenetic tree for 98 actinobacterial species whose genomes have been sequenced, based upon concatenated sequences for 35 conserved proteins. Many genera for which sequence information is available from multiple species are represented by triangles in this tree. The sizes of the triangles reflect the number of species that have been sequenced, and more detailed trees for some of these groups are presented in other figures. The tree shown is based on neighbor-joining (NJ) analysis, and the numbers at the nodes represent the bootstrap scores of the nodes. Similar branching patterns for most of these groups can also be observed in a maximum likelihood tree. The asterisks mark the Frankiales species that branch in different positions in this tree.
Remarkably, even within the same genus of Actinobacteria, the genome size differences can be significant. For example, of the 23 sequenced Mycobacterium species and strains, M. smegmatis strain MC2 155 has a genome of 7.0 Mb, while the intracellular pathogen M. leprae TN has a massively reduced genome of 3.27 Mb (Table 1) (54, 56, 287). Interestingly, the genome sizes of the 4 sequenced Frankia strains also varied from 5.43 Mb to 9.04 Mb, showing the greatest divergence yet reported for such closely related soil bacteria (97.8% to 98.9% identity in 16S rRNA genes) (216). The bacterial genome is believed to be plastic and dynamic, in which gene gains, gene losses, and lateral gene transfers (LGT) happen all the time to shape the gene repertoire (64, 181, 218, 273). The main driving force for genome expansion or reduction is niche adaptation. In the case of the Actinobacteria, most isolated species are free living, and they are from complex and densely populated soil environments. Thus, their genomes are generally large (approximately 5 to 9 Mb) in order to combat environmental changes and species competition (Table 1) (18, 170, 196, 222, 226, 314). However, some species that are parasitic or symbionts have undergone extensive genome reduction, reflecting their adaptation to the much more stable conditions within the host (56, 69, 205). Thus, while host associations favor genome contraction, host diversification leads to genome expansion. As a result, Frankia strains or Mycobacterium species that have a narrow host range or a broad host range exhibit large differences in their genome sizes (69, 216, 287, 338). Although it is debatable whether genome reduction is a strategy to reduce the energy cost of maintaining genome integrity in extreme environments (48, 91, 237), several actinobacterial species isolated under harsh conditions, such as Acidothermus cellulolyticus, Thermobifida fusca, Kocuria rhizophila, and Rubrobacter radiotolerans, etc. (14, 194, 296), have relatively small genomes (approximately 2 to 3.5 Mb) (Table 1). A number of comparative analyses suggested that selection does not act on the genome size; rather, it acts on individual genes and determines the gene repertoire, which in turn influences the genome size (92, 170, 172, 175). Thus, in order to better understand bacterial niche adaptation, it is important to study their diversified gene repertoire, especially the unique gene sets, whose products are the functional executives (regulators), workers (enzymes), and buildings (structural proteins) in the cell. A sound phylogenetic framework for the Actinobacteria should prove very helpful in these regards (231, 331).
PHYLOGENY OF ACTINOBACTERIA BASED ON COMBINED DATA SETS OF PROTEIN SEQUENCES
Detailed phylogenetic investigations of Actinobacteria have thus far been carried out mainly by using 16S rRNA sequences (3, 191, 192, 280, 283, 314, 343). Many studies have utilized alternate gene/protein sequences (e.g., RecA, RpoB, GyrB, DnaK, GrpE, GroEL, and CTP synthase, etc.) to examine actinobacterial phylogeny, but those studies employed only small numbers of Actinobacteria, and they were often limited to particular taxa (e.g., mycobacteria or Bifidobacterium) (25, 70, 97, 161, 312, 313, 315, 319, 320). The availability of genome sequences has provided new opportunities to examine actinobacterial phylogeny based upon different gene/protein sequences. With genomic sequences, many approaches have been used to infer the evolutionary relationships (10, 26, 247). These approaches include examinations of gene order (5, 173, 174) and shared gene content (29, 63, 98, 99, 118, 134, 274), the construction of supertrees based upon concatenated sequences for large numbers of proteins (49, 247, 333), the use of conserved indels to construct rooted phylogenetic trees (119, 121, 125, 126, 128), the use of character compatibility analysis based upon molecular sequences (134, 135), the construction of trees based on the protein domain content (335), and other methods or a combination of the above-mentioned approaches (5, 140, 334). Of these different approaches, the construction of phylogenetic trees based upon combined sequences of large numbers of protein sequences has proven particularly useful for an understanding of the evolutionary relationships among distantly related taxa (49, 247, 333). Phylogenetic trees based upon large numbers of characters derived from multiple conserved (or slow-evolving) genes/proteins are better able to resolve deeper-branching evolutionary relationships than those based on any single gene or protein (33, 49, 67, 139, 247). Alam et al. (5) recently reported a detailed phylogenetic analysis of 45 Actinobacteria using a number of different gene sequences (e.g., 5S rRNA, 16S rRNA, and 23S rRNA) and approaches, including a tree based upon concatenated sequences for 155 proteins. Based upon the results obtained by using different approaches, those authors drew a consensus tree for the Actinobacteria. In addition, a phylogenetic tree for the Actinobacteria based upon fragments derived from ychF, rpoB, and secY gene sequences was also constructed (3).
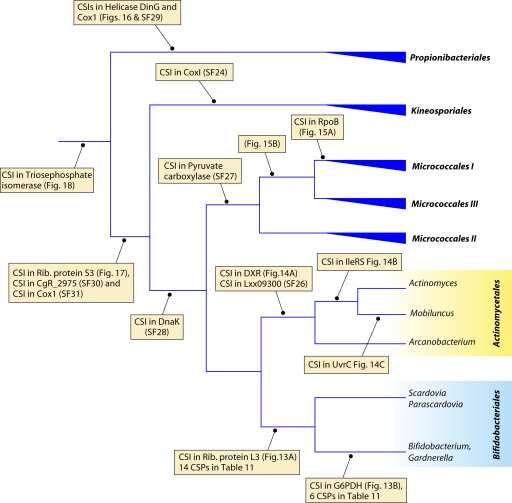
Although these trees provide good reference resources, the number of sequenced actinobacterial genomes has now greatly increased. Hence, to acquire a comprehensive view of Actinobacteria phylogeny covering different lineages, phylogenetic trees were constructed for 98 actinobacterial species (from 57 genera) whose genomes were either completely sequenced or were at the assembly stages in October 2010, when these analyses were carried out (Table 1). The species in our data set included representatives from 13 of the 15 orders of the class Actinobacteria as well as members of four of the other 5 proposed classes of this phylum (viz., Acidimicrobiia, Coriobacteriia, Rubrobacteria, and Thermoleophilia). A total of 35 universally distributed proteins, which are involved in a broad range of cellular functions, were extracted from these genomes for phylogenetic analyses (see File S1 in the supplemental material) (49). The sequence alignments for these proteins were concatenated into a single large data set, which, after the removal of all poorly aligned regions, consisted of 9,953 aligned positions. Phylogenetic trees based on this large data set of protein sequences were constructed by using the maximum likelihood (ML) and neighbor-joining (NJ) methods. Both these methods gave similar tree topologies, except for the branching points that were weakly supported in the trees. An NJ distance tree based on this data set is shown in Fig. 2. Compared to the other previously reported phylogenetic trees for Actinobacteria (3, 192, 343), where the bootstrap scores were either very low or not indicated, many of the nodes in this tree are supported by high bootstrap scores, indicating that the observed relationship is reliable and that this tree is better able to resolve the interrelationships among actinobacterial species. The important characteristics of this tree are noted below.
In contrast to the 16S rRNA tree, where Rubrobacter or a clade consisting of Rubrobacter and the Coriobacteriia was the earliest-branching lineage within the phylum Actinobacteria (343), in the tree based upon concatenated protein sequences, a clade consisting of various Coriobacteriia species constituted the deepest branch within this phylum. This clade was separated from all other Actinobacteria by a long branch. Following Coriobacteriia, a clade consisting of Rubrobacter xylanophilus and Conexibacter woesei (belonging to the classes Rubrobacteria and Thermoleophilia, respectively) as well Acidimicrobium ferrooxidans (class Acidimicrobiia) formed the next two deepest branches in the Actinobacteria tree. These species were also separated from all other species belonging to the class Actinobacteria by a long branch. Within the class Actinobacteria, a number of strongly supported clades that corresponded primarily to a number of known actinobacterial orders were observed. These clades included those corresponding to the orders Corynebacteriales, Pseudonocardiales, Micromonosporales, Propionibacteriales, Streptosporangiales, Streptomycetales, Actinomycetales, and Bifidobacteriales. In addition, this tree also supported a number of deeper-branching clades consisting of several orders of Actinobacteria. One of these clusters consisted of the orders Corynebacteriales, Pseudonocardiales, Micromonosporales, Streptosporangiales, Streptomycetales, and Frankiales. Within this large cluster, a clade consisting of the orders Corynebacteriales and Pseudonocardiales was also strongly supported by high bootstrap scores. This large clade was also observed in the consensus tree reported by Alam et al. (5). As discussed below, the existence of some of these clusters is also supported by the identification of conserved indels that are specific for them.
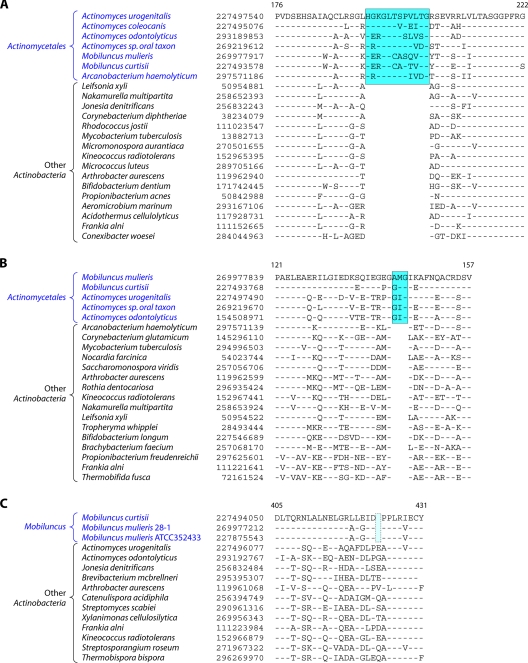
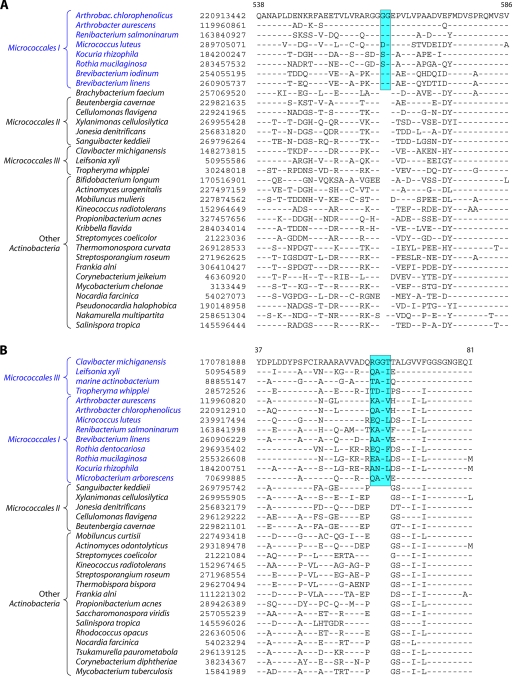
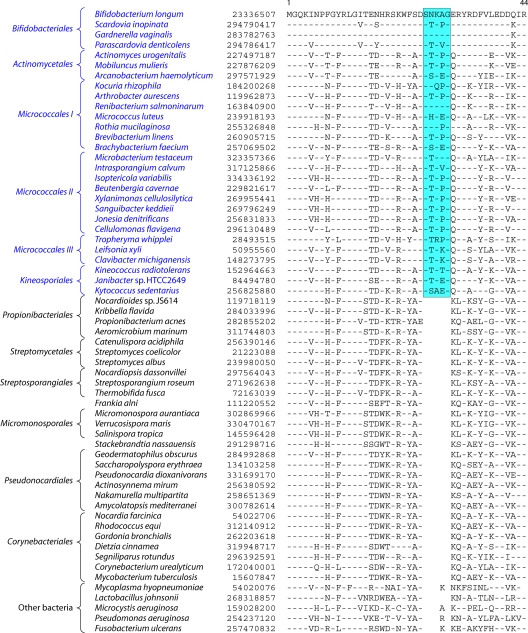
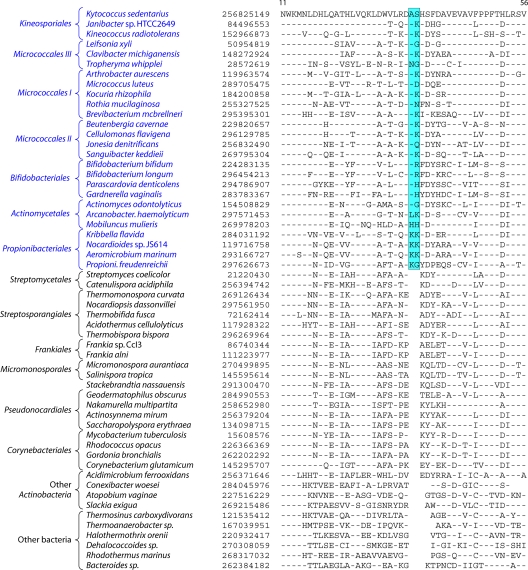
In contrast to these orders, the order Micrococcales, which is one of the largest orders of the Actinobacteria, did not form a phylogenetically coherent cluster, and Bifidobacteriales and Actinomycetales were interspersed within this order of bacteria (Fig. 2). Based upon the 16S rRNA tree and the consensus tree reported by Alam et al. (5), the order Bifidobacteriales formed the deepest-branching lineage within the class Actinobacteria (314, 343). However, in recent works by Ludwig et al. (191) and Adekambi et al. (3), it was indicated to branch in a position similar to that seen in the present work. The order Micrococcales is the most diverse group within the phylum/class Actinobacteria, and the relationships within this order cannot be resolved by the 16S rRNA tree with any degree of confidence (76, 191, 296, 343). In the phylogenetic tree shown in Fig. 2, the species of this order are split into at least three clusters. Of these, cluster I included Arthrobacter, Renibacterium, Micrococcus, Kocuria, Rothia, Brachybacterium, and Brevibacterium; cluster II consisted of Beutenbergia, Jonesia, Cellulomonas, Sanguibacter, and Xylanimonas; and cluster III consisted of Clavibacter, the marine actinobacterium PHSC20C1, Leifsonia, and the fast-evolving intracellular parasite Tropheryma. The relationships of different genera within these clusters is discussed in more detail below in conjunction with signature sequences.
In the phylogenetic tree shown in Fig. 2, species of several genera (e.g., Geodermatophilus, Nakamurella, Stackebrandtia, and Janibacter, etc.) branched within clades that do not correspond to their expected position based on the current taxonomic classification (103, 215, 343). The branching of these species in the observed positions is also independently supported by several conserved indels that are discussed in later sections. Overall, the phylogenetic tree based on combined protein sequences (Fig. 2) provides a useful reference point to interpret the species distribution patterns of various conserved indels.
USEFULNESS OF CONSERVED SIGNATURE INDELS AND SIGNATURE PROTEINS AS MOLECULAR MARKERS FOR PHYLOGENETIC/SYSTEMATIC STUDIES
The shared derived characters that are unique to particular groups or clades of organisms (i.e., synapomorphies) provide an important means for identifying various clades and also for an understanding of how these clades are related to each other. In the past, this approach has been employed largely by using morphology and other observable traits (271, 272). However, often, such traits either are plesiomorphic (i.e., a particular character is not limited to a given group) or exhibit homoplasy (the derived character state has evolved independently in the given group of organisms), which limits their utility as phylogenetic or taxonomic markers. In recent years, the availability of genome sequences has led to the discovery of large numbers of molecular characteristics that are uniquely shared by different groups of organisms and provide important means for the identification of different clades and an understanding of their evolutionary relationships (11, 13, 119, 121, 123, 126, 128, 131, 243, 246). The molecular characteristics that are ideally suited for evolutionary and systematic studies are those that are specifically present in all species belonging to certain bacterial taxa but that are not found outside those lineages. Due to their specificity for particular taxa or phylogenetically well-defined clades, the genetic events leading to the origins of these molecular characteristics likely occurred in the common ancestors of these clades, and these characteristics were then passed on to their various descendants vertically. Thus, such molecular synapomorphies act as hallmarks recording the divergence of different lineages, which can be used to delineate different taxa and clades at various phylogenetic depths (123, 124, 126, 128). The markers that are ideally suited for evolutionary and systematic studies are those that are generally not affected by factors such as multiple changes at a given site, long-branch attraction effects, differences in evolutionary rates, and lateral gene transfers, etc., which confound the inferences from phylogenetic trees (10, 81, 82, 119, 126, 130, 206). Two types of molecular markers that generally satisfy these characteristics have been identified recently for a number of bacterial phyla, and they are proving to be of great value in our understanding of bacterial phylogeny and systematics (124, 126, 127, 130, 132, 133, 179).
The first kind of these molecular markers is comprised of conserved signature indels (CSIs) of defined lengths that are present in gene/protein sequences at specific positions and which are uniquely shared by particular groups of organisms (11, 119, 121, 126, 128, 132, 243, 246). Because of the rare and highly specific nature of the genetic event that gives rise to a given conserved indel, such changes are less likely to arise independently by either convergent or parallel evolution (i.e., homoplasy) (119, 121, 126, 246). Furthermore, the presence or absence of CSIs in protein sequences should not be generally affected by factors, such as differences in evolutionary rates at different sites or among different species, that greatly influence the branching patterns of species in phylogenetic trees (82, 83, 114). Hence, when a CSI of a defined size is uniquely found in a phylogenetically defined group(s) of species, its most parsimonious explanation is that the genetic change responsible for this CSI occurred once in a common ancestor of this group and was then passed on vertically to its various descendants. Since both large as well as small CSIs (including 1-amino-acid [aa] indels) are products of single unique genetic events, they both provide reliable phylogenetic markers of a common evolutionary descent (119, 121, 126, 246, 269). Because genetic changes leading to CSIs could occur at various stages during evolution, it is possible to identify CSIs in gene/protein sequences at different phylogenetic depths corresponding to various taxonomic groupings (e.g., phylum, order, family, genus, and even single-species and subspecies levels). Additionally, based upon the presence or absence of these CSIs in the outgroup species, it is possible to infer whether a given CSI represents an insertion or a deletion in a particular clade and which of the two character states of the protein is ancestral (116, 119, 122, 132). Thus, by making use of CSIs that have been introduced at various stages in evolution, it is possible to derive a rooted evolutionary relationship among various taxa under consideration (119, 122, 129). In some cases where a given CSI is present in unrelated groups of organisms, this can be a consequence of lateral gene transfers (LGTs) or due to the independent occurrence of similar genetic events (117, 117, 119).
The second kind of molecular markers that have proven very useful for systematic and phylogenetic studies is whole proteins or conserved signature proteins (CSPs) that are confined to particular lineages (100, 128, 130). In contrast to the orphan open reading frame (ORFan) proteins that are specific for particular species or strains and are subject to rapid loss (64, 175, 265), many proteins of unknown (or even known) functions are unique and distinctive, characteristic of various species from monophyletic clades of different phylogenetic depths (74, 98, 100, 118, 130, 274). The presence of these proteins in a conserved state in all or most species and strains from these clades, but nowhere else, suggests that the genes for these proteins first evolved in a common ancestor of these clades, followed by their retention by various descendants (74, 80, 98, 100, 210). Thus, these proteins represent CSPs that are distinctive characteristics of particular lineages, and they provide useful molecular markers for defining or distinguishing those groups from other bacteria (118). However, when a CSP (or CSI) is confined to certain species or strains, based upon this information alone, it is difficult to determine whether these species form a clade in the phylogenetic sense or not. Hence, to understand the evolutionary significance of these signatures, such studies are generally performed in conjunction with phylogenetic analyses, which provide a reference point for evaluating the significance of various CSIs and CSPs (99, 118). In the work leading to this review, we carried out extensive work on actinobacterial genomes to identify CSIs and CSPs that are specific for all (or most) sequenced actinobacteria or their different groups or clades at various phylogenetic depths. The identification of these molecular markers was carried out as described in our previous work (97, 100, 130), and additional information in this regard is provided in File S1B in the supplemental material.
MOLECULAR MARKERS OF THE PHYLUM ACTINOBACTERIA
The phylum Actinobacteria is currently identified solely on the basis of the branching patterns of different species in the 16S rRNA tree (103, 110, 191, 283, 343). However, there is no known unique feature or characteristic that is commonly shared by all or most constituent taxa of this phylum. Because phylogenetic trees have no distinct boundaries, in the absence of any distinctive property of the group of species, it is difficult to delimit a group based solely on the branching in the phylogenetic trees (192, 193, 223, 234, 329). Hence, it is of central importance to determine what unique properties are shared by different species of this phylum that could be employed to more precisely define and circumscribe member species of this phylum (126, 130, 132).
CSIs That Are Uniquely Present in Most Actinobacteria
We have previously described two CSIs, consisting of a 2-aa deletion in cytochrome c oxidase subunit 1 (Cox1) and a 4-aa insert in CTP synthetase, that were uniquely present in almost all actinobacteria except for the deepest-branching genus, Rubrobacter (97). A 5-aa insert in glutamyl-tRNA synthetase (GluRS) was also identified, but it was lacking in several actinobacterial species (97). Additionally, a large insert in the 23S rRNA is also specific for most actinobacterial species (97, 248). Our recent analyses of protein sequences from actinobacterial genomes identified 6 additional CSIs in various proteins that are uniquely shared by most of the sequenced actinobacterial species. These CSIs include a 4-aa insert in the protein glucosamine-fructose-6-phosphate aminotransferase (Gft) (Fig. 3), which catalyzes the formation of glucosamine 6-phosphate and is the first and rate-limiting enzyme of the hexosamine biosynthetic pathway (290); a 3-aa insert in the enzyme glycyl-tRNA synthetase (GlyRS) that is required for protein synthesis (see File S2 in the supplemental material); a 4- to 6-aa insert in the enzyme tRNA (guanine-1)-methyltransferase (TrmD) that methylates guanosine 37 in various tRNAs (see File S3 in the supplemental material) (230); a 4-aa insert in gyrase A, which plays an essential role in DNA replication and transcription due to its ability to make transient double-strand breaks in DNA to maintain appropriate levels of supercoiling (see File S4 in the supplemental material) (185); a 9-aa insert in the enzyme S-adenosyl-l-homocysteine hydrolase (SAHH) that hydrolyzes S-adenosyl-homocysteine, which is an end product of various methylation reactions (see File S5 in the supplemental material) (143); and, finally, a 5-aa insert in the enzyme serine hydroxymethyltransferase (SHMT), which catalyzes the reversible interconversion of serine and glycine (see File S6 in the supplemental material) (117, 238).
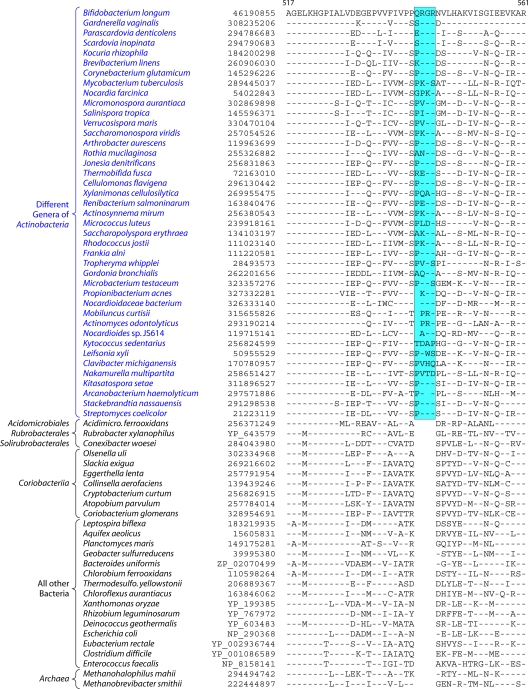
Fig 3.
Partial sequence alignment of the protein glucosamine-fructose-6-phosphate aminotransferase (GFT) showing a 4-aa insert that is uniquely present in different genera belonging to the class Actinobacteria but is not found in Coriobacteriia, Rubrobacter, Acidimicrobiia, and Thermoleophilia or any other prokaryotic organism. Sequence information for several other CSIs that are specifically found in most Actinobacteria is presented in Files S2 to S6 in the supplemental material and Table 2. The dashes in this as well as all other sequence alignments indicate identity with the amino acid on the top line. The numbers on the top lines indicate the sequence region where this CSI is found in the species shown at the top. The second column shows the GenBank accession number or GenBank identification (gi) number for the sequences. Sequence information for a limited number of Actinobacteria is shown in this alignment. However, detailed information regarding the presence or absence of this CSI in various sequenced genera of Actinobacteria is provided in the Table 2.
A partial sequence alignment of the protein glucosamine-fructose-6-phosphate aminotransferase showing the Actinobacteria-specific insert is presented in Fig. 3. The absence of this indel in the Archaea as well as other bacterial phyla provides evidence that this indel constitutes an insert in the Actinobacteria rather than a deletion in other groups. The sequence alignments for other newly identified CSIs that are uniquely present in most Actinobacteria are provided in Files S2 to S6 in the supplemental material. In all of these proteins, the identified CSIs are present in highly conserved regions. Table 2 presents information regarding the specificity as well as the presence or absence of these CSIs as well as the CSIs in Cox1, CTP synthetase, and 23S rRNA in different genera of Actinobacteria. As shown in Table 2, most of these CSIs are highly specific characteristics of the phylum Actinobacteria. The CSIs in Cox1, CTP synthase, Gft, TrmD, and 23S rRNA are not found in any other bacteria except Actinobacteria, whereas for the other signatures, CSIs of similar lengths were also present in a small number of distantly related organisms, which could be due to either LGT or the independent occurrence of similar genetic changes in these lineages. From the species distribution profiles of these CSIs, it is clear that while most of these CSIs are commonly shared by virtually all sequenced genera belonging to the class Actinobacteria, they are generally not found in the deeper-branching lineages of Actinobacteria. Of these CSIs, only the Cox1 CSI was present in the genus Acidimicrobium, while the genus Conexibacter contained CSIs in the SAHH and SHMT proteins. However, none of these CSIs were detected in Rubrobacter or the Coriobacteriia. For some of these proteins, their homologs were also not detected in most of the Coriobacteriia (Table 2).
Table 2.
Presence or absence of various CSIs in different genera of Actinobacteriaa
| Genus | Presence of CSI in: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CoxI | CTPS | Gft | GlyRS | TrmD | Gyrase A | SAHH | SHMT | 23S rRNA | |
| Mycobacterium | + | + | + | + | + | + | + | + | + |
| Tsukamurella | + | + | − | + | + | + | + | + | + |
| Gordonia | + | + | + | + | + | + | + | + | + |
| Nocardia | + | + | + | + | + | + | + | + | + |
| Rhodococcus | + | + | + | + | + | + | + | + | + |
| Segniliparus | + | + | + | + | + | + | + | + | + |
| Amycolicicoccus | + | + | + | + | + | + | + | + | + |
| Corynebacterium | + | + | + | + | + | + | + | + | + |
| Nakamurella | + | + | + | + | + | + | + | + | + |
| Pseudonocardia | + | + | + | + | + | + | + | + | + |
| Saccharopolyspora | + | + | + | + | + | + | + | + | + |
| Actinosynnema | + | + | + | + | + | + | + | + | + |
| Saccharomonospora | + | + | + | + | + | + | + | + | + |
| Amycolatopsis | + | + | + | + | + | + | + | + | + |
| Geodermatophilus | + | + | + | + | + | + | + | + | + |
| Stackebrandtia | + | + | + | + | + | + | + | 0 | + |
| Verrucosispora | + | + | + | + | + | + | + | 0 | + |
| Micromonospora | + | + | + | + | + | + | + | − | + |
| Salinispora | + | + | + | + | + | + | + | − | + |
| Frankia | + | + | + | 0 | 0 | + | + | − | + |
| Acidothermus | + | + | + | + | + | + | + | − | + |
| Streptosporangium | + | + | + | + | + | + | + | − | + |
| Thermomonospora | + | + | + | + | + | + | + | + | + |
| Thermobifida | + | + | + | + | + | + | + | + | + |
| Nocardiopsis | + | + | + | + | + | + | + | + | + |
| Catenulispora | + | + | + | + | + | + | + | − | + |
| Streptomyces | + | + | + | + | + | + | + | + | + |
| Propionibacterium | + | + | + | + | + | + | 0 | − | + |
| Kribbella | + | + | − | + | + | + | + | − | + |
| Nocardioides | + | + | + | + | + | + | + | − | + |
| Kineococcus | + | + | + | + | + | + | + | + | + |
| Janibacter | + | + | + | + | + | + | + | − | + |
| Kytococcus | + | + | + | + | + | + | 0 | − | + |
| Brachybacterium | + | + | + | + | + | + | 0 | + | + |
| Brevibacterium | + | + | + | + | + | + | + | + | + |
| Intrasporangium | + | + | + | + | + | + | + | + | + |
| Isoptericola | + | + | + | + | + | + | + | + | + |
| Microbacterium | + | + | + | + | + | + | 0 | + | + |
| Rothia | + | + | + | + | + | + | 0 | + | + |
| Kocuria | + | + | + | + | + | + | 0 | + | + |
| Micrococcus | + | + | + | + | + | + | + | + | + |
| Renibacterium | + | + | + | + | + | + | + | + | + |
| Arthrobacter | + | + | + | + | + | + | + | + | + |
| Beutenbergia | + | + | + | + | + | + | + | + | + |
| Cellulomonas | + | + | + | + | + | + | 0 | + | + |
| Xylanimonas | + | + | + | + | + | + | + | + | + |
| Jonesia | + | + | + | + | + | + | + | + | + |
| Sanguibacter | + | + | + | + | + | + | + | + | + |
| Mobiluncus | 0 | 0 | + | + | + | + | 0 | + | + |
| Actinomyces | 0 | + | + | + | + | + | 0 | + | + |
| Arcanobacterium | 0 | 0 | + | + | + | + | − | + | + |
| Gardnerella | 0 | + | + | + | + | + | 0 | 0 | + |
| Bifidobacterium | 0 | + | + | + | + | + | 0 | + | + |
| Tropheryma | + | + | + | 0 | + | + | 0 | + | + |
| Leifsonia | + | + | + | + | + | + | + | + | + |
| Clavibacter | + | + | + | + | + | + | + | + | + |
| Marine actinobacterium | + | + | + | + | + | + | 0 | + | + |
| Acidimicrobium | + | − | − | − | 0 | − | − | − | − |
| Conexibacter | − | − | − | − | − | − | + | + | − |
| Rubrobacter | 0 | − | − | − | − | − | − | − | − |
| Olsenella | 0 | − | − | 0 | 0 | − | 0 | 0 | − |
| Slackia | 0 | − | − | 0 | 0 | − | − | − | − |
| Eggerthella | 0 | − | − | 0 | 0 | − | 0 | − | − |
| Cryptobacterium | 0 | 0 | − | 0 | 0 | − | 0 | 0 | − |
| Coriobacterium | 0 | − | − | 0 | 0 | − | 0 | 0 | − |
| Non-Actinobacteria | None | None | None | Magnetospirillum + few planctomycetes | None | Some Firmicutes, 1 Bacteroides sp., 1 Agrobacterium sp. | Anaeromyxobacter, Fibrobacter succinogenes | Some fungi | None |
The presence or absence of various CSIs in different genera of genome-sequenced Actinobacteria was determined by means of Blastp searches. The symbols + and − indicate whether the indicated CSI is present or absent in the species of various genera. The symbol “0” indicates that no homologs of these proteins were detected in these genera. The abbreviations for the proteins are as follows: Cox1, cytochrome oxidase subunit 1; CTPS, CTP synthetase; GFT, glucose fructose 6-PO4 aminotransferase; GlyRS, glycyl-tRNA synthetase; TrmD, tRNA (guanine-1)-methyltransferase; SAHH, S-adenosyl-l-homocysteine hydrolase; SHMT, serine hydroxymethyltransferase. The sequence alignments for Cox1, CTP synthetase, and 23S rRNA showing the presence of the CSIs in these genes/proteins were described in previous work (97, 100). Information for other CSIs is provided in the Fig. 2 and Files S2 to S6 in the supplemental material. Besides Actinobacteria, in some cases, CSIs of similar lengths can also be found in an isolated or limited number of species of other groups of organisms. This could be due to LGT, or it could also result from independent genetic events.
CSPs That Are Specific for the Phylum Actinobacteria
In addition to these CSIs, our Blastp analyses of several actinobacterial genomes (viz., M. leprae TN, Leifsonia xyli subsp. xyli strain CTCB07, Bifidobacterium longum NCC2705, and T. fusca YX) previously identified 29 CSPs that were indicated to be specific for either all or most genome-sequenced Actinobacteria (100). Since the number of sequenced actinobacterial genomes has increased from 25 at that time to >150 at present, the Actinobacteria specificities of these proteins were reexamined. Of the 29 proteins that were reported to be specific for all (or most) of the Actinobacteria, 24 are still specifically present in all of the sequenced actinobacterial genera, except for a few of the deepest-branching lineages (see File S7 in the supplemental material). A summary of the properties of these proteins and information regarding their Actinobacteria specificities are provided in Table 3. Except for Actinobacteria, homologs showing significant similarity to these proteins are not found in any other bacterial phyla. Five proteins that were previously retained despite their presence in some other bacterial groups are now excluded from Table 3. The 24 proteins listed in Table 3 are present in virtually all sequenced genera (total of 57) belonging to the class Actinobacteria (see File S7 in the supplemental material). The homologs of two of them, viz., ML1306 (GenBank accession number NP_301939.1) and ML1009 (accession number NP_301746.1), were also found in Rubrobacter xylanophilus, Conexibacter woesei, and Acidimicrobium ferrooxidans, belonging to the classes Rubrobacteria, Thermoleophilia, and Acidimicrobiia, respectively. Homologs of four additional proteins (viz., ML0642, ML1029, ML0760, and ML0804) were also present in one or two of these three classes (see File S7 in the supplemental material). However, significantly, of all the CSPs identified by comparative genomic analyses, the homologs of none of them were detected in any of the members of the class Coriobacteriia.
Table 3.
Signature proteins that are uniquely found in all (or most) Actinobacteriaa
| Gene | GenBank accession no. | Protein function (reference[s]) | Length (aa) | Species specificity |
|---|---|---|---|---|
| ML1306 | NP_301939.1 | ParJ, chromosome segregation (71) | 274 | All except Coriobacteriia |
| ML1009 | NP_301746.1 | Hypothetical | 326 | All except Coriobacteriia |
| ML0642 | NP_301530.1 | Hypothetical | 479 | All except Acidimicrobiia and Coriobacteriia |
| ML1029 | NP_301762.1 | Hypothetical | 273 | All except Acidimicrobiia and Coriobacteriia |
| ML0760 | NP_301589.1 | whiB-like, sporulation (31, 90) | 89 | All except Coriobacteriia and Rubrobacter |
| ML0804 | NP_301614.1 | whiB-like, sporulation(31, 90) | 84 | All except Coriobacteriia and Rubrobacter |
| ML0857 | NP_301645.1 | Hypothetical | 250 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML0869 | NP_301656.1 | Hypothetical | 124 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML1016 | NP_301752.1 | Hypothetical | 107 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML1026 | NP_301759.1 | Hypothetical | 100 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML2137 | NP_302410.1 | Hypothetical | 251 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML2204 | NP_302445.1 | Hypothetical | 62 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML0013 | NP_301140.1 | Septation inhibitor protein (31) | 93 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML0007 | NP_301135.1 | Hypothetical | 303 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML0580 | NP_301492.1 | Hypothetical | 265 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML0921 | NP_301704.1 | Hypothetical | 96 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML1439 | NP_302017.1 | Hypothetical | 111 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML1610 | NP_302109.1 | Hypothetical | 101 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML2207 | NP_302448.1 | Hypothetical | 131 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML0775 | NP_301599.1 | LpqB, cell wall-related process (212) | 589 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML0761 | NP_301590.1 | Hypothetical | 167 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML0814 | NP_301620.1 | Hypothetical | 82 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML1649 | NP_302131.1 | Hypothetical | 140 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
| ML2142 | NP_302413.1 | Hypothetical | 269 | All except Coriobacteriia, Rubrobacter, and Acidimicrobiia |
All significant Blast hits for these proteins (barring an isolated exception) were observed for Actinobacteria. The first and second columns indicate the gene identifications for these proteins from M. leprae and their accession numbers. Most proteins are of unknown functions; however, in a few cases where some information is available, it is noted in the third column. The last column indicates the different classes of Actinobacteria where these proteins are found. Homologs of most of these proteins are present in virtually all genome-sequenced species of the class Actinobacteria. However, their presence or absence in other classes of Actinobacteria is noted in the last column. As noted, none of these proteins are found in any of the species of the class Coriobacteriia.
Predictive Value and Usefulness of the Identified CSIs and CSPs for Delimiting the Phylum Actinobacteria
The results obtained with various CSIs and CSPs are significant in a number of respects. First, they provide important information for validating the specificity and reliability of these signatures. Many of these CSIs and all of these CSPs were identified when the number of sequenced actinobacterial genomes was very limited (97, 100). However, despite a large increase in the number of sequenced genomes (between 6- and 10-fold) for both Actinobacteria as well as other bacteria, most of these signatures are still specific for Actinobacteria. Additionally, most of these signatures are present in virtually all sequenced genera of Actinobacteria, except those from the deepest-branching lineages. Thus, the presence of these signatures can be used to distinguish member species belonging to the class/phylum Actinobacteria from all other bacteria with a high degree of predictive ability. This inference is further strongly supported by our previous work, where the presence of CSIs in Cox1, CTP synthase, and 23S rRNA was examined in a large number of other Actinobacteria belonging to different families, whose genomes have not been sequenced, by PCR amplification of the corresponding fragments (97). The results of those studies showed that of the 50 gene fragments for these three genes that were sequenced from diverse members of the Actinobacteria, all contained the indicated indels, thereby providing strong evidence that these CSIs are distinctive characteristics of various Actinobacteria, even those for whom sequence information is not available at present (97).
Based upon the presence of these CSIs and CSPs, the class Actinobacteria, which comprises more than 90% of the known actinobacterial genera, can now be delimited and circumscribed in clear molecular terms based upon large numbers of independent molecular markers that are unique characteristics of different members of this class (Fig. 3 and Tables 2 and 3, and see Files S2 to S6 in the supplemental material). Based upon the two CSPs that are uniquely found in the class Actinobacteria and members of the classes Acidimicrobiia, Rubrobacteria, and Thermoleophilia, a case can also be made that these bacterial groups are specifically related to the class Actinobacteria and that they should thus be part of the phylum Actinobacteria. However, detailed analyses of the genomes of Actinobacteria have not identified any CSP or CSI that is commonly shared by the above-mentioned classes Actinobacteria and Coriobacteriia, which is now represented by five sequenced genomes (Table 1) (108, 195, 251). This observation in conjunction with the fact that the Coriobacteriia are separated from all other members of the Actinobacteria by a long branch in the phylogenetic tree (Fig. 2) makes a strong case for the exclusion of the Coriobacteriia from the phylum Actinobacteria. It should be noted in this context that the absence of various CSIs and CSPs in Symbiobacterium thermophilum, which was previously placed into the phylum Actinobacteria, argued against its inclusion within this phylum (97, 100). This inference was later strongly supported by its genome sequence and other lines of evidence (174, 310), and this species is now grouped with the Firmicutes (77). No sequences are available at present for the two genera (viz., Euzebya and Nitriliruptor) that make up the class Nitriliruptoria (77, 176, 191). Hence, the affiliation of Nitriliruptoria with other classes of the phylum Actinobacteria (viz., Actinobacteria, Acidimicrobiia, Rubrobacteria, and Thermoleophilia) cannot be confirmed at present.
MOLECULAR SIGNATURES OF THE ORDER CORYNEBACTERIALES AND SOME OF ITS FAMILIES
The order Corynebacteriales represents one of the largest groups within the actinobacteria in terms of the numbers of genomes that have been sequenced (Table 1). Forty-eight of the sequenced genomes, representing about one-third of the total actinobacterial genomes, are from this order. This is also due to the fact that species of many genera within this order (viz., Mycobacterium, Nocardia, Corynebacterium, and Gordonia) are important human and animal pathogens (39, 53, 54, 56, 72, 204, 252, 264, 342). Members of this order form a strongly supported clade in phylogenetic trees based on 16S rRNA and other gene/protein sequences (Fig. 2) (3, 5, 192, 314, 343). The species of this order, similar to those of the Pseudonocardiales, have cell wall chemotype IV, defined by the presence of meso-diaminopimelic acid, arabinose, and galactose in their cell walls (111, 182, 295). However, unlike species of the order Pseudonocardiales, which lack mycolic acids, mycolic acids are an important component of the cell envelopes of all species (with the few exceptions noted below) of the order Corynebacteriales (111, 187). Although the presence of mycolic acids in the cell wall is considered to be a defining characteristic of members of the order Corynebacteriales, a number of genera (viz., Turicella and Amycolicicoccus) as well as Corynebacterium amycolatum and C. kroppenstedtii lack mycolic acids (111, 178, 187, 191). Other than the presence of mycolic acids, very few reliable markers that are distinctive characteristics of various species of this order are known.
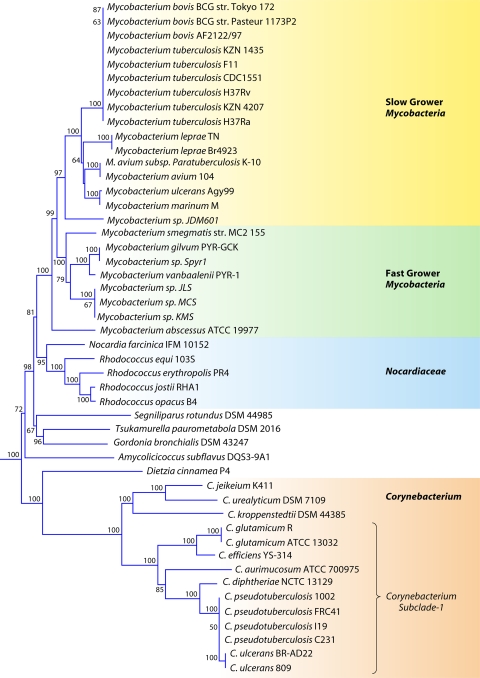
The order Corynebacteriales is currently divided into six families: Corynebacteriaceae, Mycobacteriaceae, Nocardiaceae, Dietziaceae, Segniliparaceae, and Tsukamurellaceae (77, 103, 191). Since genome sequences are now available for species of each of these families, a phylogenetic tree for species from the sequenced genomes was constructed based upon the concatenated sequences of three large and conserved proteins (RpoB, RpoC, and gyrase B) (Fig. 4). In this tree, and also in previous studies (111), species of the families Corynebacteriaceae and Mycobacteriaceae formed strongly supported clades and were clearly distinguished. The genera Rhodococcus and Nocardia, which until recently were the only two genera that constituted the family Nocardiaceae (103), also formed a well-supported clade in the tree. This clade branched distinctly from Gordonia bronchialis, which is now proposed to be a part of the family Nocardiaceae (191). A clade consisting of Gordonia and Tsukamurella was supported both in this phylogenetic tree as well as in the tree shown in Fig. 2.
Fig 4.
Bootstrapped neighbor-joining tree for Corynebacteriales species based upon concatenated sequences for the RpoB, RpoC, and gyrase B proteins. The distinctness of a number of clades seen in this tree is independently supported by many identified CSIs and CSPs.
CSIs and CSPs That Are Specific for the Order Corynebacteriales
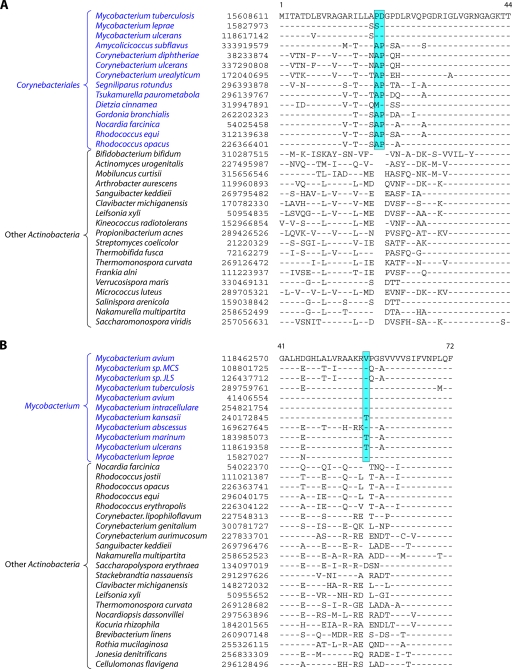
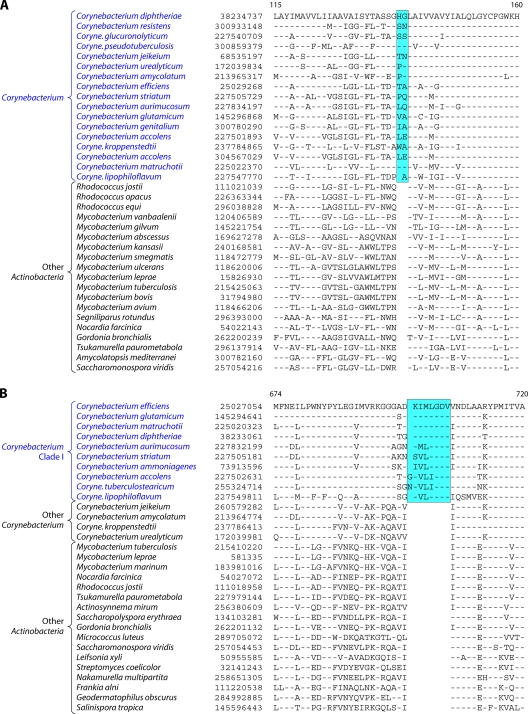
Analyses of protein sequences from Corynebacteriales genomes have identified many CSIs and CSPs that are specific for members of this order. In a macrolide transporter ATP-binding protein, a 2-aa insert in a conserved region is specifically present in all of the Corynebacteriales but no other Actinobacteria (Fig. 5A). Likewise, in the enzyme alpha-ketoglutarate decarboxylase (KGD), which is a part of the tricarboxylic acid cycle (301), a 1-aa deletion in a conserved region is uniquely present in all available Corynebacteriales sequences (see File S8 in the supplemental material). Although sequence information is shown for only a limited number of Corynebacteriales and other Actinobacteria, these indels are highly specific characteristics of all Corynebacteriales and are not found in any other Actinobacteria. Another CSI consisting of a 1-aa insert that is largely specific for the order Corynebacteriales is found in the chromosome segregation DNA-binding protein (ParB) (see File S9 in the supplemental material), which binds to DNA at the origin of replication and is involved in chromosome partitioning (156). The conserved insert in ParB is again present in all of the sequenced genera of Corynebacteriales, and with the sole exception of Leifsonia xyli, it is not found in any other Actinobacteria or in other phyla of bacteria. The presence of this indel in L. xyli could be due to LGT or could result from other possibilities that we cannot distinguish at present. Interestingly, the insert in ParB, although it is present in most of the genome-sequenced Corynebacterium species, is not found in C. aurimucosum and a number of other Corynebacterium species (viz., C. ammoniagenes, C. pseudogenitalium, C. tuberculostearicum, C. accolens, C. striatum, and C. glucuronolyticum), whose genomes are not sequenced and which are not shown in the phylogenetic tree in Fig. 4. In a phylogenetic tree based on ParB protein sequences, the Corynebacterium species lacking this insert formed a distinct clade (see File S10 in the supplemental material). Hence, the most plausible way to explain the species distribution of this indel is that the genetic change leading to this occurred in a common ancestor of the Corynebacteriales, followed by the loss of this CSI from this gene, or LGT of this gene, in this particular subclade of Corynebacterium.
Fig 5.
(A) Partial sequence alignment of a macrolide ABC transporter ATP-binding protein showing a 2-aa conserved indel that is uniquely present in various Corynebacteriales species. Information for two other CSIs that are specific for Corynebacteriales is provided in Files S8 and S9 in the supplemental material. Sequence information for most of the CSIs is shown for a limited number of species; however, unless otherwise indicated, they are specific for the indicated groups. (B) Excerpt from the sequence alignment of the pantoate beta-alanine ligase (PanC) protein showing a 1-aa conserved insert that is specific for Mycobacterium species but not found in any other Actinobacteria. Sequence information for another Mycobacterium-specific CSI in the protein OMP-decarboxylase is presented in File S11 in the supplemental material.
In addition to these CSIs, our Blast analysis of various proteins from the genome of Corynebacterium glutamicum ATCC 13032 identified four CSPs (Table 4), for which homologs showing significant sequence similarity are restricted to all of the sequenced Corynebacteriales species but are not detected in other bacteria. Two of these proteins, viz., arabinosyltransferase (EmbB) and AftA, are involved in the synthesis of cell wall arabinan (6, 24, 259), whereas the other proteins are of unknown functions.
Table 4.
Signature proteins that are specific for the order Corynebacterialesa
| Gene or protein | GenBank accession no. | Protein function (reference) | Length (aa) |
|---|---|---|---|
| ML0099 | NP_301197 | Hypothetical | 336 |
| Arabinosyl transferase (EmbB) | NP_301201 | Mycobacterial cell wall arabinan synthesis protein (300) | 1,083 |
| AftA (ML0107) | NP_301204 | Cell wall arabinan biosynthesis (6) | 632 |
| ML1270 | NP_301915 | Tryptophan-associated transmembrane protein | 265 |
These signature proteins were identified by Blastp searches for different proteins from the genome of Mycobacterium leprae TN. For these proteins, all significant Blast hits were observed for the order Corynebacteriales.
Molecular Signatures of Mycobacteriaceae/Mycobacterium
The family Mycobacteriaceae contains a single genus, Mycobacterium, which harbors some of the most important human pathogens, including those responsible for tuberculosis and leprosy (103, 142, 191, 252, 264). Sequence information for large numbers of species of this genus is now available (viz., M. tuberculosis, M. abscessus, M. avium, M. bovis, M. gilvum, M. leprae, M. marinum, M. ulcerans, and M. vanbaalenii) (Table 1) (1, 32, 55, 56, 88, 101, 186, 204, 242, 260, 288, 342). Multiple strains have been sequenced for a number of species. Mycobacterium species have been divided into two major groups (slow growers and fast growers) depending upon their growth rates (142). The species of these two groups generally branch distinctly in phylogenetic trees (232, 285). Their distinctness is also supported by the presence of a longer helix between positions 451 and 482 in the 16S rRNA gene in the slow growers than in the fast growers (232). Of these, the slow-growing Mycobacterium species/strains are clinically important, whereas the fast growers are ecologically important (142). In the phylogenetic tree shown in Fig. 4, all of the sequenced Mycobacterium species/strains formed a strongly supported clade, and within it, a cluster consisting of the slow-growing Mycobacterium species was also strongly supported. We have identified a number of CSIs and CSPs that are specific for either all sequenced Mycobacterium species or the slow-growing clade. Sequence information for one of the CSIs that is specific for the genus Mycobacterium is presented in Fig. 5B. In the enzyme pantoate-beta-alanine ligase, which is involved in the metabolism of beta-alanine (200), a 1-aa insert in a conserved region is uniquely present in all of the sequenced Mycobacterium species but is not found in any other bacteria (Fig. 5B). Similarly, in the enzyme orotidine-5′-phosphate-decarboxylase (OMP-decarboxylase), which catalyzes the last essential step in the de novo biosynthesis of pyrimidines (199), a 1-aa deletion is specifically present in all Mycobacterium species (see File S11 in the supplemental material). Both these signatures are highly specific for the sequenced Mycobacterium species and provide novel molecular markers for this genus.
In our earlier work, Blastp searches for various proteins from the genome of M. leprae TN led to the identification of 24 CSPs that were indicated to be specific for the genus Mycobacterium (100). A reevaluation of the specificity of these proteins by Blastp searches revealed that all of these proteins are still specific for the genus Mycobacterium. However, of these, the first 18 proteins listed in Table 5 are specifically present in all of the sequenced Mycobacterium genomes (with isolated exceptions as noted), whereas the last 6 proteins are limited to the subclade of slow-growing Mycobacterium species (viz., Mycobacterium bovis, M. tuberculosis, M. ulcerans, M. marinum, M. avium, M. paratuberculosis, M. leprae, and Mycobacterium sp. strain JDM601), which are clinically important members of this genus. Although the exact cellular functions of most of these proteins remain to be determined, some of them are putative virulence factors belonging to the PE/PPE or Lpq family of proteins (158, 289).
Table 5.
Signature proteins that are specific for the genus Mycobacterium or its subcladea
| Gene or protein | GenBank accession no. | Function (references) | Length (aa) | Species specificity |
|---|---|---|---|---|
| PE family protein | YP_879413.1 | Hypothetical | 101 | Genus Mycobacteriumb |
| MAP0046c | NP_958980.1 | Hypothetical | 113 | Genus Mycobacterium |
| PPE family protein | YP_879414.1 | Hypothetical | 557 | Genus Mycobacterium |
| MAV_1008 | YP_880267.1 | Hypothetical | 91 | Genus Mycobacterium |
| Proline-rich 28-kDa antigen | YP_879354.1 | Lipoprotein LpqN (55, 294) | 366 | Genus Mycobacterium |
| MAV_0378 | YP_879665.1 | Hypothetical | 277 | Genus Mycobacteriumb |
| MAV_0398 | YP_879683.1 | Hypothetical | 220 | Genus Mycobacterium |
| MAV_1034 | YP_880290.1 | Hypothetical | 129 | Genus Mycobacteriumb |
| 34 kDa antigenic protein | YP_880332.1 | Hypothetical | 302 | Genus Mycobacterium |
| MAV_1122 | YP_880374.1 | Hypothetical | 220 | Genus Mycobacteriumb |
| LpqT protein | YP_880404.1 | Lipoprotein LpqT (55, 293) | 219 | Genus Mycobacteriumb |
| LprE protein | YP_880642.1 | Hypothetical | 195 | Genus Mycobacterium |
| MAV_1668 | YP_880900.1 | Hypothetical | 253 | Genus Mycobacterium |
| MAV_1760 | YP_880985.1 | Hypothetical | 376 | Genus Mycobacterium |
| MAV_2294 | YP_881498.1 | Hypothetical | 210 | Genus Mycobacterium |
| MAV_2346 | YP_881550.1 | Hypothetical | 131 | Genus Mycobacterium |
| ModD protein | YP_882045.1 | Fibronectin attachment protein | 385 | Genus Mycobacterium |
| MAV_3078 | YP_882262.1 | Hypothetical | 61 | Genus Mycobacterium |
| PPE family protein | YP_883994.1 | Hypothetical | 488 | Mycobacterium subcladec,d |
| PE family protein | YP_882101.1 | Hypothetical | 99 | Mycobacterium subcladec |
| MAV_1177 | YP_880425.1 | Hypothetical | 94 | Mycobacterium subcladec |
| PPE family protein | YP_880574.1 | Hypothetical | 555 | Mycobacterium subcladec |
| PPE family protein | YP_883484.1 | Hypothetical | 529 | Mycobacterium subcladec |
| PPE family protein | YP_884001.1 | Hypothetical | 527 | Mycobacterium subcladec |
These CSPs were identified by Blastp searches for proteins from the genome of M. leprae TN as described previously (100).
A significant Blast hit was also observed for 1 to 2 other species of the suborder Corynebacteriales.
Specific for a subclade consisting of the slow-growing mycobacteria Mycobacterium bovis, M. tuberculosis, M. ulcerans, M. marinum, M. avium, M. paratuberculosis, M. leprae, and Mycobacterium sp. JDM601.
Also found in M. abscessus.
Molecular Signatures of Rhodococcus and Nocardia
The species of the genera Rhodococcus and Nocardia form a strongly supported clade in various phylogenetic trees (Fig. 2) (3, 5, 111, 136, 178). The distinctness of species of these two genera from all other genera or families of the order Corynebacteriales is also strongly supported by several CSIs and CSPs that we have identified. A partial sequence alignment of one protein showing a CSI that is specific for Rhodococcus and Nocardia species is presented in Fig. 6. In a protein annotated as an ATP-binding protein, a 3-aa insert in a conserved region is specifically present in species of these two genera. Similarly, another CSI consisting of a 1-aa deletion in a conserved region is found in the alpha-subunit of the enzyme acetyl coenzyme A (acetyl-CoA) carboxylase (ACC), which catalyzes the irreversible carboxylation of acetyl-CoA to produce malonyl-CoA (51) (see File S12 in the supplemental material). Both these indels are not found in Gordonia bronchialis or any other Actinobacteria. Our Blastp searches of the genome of Rhodococcus jostii RHA1 have led to the identification of 14 CSPs whose homologs are specifically found in Rhodococcus and Nocardia species (Table 6), except for isolated exceptions. However, in our analyses, we have not come across any CSI or CSP that is commonly shared by Rhodococcus and Nocardia species as well as by Gordonia bronchialis, whose genome has been sequenced (153). These observations make a strong case that the family Nocardiaceae should be limited to the genera Rhodococcus and Nocardia, as it was in the past (103, 136), and that the genus Gordonia, which was part of the family Gordoniaceae, should not be merged with it, as was recently proposed (191). There is no sequence information available at present for the genera Skermania and Williamsia to determine if they are specifically related to Rhodococcus and Nocardia.
Fig 6.
Partial sequence alignments of an ATP-binding protein showing a 3-aa CSI that is uniquely found in Rhodococcus-Nocardia species. Another CSI that is specific for Rhodococcus-Nocardia is shown in File S12 in the supplemental material.
Table 6.
Signature proteins that are specific for the family Nocardiaceaea
| Gene | GenBank accession no. | Protein function | Length (aa) | Species specificity |
|---|---|---|---|---|
| RHA1_ro00267 | YP_700261.1 | Hypothetical | 108 | Rhodococcus and Nocardia |
| RHA1_ro00333 | YP_700327.1 | Hypothetical | 172 | Rhodococcus and Nocardia |
| RHA1_ro01075 | YP_701060.1 | Hypothetical | 250 | Rhodococcus and Nocardiab |
| RHA1_ro01170 | YP_701155.1 | Hypothetical | 151 | Rhodococcus and Nocardia |
| RHA1_ro02067 | YP_702032.1 | Hypothetical | 111 | Rhodococcus and Nocardia |
| RHA1_ro02254 | YP_702219.1 | Hypothetical | 207 | Rhodococcus and Nocardia |
| RHA1_ro02467 | YP_702430.1 | Hypothetical | 109 | Rhodococcus and Nocardia |
| RHA1_ro02848 | YP_702811.1 | Hypothetical | 97 | Rhodococcus and Nocardiab |
| RHA1_ro04046 | YP_704001.1 | Hypothetical | 275 | Rhodococcus and Nocardiab |
| RHA1_ro04254 | YP_704203.1 | Hypothetical | 389 | Rhodococcus and Nocardia |
| RHA1_ro05348 | YP_705286.1 | Hypothetical | 201 | Rhodococcus and Nocardiab |
| RHA1_ro05515 | YP_705453.1 | Hypothetical | 311 | Rhodococcus and Nocardia |
| RHA1_ro05936 | YP_705871.1 | Hypothetical | 52 | Rhodococcus and Nocardia |
| RHA1_ro05750 | YP_705686.1 | Hypothetical | 141 | Rhodococcus and Nocardia |
These CSPs were identified by Blastp searches of proteins from the genome of Rhodococcus jostii RHA1.
Significant hits were also observed for other isolated actinobacterial species.
In addition to these CSIs and CSPs that are shared by both Rhodococcus and Nocardia species, we have also identified a 3-aa insert in a hypothetical protein, BlinB_00480, that is specifically shared by all four sequenced Rhodococcus species (viz., R. jostii, R. opacus, R. equi, and R. erythropolis), providing a molecular marker for this genus (see File S13 in the supplemental material).
Molecular Signatures of Corynebacterium and the Corynebacteriaceae
The genus Corynebacterium contains numerous species that are of much interest due to their involvement in human and animal diseases (viz., C. diphtheriae, C. striatum, C. jeikeium, C. urealyticum, C. ulcerans, and C. pseudotuberculosis) and also for large numbers of industrial applications, including the production of amino acids, nucleotides, and other nutritional factors; hydrocarbon degradation; and the bioconversion of steroids, etc. (27, 37, 57, 94, 113, 149, 166, 224, 267, 327). As a result, large numbers of genomes of Corynebacterium species and strains, including C. aurimucosum, C. diphtheriae, C. efficiens, C. glutamicum, C. jeikeium, C. kroppenstedtii, C. pseudotuberculosis, C. ulcerans, and C. urealyticum, have been sequenced, and many are in the process of being sequenced (39, 159, 213, 267, 298, 299, 304, 339). The family Corynebacteriaceae contains two genera, Corynebacterium and Turicella (57, 103, 187, 191). However, currently, no sequences are available for the latter genus, which also lacks mycolic acids, which is a uniquely shared characteristic of most other members of the Corynebacteriales (58, 111, 187, 191). In phylogenetic trees based upon 16S rRNA (187, 227, 343) or concatenated protein sequences (Fig. 2 and 4), Corynebacterium species formed a strongly supported clade, and it was separated from other Corynebacteriales species by a long branch. In the tree shown in Fig. 4, Dietzia was its closest relative, and a clade consisting of these two genera was also strongly supported. Within the clade consisting of Corynebacterium species, a number of distinct clusters or subclades were also well resolved (Fig. 4). The distinctness of Corynebacterium species from all other members of the Corynebacteriales and the existence of a number of discrete clades within this genus are also independently supported by many CSIs and CSPs that we have identified.
The presence of an arabinogalactan polymer in the cell wall is a unique characteristic of members of the orders Corynebacteriales and Pseudonocardiales (111, 182, 187). The enzyme phosphoribose diphosphate:decaprenyl-phosphate phosphoribosyltransferase (UbiA) plays an essential role in this process by catalyzing the transfer of ribose-5-phosphate from phosphoribose diphosphate to decaprenylphosphate to form decaprenylphosphoryl-5-phosphoribose (198). In this enzyme, we have identified a 2-aa insert in a conserved region that is uniquely present in all of the sequenced Corynebacterium species (Fig. 7A) but not in any other Actinobacteria. Similarly, in the enzyme acetate kinase, which carries out the phosphorylation of acetate to produce acetyl phosphate, a 3-aa insert in a conserved region is specifically present in all available sequences of Corynebacterium species (see File S14 in the supplemental material). Another CSI that is specific for Corynebacterium is present in the enzyme protoheme IX farnesyltransferase (CyoE), which is involved in the biosynthesis of heme A (250). Most Corynebacterium species have a 7-aa insert in this protein; however, C. jeikeium and C. urealyticum, which form a distinct clade, contain a longer insert (10 aa) in the same position (see File S15 in the supplemental material). Blast searches for various proteins from the genome of Corynebacterium glutamicum ATCC 13032 have also identified 20 CSPs that are uniquely present in all or most of the sequenced Corynebacterium species (Table 7). While 16 of these 20 CSPs are entirely specific for the genus Corynebacterium, the homologs of three of them are also present in Dietzia cinnamea (belonging to the family Dietziaceae), which forms the outgroup of the Corynebacterium clade in the phylogenetic tree (Fig. 4). The shared presence of these CSPs in Corynebacterium and Dietzia cinnamea supports the inference from the phylogenetic tree (Fig. 4) that species of these two families are distantly but specifically related to each other.
Fig 7.
(A) Partial sequence alignments of the protein phosphoribose diphosphate:decaprenyl-phosphate phosphoribosyltransferase acyl-CoA carboxylase acetate kinase showing a 2-aa conserved insert that is uniquely found in various Corynebacterium species but not in any other bacteria. The acetate kinase and CyoE proteins also contain CSIs that are specific for the genus Corynebacterium (see Files S14 and S15 in the supplemental material). (B) Partial sequence alignments of the RNA polymerase β′-subunit (RpoC) showing a 7- to 8-aa conserved insert that is specifically found in clade I Corynebacterium species (Fig. 4). Another CSI that is specific for clade I Corynebacterium species is present in the GTP-binding protein LepA (see File S16 in the supplemental material).
Table 7.
Signature proteins that are specific for the genus Corynebacteriuma
| Gene | GenBank accession no. | Protein function | Length (aa) | Species specificity |
|---|---|---|---|---|
| NCgl0188 | NP_599444.1 | Hypothetical | 75 | Genus Corynebacteriumb |
| NCgl0238 | NP_599494.1 | Hypothetical | 183 | Genus Corynebacterium |
| NCgl0362 | NP_599621.1 | Hypothetical | 109 | Genus Corynebacterium |
| NCgl0481 | NP_599742.1 | Hypothetical | 233 | Genus Corynebacterium |
| NCgl0588 | NP_599849.1 | Hypothetical | 147 | Genus Corynebacterium |
| NCgl1056 | NP_600329.1 | Hypothetical | 137 | Genus Corynebacterium |
| NCgl1090 | NP_600363.1 | Hypothetical | 267 | Genus Corynebacterium |
| NCgl1456 | NP_600729.1 | Hypothetical | 126 | Genus Corynebacterium |
| NCgl1866 | NP_601148.1 | Hypothetical | 252 | Genus Corynebacterium |
| NCgl2043 | NP_601325.1 | Hypothetical | 77 | Genus Corynebacteriumb |
| NCgl2214 | NP_601494.1 | Hypothetical | 226 | Genus Corynebacterium |
| NCgl2224 | NP_601505.1 | Hypothetical | 585 | Genus Corynebacterium |
| NCgl2534 | NP_601824.1 | Hypothetical | 109 | Genus Corynebacterium |
| NCgl2641 | NP_601932.1 | Hypothetical | 221 | Genus Corynebacteriumc |
| NCgl2776 | NP_602066.1 | Hypothetical | 166 | Genus Corynebacterium |
| NCgl2836 | NP_602124.1 | Hypothetical | 183 | Genus Corynebacteriumb,c |
| NCgl2882 | NP_602180.1 | Hypothetical | 63 | Genus Corynebacterium |
| NCgl2888 | NP_602186.1 | Hypothetical | 165 | Genus Corynebacterium |
| NCgl2197 | NP_601477.1 | Hypothetical | 194 | Genus Corynebacterium |
| NCgl0807 | NP_600070.1 | Hypothetical | 89 | Genus Corynebacterium |
These CSPs were identified by Blastp searches for proteins from the genome of C. glutamicum ATCC 13032.
Also found in Dietzia cinnamea.
Also present in 1 to 2 Pseudonocardiales species.
In the phylogenetic tree shown in Fig. 4, C. diphtheriae, C. pseudotuberculosis, C. ulcerans, C. aurimucosum, C. glutamicum, and C. efficiens formed a distinct cluster (marked as cluster I) within the genus Corynebacterium. The existence of this clade is also strongly supported by a number of identified CSIs and CSPs. One example of a CSI that is specific for cluster I Corynebacterium species is shown in Fig. 7B. In this case, in the β′-subunit of RNA polymerase (RpoC), which is highly conserved and universally distributed, a 7- to 8-aa insert in a conserved region is specifically present in all of the cluster I Corynebacterium species, but it is not found in other species, such as C. jeikeium, C. urealyticum, and C. kroppenstedtii, that are not part of this clade. Another 2-aa insert that is specific for cluster I species is present in a conserved region of the GTP-binding protein LepA (see File S16 in the supplemental material), which plays an important role in protein synthesis, particularly under stress conditions (236). For some species that contain the RpoC and LepA inserts (viz., C. matruchotii, C. striatum, C. ammoniagenes, C. accolens, C. lipophiloflavum, C. tuberculostearicum, and C. glucuronolyticum), because their genomes were not sequenced, sequence information is not present in the phylogenetic tree (Fig. 4). However, based upon the shared presence of CSIs in both the RpoC and LepA proteins, it is predicted that these species will also group with cluster I Corynebacterium species. The genetic distinctness of cluster I is also strongly supported by 21 CSPs that are uniquely present in all of the sequenced species of this cluster (see the first 21 entries in File S17 in the supplemental material). Additionally, File S17 in the supplemental material lists 19 other CSPs that are uniquely found in C. jeikeium and C. urealyticum, which form another strongly supported cluster in the phylogenetic tree (Fig. 4) (187). These CSIs and CSPs provide novel molecular markers for the identification and circumscription of the genus Corynebacterium and two of its clades. It should be noted that the genetic distances between these subclades of the genus Corynebacterium are greater than those observed among or between species of the families Mycobacteriaceae and Nocardiaceae. Hence, it can be argued that species of these subclades should be recognized as distinct genera rather than being part of the same genus. It should also be noted that the various CSIs or CSPs that are specific for cluster I Corynebacterium species or for C. jeikeium and C. urealyticum are not found in C. kroppenstedtii, which is separated from both of these clusters by a long branch (Fig. 4). Unlike other Corynebacterium species, C. kroppenstedtii lacks mycolic acid (298), and its phylogenetic position and the absence of signatures for the other two clusters suggest that it forms a distinct subgroup of Corynebacterium species.
Molecular Signatures Supporting the Deeper Branching of Corynebacterium and Dietzia within the Order Corynebacteriales
Within the Order Corynebacteriales, a clade consisting of Corynebacterium species and Dietzia shows the deepest branching, and it is separated from other Corynebacteriales by a long branch (Fig. 4). Within this clade, the species belonging to the families Mycobacteriaceae and Nocardiaceae generally group together, and the other Corynebacteriales species branch in between these two clades. Our analyses of Corynebacteriales genomes have identified a number of CSPs that further strongly support these relationships. Table 8 lists a number of CSPs that are present in most other Corynebacteriales species except Corynebacterium species and Dietzia. The genes for these proteins likely originated from a common ancestor(s) of the other Corynebacteriales following the divergence of Corynebacterium and Dietzia. Of the proteins listed in Table 8, the first four are found mainly in Mycobacteriaceae and Nocardiaceae species, supporting a closer relationship between these two families. The homologs of the remainder of these CSPs are found in Gordonia bronchialis and also, in some cases, in Segniliparus rotundus, Tsukamurella paurometabola, and Amycolicicoccus subflavus, supporting their branching in between the Corynebacterium-Dietzia clade and the Mycobacteriaceae-Nocardiaceae clade.
Table 8.
CSPs that are present in most members of the Corynebacteriales except Corynebacterium
| Gene or protein | GenBank accession no. | Protein function | Length (aa) | Species specificitya |
|---|---|---|---|---|
| MAV_0513 | YP_879795.1 | Hypothetical | 328 | Mycobacterium and Nocardiaceae |
| MAV_1758 | YP_880983.1 | Hypothetical | 216 | Mycobacterium and Nocardiaceae |
| MAV_3193 | YP_882377.1 | Hypothetical | 225 | Mycobacterium and Nocardiaceae |
| MAV_0614 | YP_879894.1 | Hypothetical | 133 | Mycobacterium, Nocardiaceae, and Gordonia |
| MAV_0754 | YP_880029.1 | Hypothetical | 32 | Mycobacterium, Nocardiaceae, and Gordonia |
| LysM domain-containing protein | YP_882790.1 | Bacterial cell wall degradation | 164 | Mycobacterium, Nocardiaceae, and Gordonia |
| MAV_4251 | YP_883392.1 | Hypothetical | 86 | Mycobacterium, Nocardiaceae, and Gordonia |
| MAV_0454 | YP_879736.1 | Hypothetical | 126 | Mycobacterium, Rhodococcus, and Gordoniab |
| MAV_4261 | YP_883402.1 | Hypothetical | 108 | Mycobacterium, Nocardiaceae, and Gordoniab |
| MAV_5300 | YP_884410.1 | Hypothetical | 254 | Mycobacterium, Nocardiaceae, and Gordoniab |
| MAV_2940 | YP_882126.1 | Hypothetical | 186 | Mycobacterium, Nocardiaceae, and Gordoniab |
| MAV_4016 | YP_883169.1 | Hypothetical | 117 | Mycobacterium, Nocardiaceae, and Gordoniab |
These CSPs were identified by Blastp searches for proteins from the genome of Mycobacterium avium 104. Although these CSPs are found mainly in the indicated families of Corynebacteriales, isolated hits for a few of them may also be present in 1 to 2 other species.
Also present in one or more of the following Corynebacteriales species: Segniliparus rotundus, Tsukamurella paurometabola, and Amycolicicoccus subflavus.
MOLECULAR SIGNATURES SHOWING THAT CORYNEBACTERIALES AND PSEUDONOCARDIALES ARE CLOSELY RELATED
The orders Pseudonocardiales and Corynebacteriales are the only two orders within the phylum Actinobacteria that have cell walls containing meso-diaminopimelic acid, arabinose, and galactose (cell wall chemotype IV) (58, 111, 187). However, unlike the Corynebacteriales, mycolic acids are absent from the cell walls of Pseudonocardiales species. In phylogenetic trees based upon 16S rRNA or other gene/protein sequences, Pseudonocardiales species generally cluster with species of the order Corynebacteriales (3, 5, 343), but this clade is not strongly supported. The order Pseudonocardiales was until recently comprised of two families, Pseudonocardiaceae and Actinosynnemataceae (103). However, both these families are now combined into the family Pseudonocardiaceae (191). Genome sequences are now available for a number of genera of this order, including Saccharomonospora, Saccharopolyspora, Actinosynnema, and Amycolatopsis (Table 1) (222, 228).
In the phylogenetic tree based upon concatenated protein sequences (Fig. 2), the sequenced Pseudonocardiaceae species formed a strongly supported clade. Nakamurella multipartite, which is currently a part of the order Frankiales (77, 103), formed an outgroup of the Pseudonocardiaceae clade, and a clade consisting of N. multipartite and Pseudonocardiaceae species was also strongly supported (100% bootstrap score). However, other Frankiales species did not branch with N. multipartite. We have also identified a CSI consisting of a 1-aa insert in the enzyme uridylate kinase, which catalyzes the reversible phosphorylation of UMP to UDP, which is uniquely shared by most of the Pseudonocardiaceae species (all except Saccharomonospora) and also by N. multipartite (Fig. 8A). This CSI, in addition to providing a molecular marker for most of the Pseudonocardiales, also provides evidence that N. multipartite is closely related to this group.
Fig 8.
Excerpts from sequence alignments of the uridylate kinase protein (A) and the hypothetical protein Lxx093000 (B) showing two conserved inserts that are specific for species of the orders Pseudonocardiales and Micromonosporales, respectively.
A number of additional identified CSIs and CSPs provide evidence that species of the orders Corynebacteriales and Pseudonocardiales are specifically related to each other. In the enzyme UDP-galactopyranose mutase (UGM), which catalyzes the interconversion of UDP-galactopyranose (UDP-Galp) and UDP-galactofuranose (UDP-Galf) (303), a 3-aa insert in a conserved region is uniquely present in various Corynebacteriales and Pseudonocardiales species (Fig. 9A). This insert is also present in N. multipartite and also Geodermatophilus obscurus (another member of the Frankiales), which forms an outgroup of the clade consisting of the above-described two orders, but it is not found in any other Actinobacteria. The enzyme UDP-galactopyranose mutase plays an important role in the biosynthesis of cell wall arabinogalactan, and inhibitors of this enzyme are growth inhibitory to M. tuberculosis (75, 275). Another CSI, consisting of a 2-aa deletion, that is uniquely shared by most of the species of these two orders is present in translation initiation factor 2 (IF-2) (see File S18 in the supplemental material), which plays an essential role in the process of protein biosynthesis (177). In this case, the identified CSI is commonly present in all of the Corynebacteriales as well as Pseudonocardiales species, but it is not found in any other bacteria except G. obscurus, which also contains the UGM insert (Fig. 9A). The shared presence of these CSIs in all of the Corynebacteriales as well as Pseudonocardiales species but not in any other Actinobacteria (except G. obscurus and N. multipartite, which branch with them or between them) strongly supports the inference from phylogenetic studies that these two orders are closely related and that they shared a common ancestor exclusive of other Actinobacteria. A number of studies indicated that species of the order Frankiales do not form a coherent phylogenetic lineage, and the taxonomy of this order needs to be emended (191, 215, 343). In this context, our observations that N. multipartite and G. obscurus consistently branch with Pseudonocardiales species in a phylogenetic tree based upon concatenated protein sequences and that they share several CSIs in common with them support the placement of these species into this order of the Actinobacteria.
Fig 9.
(A) Partial sequence alignments of the protein UDP-galactopyranose mutase showing a 3-aa CSI that is uniquely shared by various species of the orders Corynebacteriales and Pseudonocardiales but that is not found in other Actinobacteria. (B) Partial sequence alignments of DNA polymerase HolB showing a CSI that is uniquely shared by various species of the orders Corynebacteriales, Pseudonocardiales, Micromonosporales, and Glycomycetales, indicating that species from these groups shared a common ancestor exclusive of other Actinobacteria. Sequence information for other CSIs that are specific for these actinobacterial orders is presented in Files S18 to S20 and S22 in the supplemental material.
In addition to these CSIs, we have also identified 5 CSPs that are uniquely present in most of the species of the orders Corynebacteriales and Pseudonocardiales (Table 9). In addition to the species of these two orders, homologs of these proteins are also generally present in G. obscurus and N. multipartite, providing further evidence that they are closely related to species of these orders, particularly those of the Pseudonocardiales. Of these five CSPs, two (EmbA and EmbC) are involved in the synthesis of cell wall arabinan, which is a uniquely shared biochemical characteristic of the cell walls of these two orders of the Actinobacteria (58, 106, 178, 182, 263). In contrast to these two proteins, two other proteins involved in the synthesis of cell wall arabinan (viz., EmbB and AftA) are limited to only various Corynebacteriales (Table 4). All four of these genes are part of the Emb operon, and they provide important targets for antitubercular drugs (24, 300). The antimycobacterial drug ethambutol inhibits the growth of M. tuberculosis through the inhibition of arabinofuranosyltransferases EmbA and EmbB (6, 24, 300). The other 3 CSPs listed in Table 9 are of unknown functions.
Table 9.
Signature proteins that are specific for the orders Corynebacteriales and Pseudonocardialesa
| Gene | GenBank accession no. | Protein function (references) | Length (aa) |
|---|---|---|---|
| ML0105 | NP_301202 | EmbA, arabinosyl transferase (7, 259) | 1,111 |
| ML0106 | NP_301203 | EmbC, arabinosyl transferase (112, 259) | 1,070 |
| ML0281 | NP_301322 | Hypothetical | 229 |
| ML0810 | NP_301617 | Hypothetical | 407 |
| ML0990 | NP_301735 | Hypothetical | 209 |
These CSPs were identified by Blastp searches of the genome of M. leprae TN (100). Significant Blast hits for some of these proteins have also been observed for G. obscurus.
Molecular Signatures of Micromonosporales and Identification of a Higher Clade Consisting of the Orders Corynebacteriales, Pseudonocardiales, Glycomycetales, and Micromonosporales
The order Micromonosporales contains a single family, Micromonosporaceae, that is made up of 20 genera (77, 103, 110, 283, 309). However, these genera do not form a distinct clade in the 16S rRNA trees, and species from other groups are interspersed within this order. Hence, this order is presently poorly defined in a phylogenetic or taxonomic sense (191). Genome sequences are now available for this order, from Micromonospora aurantiaca, Micromonospora sp. strain L5, Salinispora tropica, and Salinispora arenicola (Table 1) (229, 309). In the phylogenetic tree constructed based upon concatenated sequences for 35 broadly distributed proteins (Fig. 2), the sequenced Micromonosporaceae species formed a strongly supported clade branching in the neighborhood of Pseudonocardiales. Stackebrandtia nassauensis, which is the only species in our data set belonging to the order Glycomycetales, was most closely related to this group, and a clade consisting of the Micromonosporaceae species and S. nassauensis was strongly supported by the bootstrap score. A clade consisting of these species in turn was part of a larger clade that included all species of the orders Corynebacteriales and Pseudonocardiales.
A number of identified CSIs provide useful information regarding the Micromonosporaceae species and their relationships to other orders of the Actinobacteria. First, in a protein of unknown function, Lxx09300, we identified a 1-aa insert in a conserved region that is specifically present in all sequenced Micromonosporaceae species (Fig. 8B). This CSI provides a potential molecular marker for distinguishing species of this order from those of other Actinobacteria. Second, we have also identified 3 CSIs in important proteins that are uniquely shared by all sequenced species of the orders Corynebacteriales, Pseudonocardiales, Micromonosporales, and Glycomycetales (represented by S. nassauensis). The first of these CSIs consists of a 2-aa insert in the delta subunit of DNA polymerase III (HolB), which is involved in replicative DNA synthesis in bacteria (73) (Fig. 9B). This insert is uniquely shared by all species of these orders but is not found in any other Actinobacteria (except Jonesia denitrificans) or bacteria. Another CSI showing a similar species distribution is present in a highly conserved region of the ribosomal protein S3 (see File S19 in the supplemental material). Lastly, in the enzyme alpha-ketoglutarate decarboxylase (KGD), which is involved in the decarboxylation of alpha-ketoglutarate, a 1-aa insert in a conserved region is commonly present in all species of these orders, but except for Acidothermus cellulolyticus, it is not found in any other Actinobacteria (see File S20 in the supplemental material). The shared presence of these CSIs in these important housekeeping proteins by the above-described orders of Actinobacteria, which also cluster together in the phylogenetic tree, strongly indicates that these orders of Actinobacteria shared a common ancestor exclusive of all other Actinobacteria.
Molecular Signatures of Frankia and Identification of a Clade Consisting of the Orders Corynebacteriales, Pseudonocardiales, Glycomycetales, Micromonosporales, and Frankiales
The order Frankiales is presently comprised of six families: Frankiaceae, Acidothermaceae, Nakamurellaceae, Cryptosporangiaceae, Geodermatophilaceae, and Sporichthyaceae (77, 103, 216). Genome sequences are now available for a number of Frankia species (216) as well as a number of other genera (viz., Acidothermus, Nakamurella, and Geodermatophilus) covering three other families (14, 154, 302) (Table 1). As noted above, species of the order Frankiales do not form a coherent phylogenetic lineage, and they branch in a number of independent positions in the 16S rRNA tree and other phylogenetic trees (283, 343). This is clearly seen from the branching positions of G. obscurus, N. multipartite, Acidothermus cellulolyticus, and Frankia species in the tree shown in Fig. 2. As discussed above, G. obscurus and N. multipartite, based upon their branching in the tree and a number of CSIs, are more closely related to the Pseudonocardiales than to the type genus of this order, Frankia, which contains the type species F. alni. Furthermore, although a clade consisting of different sequenced strains of Frankia branches in the proximity of A. cellulolyticus (5), a specific relationship between these species was not supported by our tree. Thus, the order Frankiales, as described currently, cannot be delimited by any means, and its taxonomy needs to be emended. However, we have identified a 7-aa insert in a highly conserved region of the DNA gyrase B protein that is uniquely present in various Frankia species (Fig. 10). In addition to the sequenced genomes, partial information for gyrase B covering this region is available for a large number of other Frankia species and strains (217), and this insert is present in all of them, thus providing a highly specific molecular marker for this genus. In addition to the CSI in gyrase B, sequence information for two additional CSIs that are also specific mainly for Frankia species is provided in File S21 in the supplemental material. These CSIs include a 4- to 5-aa insert in the DNA repair protein RadA and a 3-aa insert in a hypothetical protein, Ncg1. However, besides Frankia, the latter CSIs are also present in a few other Actinobacteria, which could be due to LGTs.
Fig 10.
Excerpts from the sequence alignment of the gyrase B protein showing a 7-aa insert in a conserved region that is uniquely present in various Frankia species but is not found in other Actinobacteria. Two other CSIs that are also largely specific for the genus Frankia are shown in File S21 in the supplemental material.
In the phylogenetic tree based on concatenated protein sequences (Fig. 2), although the clade consisting of Frankia spp. branched in the proximity of Micromonosporales, it was not specifically related to this order or to the larger clade consisting of the orders Corynebacteriales, Pseudonocardiales, Micromonosporales, and Glycomycetales (Fig. 2). However, we have identified one CSI consisting of a 1-aa insert in a conserved region of the protein glutamine phosphoribosylpyrophosphate amidotransferase that is uniquely shared by all species of the orders Corynebacteriales, Pseudonocardiales, Micromonosporales, and Glycomycetales and also by various Frankia spp. (see File S22 in the supplemental material). Except for species of these orders, this insert is found only in Propionibacterium acnes and Micrococcus luteus and not in other Propionibacteriales or Micrococcales or other orders of Actinobacteria. This CSI provides suggestive evidence that Frankia spp. may also have shared a common ancestor with the clade consisting of Corynebacteriales, Pseudonocardiales, Micromonosporales, and Glycomycetales.
Based upon the species distribution patterns of various identified CSIs and CSPs that are discussed above, the evolutionary stages in which the genes for these CSPs, or the genetic changes responsible for the observed CSIs, are postulated to have evolved are depicted in Fig. 11. Most of the nodes in this diagram are supported by phylogenetic analysis and independently by many identified molecular markers indicating that these branching patterns are reliable.
Fig 11.
Summary diagram showing the evolutionary relationships among the orders Corynebacteriales, Pseudonocardiales, Micromonosporales, Glycomycetales, Frankiales, Streptosporangiales, Streptomycetales, and Catenulisporales based upon phylogenetic trees (Fig. 2 and 4, and see File S10 in the supplemental material) and various identified CSIs and CSPs. OMPdecase, OMP-decarboxylase; GPAT, glutamine phosphoribosyl amidotransferase; SF11, File S11 in the supplemental material.
MOLECULAR SIGNATURES OF THE STREPTOMYCETALES AND EVIDENCE FOR ITS RELATEDNESS TO THE CATENULISPORALES
The order Streptomycetales consists of a single family, Streptomycetaceae, that is comprised of three genera, Streptomyces, Kitasatospora, and Streptacidiphilus (77, 103, 160). This group of species, particularly Streptomyces, has been extensively studied since the discovery of the earliest antibiotics from species of this genus in the 1940s (12, 21, 68, 197). Streptomyces spp. are now the source of nearly two-thirds of all known antibiotics, and they also produce numerous other biologically important compounds, including herbicides, antiparasitic agents, immunosuppressants, and other compounds that are of industrial interest (16, 21, 36, 41, 45, 86, 107). Streptomyces spp. in particular, and the Actinobacteria as a whole, are now recognized as the richest source of small-molecule diversity on the planet (12, 21, 36, 45, 86, 87, 220, 249). The genome sequences of these bacteria are among the largest of the prokaryotes (Table 1), and they contain the largest numbers of gene clusters involved in the synthesis of known or predicted novel small molecules (18, 36, 40, 45, 87, 219, 220, 222, 257, 314, 322, 332). Streptomyces species also possess a complex but well-studied developmental cycle, and of these species, S. coelicolor has provided a good model system for different types of studies (41–43, 137, 163, 257, 314). Methods for the genetic manipulation of S. coelicolor (viz., gene expression and gene knockout and replacement, etc.) are also now well established (138, 163). Due to huge interest in the bioprospecting of Streptomyces and related bacteria for the discovery of novel biological compounds, >500 species of Streptomyces have now been identified (77, 160). The genomes of several Streptomyces species (viz., S. avermitilis, S. bingchenggensis, S. coelicolor, S. flavogriseus, S. griseus, and S. scabiei) have been sequenced, and the sequencing of numerous other genomes is in progress (18, 23, 144, 148, 322). These genomes provide a valuable resource for the identification of molecular signatures that are specific for the order Streptomycetales and provide information regarding its evolutionary relationship to other orders of Actinobacteria.
The order Catenulisporales contains a total of 5 species that are placed into two monogeneric families (viz., Catenulisporaceae and Actinospicaceae) (77, 191). Very little work has been carried out on species of this order, and their phylogenetic relationships to other orders of Actinobacteria are presently unclear. The genome sequence of Catenulispora acidiphila of this order is now available (59). In the phylogenetic tree for Actinobacteria based upon concatenated protein sequences, the sequenced Streptomyces spp. formed a tight cluster, and C. acidiphila formed an outgroup of this cluster (Fig. 2). A clade consisting of Streptomyces species and C. acidiphila had a bootstrap score of 100%. No other actinobacterial groups showed a close or specific relationship to this cluster. A more detailed tree for Streptomycetales species based upon concatenated sequences for three large proteins (RpoB, RpoC, and gyrase B) that includes information for many additional Streptomyces species as well as Kitasatospora setae is presented in File S23 in the supplemental material. K. setae formed the immediate outgroup of the Streptomyces species in this tree.
CSIs and CSPs That Are Specific for the Order Streptomycetales
The sequence alignments of actinobacterial genomes have led to the identification of 3 CSIs that are of interest. In the enzyme porphobilinogen deaminase (PBGD), which converts porphobilinogen into hydroxymethylbilane and is the third enzyme in the heme biosynthetic pathway (190), a 4-aa insert in a conserved region is specifically present in all sequences of Streptomyces species and also Kitasatospora setae, but it is not found in any other Actinobacteria (Fig. 12A). Similarly, in the enzyme adenylate kinase, which catalyzes the interconversion of adenine nucleotides and plays an important role in cellular energy homeostasis, a 1-aa insert in a conserved region is specifically present in various Streptomyces species and K. setae but not in any other Actinobacteria (see File S24 in the supplemental material). Blastp searches for proteins of the genome of S. coelicolor A3(2) have also identified a number of CSPs, all significant hits of which are present in various sequenced Streptomyces species but not in any other bacteria (Table 10). For the first 5 proteins in Table 10, homologs showing significant similarity were detected in various Streptomyces species but not in K. setae or other bacteria. These proteins could be specific for the genus Streptomyces; however, as the complete genome of K. setae is not yet available, it is possible that homologs of these proteins will also be found in this species. For the next 11 entries of Table 10, homologs were detected in both Streptomyces species and K. setae but not in other Actinobacteria. These CSPs thus could be specific for the entire order Streptomycetales. Due to their specificity for species of the order Streptomycetales, they provide novel molecular markers for distinguishing this group of bacteria from all other Actinobacteria.
Fig 12.
Excerpts from sequence alignments of two proteins showing CSIs that are specific for the order Streptomycetales or shared with Catenulisporales. (A) A 4-aa insert in the porphobilinogen deaminase (PBGD) protein that is specific for various Streptomycetales species, including Kitasatospora setae. The adenylate kinase protein also contains a CSI that is specific for the Streptomycetales (see File S24 in the supplemental material). (B) A 1-aa insert in the lipid A biosynthesis lauroyl acyltransferase (MsbB) protein that is uniquely shared by various Streptomycetales species and Catenulispora acidiphila.
Table 10.
Signature proteins that are specific for Streptomyces (Streptomycetales)a
| Gene or protein | GenBank accession no. | Protein function | Length (aa) | Species specificity |
|---|---|---|---|---|
| Small membrane protein | NP_625909.1 | Small membrane protein | 64 | Genus Streptomyces |
| SCO2919 | NP_627145.1 | Hypothetical | 114 | Genus Streptomyces |
| SCO4335 | NP_628506.1 | Hypothetical | 62 | Genus Streptomyces |
| Secreted serine-rich protein | NP_627511.1 | Secreted serine-rich protein | 327 | Genus Streptomyces |
| SCO3544 | NP_627742.1 | Hypothetical | 132 | Genus Streptomyces |
| SCO1392 | NP_625675.1 | Hypothetical | 300 | Streptomycetaceae |
| SCO1529 | NP_625808.1 | Hypothetical | 551 | Streptomycetaceae |
| Secreted protein | NP_626808.1 | Secreted protein | 258 | Streptomycetaceae |
| Membrane protein | NP_626821.1 | Membrane protein | 356 | Streptomycetaceae |
| SCO2621 | NP_626857.1 | Hypothetical | 64 | Streptomycetaceae |
| Lipoprotein | NP_627319.1 | Lipoprotein | 215 | Streptomycetaceae |
| SCO3905 | NP_628091.1 | Hypothetical | 101 | Streptomycetaceae |
| Transmembrane protein | NP_628124.1 | Transmembrane protein | 290 | Streptomycetaceae |
| Integral membrane protein | NP_628309.1 | Integral membrane protein | 102 | Streptomycetaceae |
| Integral membrane protein | NP_627868.1 | Integral membrane protein | 350 | Streptomycetaceae |
| SCO4669 | NP_628829.1 | Hypothetical | 379 | Streptomycetaceae |
| SCO3799 | NP_627989.1 | Hypothetical | 156 | Streptomycetaceae and Catenulispora acidiphila |
| Integral membrane protein | NP_628308.1 | Integral membrane protein | 266 | Streptomycetaceae and Catenulispora acidiphila |
| SCO3624 | NP_627818.1 | Hypothetical | 221 | Streptomycetaceae and Catenulispora acidiphilab |
These CSPs were identified by Blastp searches of the genome of Streptomyces coelicolor A3(2).
Also found in Variovorax paradoxus and Cellulophaga lytica.
CSIs and CSPs That Are Uniquely Shared by the Orders Streptomycetales and Catenulisporales
In the phylogenetic tree shown in Fig. 2, C. acidiphila formed an outgroup of the Streptomyces cluster, indicating that the orders Streptomycetales and Catenulisporales are closely related. This inference is also supported by a 1-aa CSI in the lipid A biosynthesis lauroyl acyltransferase (MsbB) protein, which is uniquely shared by all Streptomycetaceae species, including K. setae, as well as by C. acidiphila but not any other Actinobacteria (Fig. 12B). Further evidence that these two orders of Actinobacteria are closely related is provided by our identification of 3 CSPs listed in Table 10, whose homologs are specifically present in various Streptomycetaceae species as well as C. acidiphila. Based upon phylogenetic evidence as well as the identification of a number of molecular markers that are uniquely shared by species of the orders Streptomycetales and Catenulisporales, these observations make a strong case that the order Catenulisporales, which contains only a limited number of species, should be merged with the order Streptomycetales.
In the phylogenetic tree shown in Fig. 2, a superclade consisting of the orders Corynebacteriales, Pseudonocardiales, Glycomycetales, Micromonosporales, Frankiales, Streptosporangiales, Streptomycetales, and Catenulisporales is strongly supported by its observed bootstrap score (97%). The consensus tree reported previously by Alam et al. (5) also supported a clade consisting of these orders. Although we have not come across any CSI that is specifically shared by species of all of these orders, the placement of the orders Streptosporangiales, Streptomycetales, and Catenulisporales as the outer branches of the large clade shown in Fig. 11 is strongly supported by phylogenetic analyses.
MOLECULAR SIGNATURES OF THE ORDERS BIFIDOBACTERIALES, ACTINOMYCETALES, AND MICROCOCCALES
Molecular Signatures of the Bifidobacteriales and Bifidobacteriaceae
Species of the order Bifidobacteriales are generally found in the human gastrointestinal tract, and they are important for establishing and maintaining the homeostasis of the intestinal ecosystem to allow for normal digestion (61, 183, 307, 312, 318). The order Bifidobacteriales is comprised of a single family, the Bifidobacteriaceae, which in turn consists of seven genera, Bifidobacterium, Gardnerella, Scardovia, Parascardovia, Alloscardovia, Metascardovia, and Aeriscardovia (22, 191). Except for the genus Bifidobacterium, which contains 29 species, all other genera are monospecific and contain only a single species (77, 191). Due to the importance of bifidobacteria for human health and also due to their probiotic potential, the genomes of large numbers of Bifidobacterium species and strains as well as Gardnerella vaginalis have been sequenced (15, 93, 102, 141, 164, 183, 254, 262, 292, 306, 313, 314, 318, 326, 337, 344). The genetic, biochemical, and genomic characteristics of Bifidobacterium species were reviewed previously by Ventura and coworkers (28, 61, 307, 308, 313, 314). In addition to Bifidobacterium, sequence information for most of the genes and proteins from the genomes of Scardovia inopinata and Parascardovia denticolens, whose genomes are at assembly stages, is also now available in public databases.
In a phylogenetic tree for the Bifidobacteriales, G. vaginalis was found to branch in between different Bifidobacterium species, making this genus polyphyletic (see File S25 in the supplemental material). In particular, Bifidobacterium animalis was found to branch more deeply than G. vaginalis. Hence, the relationship of G. vaginalis to other Bifidobacterium species and its possible placement in this genus should be considered. Alignments of protein sequences of Actinobacteria species have identified two CSIs that are specific for the Bifidobacteriales. One of these CSIs, consisting of a 1-aa deletion in the ribosomal protein L13, is present in all Bifidobacteriales species, including S. inopinata and P. denticolens, but it is not found in any other Actinobacteria (or other bacteria) (Fig. 13A). Thus, this CSI provides a potential molecular marker for the entire order Bifidobacteriales. Another 1-aa insert in the enzyme glucose-6-phosphate dehydrogenase, which is a part of the pentose phosphate pathway, is uniquely present in various Bifidobacterium species and also in G. vaginalis, but it is not found in Scardovia, Parascardovia, or any other Actinobacteria (Fig. 13B). Thus, this CSI distinguishes the clade consisting of the genera Bifidobacterium and Gardnerella from other genera of this order. Blastp searches for various proteins of the genome of Bifidobacterium dentium Bd1 (318) also identified 16 proteins that are uniquely found in various Bifidobacteriales species as well as 6 CSPs for which all significant Blast hits are from the genera Bifidobacterium and Gardnerella (Table 11). Previously, many CSPs that were specific for B. dentium were also identified (316, 318). These CSPs provide additional markers for distinguishing Bifidobacteriales species from other Actinobacteria.
Fig 13.
Partial sequence alignments of ribosomal protein L3 (A) and glucose-6-phosphate dehydrogenase (G6PDH) (B) showing two CSIs consisting of a 1-aa deletion and a 1-aa insert, respectively, that are specific for Bifidobacteriales species. The CSI in the ribosomal protein is present in all sequenced Bifidobacteriales species, whereas that in G6PDH is found only in Bifidobacterium and Gardnerella species.
Table 11.
Signature proteins that are specific for the Bifidobacteriaceaea
| Gene | GenBank accession no. | Protein function | Length (aa) | Species specificity |
|---|---|---|---|---|
| BIFDEN_00796 | ZP_02917515.1 | Hypothetical | 124 | Bifidobacteriaceae |
| BIFDEN_00793 | ZP_02917512.1 | Hypothetical | 73 | Bifidobacteriaceae |
| BIFDEN_00600 | ZP_02917322.1 | Hypothetical | 275 | Bifidobacteriaceae |
| BIFDEN_00594 | ZP_02917316.1 | Hypothetical | 119 | Bifidobacteriaceae |
| BIFDEN_00539 | ZP_02917261.1 | Hypothetical | 336 | Bifidobacteriaceae |
| BIFDEN_00419 | ZP_02917147.1 | Hypothetical | 228 | Bifidobacteriaceae |
| BIFDEN_00378 | ZP_02917106.1 | Hypothetical | 399 | Bifidobacteriaceae |
| BIFDEN_00301 | ZP_02917034.1 | Hypothetical | 204 | Bifidobacteriaceaeb |
| BIFDEN_02476 | ZP_02919152.1 | Hypothetical | 201 | Bifidobacteriaceae |
| BIFDEN_02473 | ZP_02919149.1 | Hypothetical | 174 | Bifidobacteriaceae |
| BIFDEN_02131 | ZP_02918813.1 | Hypothetical | 121 | Bifidobacteriaceae |
| BIFDEN_00191 | ZP_02916931.1 | Hypothetical | 84 | Bifidobacteriaceae |
| BIFDEN_01066 | ZP_02917770.1 | Hypothetical | 76 | Bifidobacteriaceae |
| BIFDEN_02253 | ZP_02918933.1 | Hypothetical | 321 | Bifidobacteriaceae |
| BIFDEN_00382 | ZP_02917110.1 | Hypothetical | 213 | Bifidobacterium and Gardnerella |
| BIFDEN_00315 | ZP_02917048.1 | Hypothetical | 222 | Bifidobacterium and Gardnerella |
| BIFDEN_02465 | ZP_02919141.1 | Hypothetical | 299 | Bifidobacterium and Gardnerella |
| BIFDEN_02410 | ZP_02919088.1 | Hypothetical | 260 | Bifidobacterium and Gardnerella |
| BIFDEN_01330 | ZP_02918031.1 | Hypothetical | 283 | Bifidobacterium and Gardnerella |
| BIFDEN_02361 | ZP_02919040.1 | Hypothetical | 189 | Bifidobacterium and Gardnerella |
These CSPs were identified by Blastp searches of the genome of B. dentium Bd1.
Also found in Isoptericola variabilis and Xylanimonas cellulosilytica.
Molecular Signatures of the Actinomycetales
The order Actinomycetales, which corresponds to the suborder Actinomycineae in the earlier taxonomic scheme (103, 343), contains only one family, the Actinomycetaceae, which is comprised of several medically important genera, such as Actinomyces, Arcanobacterium, Actinobaculum, Mobiluncus, and Varibaculum (103, 191, 253, 343). Of these genera, the genus Actinomyces has been indicated to be quite diverse, and in a phylogenetic tree based upon 16S rRNA, it showed polyphyletic branching into a number of different clusters (253). The genome sequences of Arcanobacterium haemolyticum (336) and Mobiluncus curtisii are now available, and sequence information for most of the proteins from a number of other species (viz., Actinomyces odontolyticus, Actinomyces urogenitalis, Actinomyces coleocanis, and Mobiluncus mulleris) is also available in the NCBI database. Our analyses have identified a number of CSIs that are specific for species of this order. The enzyme deoxy-d-xylulose 5-phosphate reductoisomerase (DXR), which is a part of the nonmevalonate pathway of isoprenoid biosynthesis (245), contains a 12-aa insert in a highly conserved region that is uniquely present in all available sequences of Actinomycetales species, including those of the genera Actinomyces, Arcanobacterium, and Mobiluncus (Fig. 14A). Another CSI consisting of a 6-aa insert that is specific for all sequenced Actinomycetales species is present in the integral membrane protein Lxx09300 (see File S26 in the supplemental material). The high degrees of conservation and specificity of these signatures for species of this order indicate that they provide good and reliable molecular markers for this order of Actinobacteria. Isoleucine tRNA synthetase (IleRS), which is essential for protein synthesis, also contains a 3-aa insert in a conserved region that is specifically present in all available sequences of the genera Actinomyces and Mobiluncus but which is lacking in Arcanobacterium haemolyticum as well as all other Actinobacteria (Fig. 14B). In the phylogenetic tree for Actinobacteria based on protein sequences, A. haemolyticum showed the deepest branching of the three available genera, and a clade consisting of Actinomyces and Mobiluncus species was strongly supported (Fig. 2, and see File S25 in the supplemental material). Thus, it is likely that the genetic change responsible for this CSI took place in a common ancestor of these two genera after the divergence of Arcanobacterium. Lastly, we have also identified a 1-aa deletion in the excision endonuclease UvrC that is specifically present in the two Mobiluncus species (Fig. 14C), providing a molecular marker for this genus.
Fig 14.
(A and B) Partial sequence alignments of the deoxy-d-xylulose 5-phosphate reductoisomerase (DXR) (A) and isoleucine tRNA synthetase (IleRS) (B) proteins depicting 12-aa and 3-aa inserts, respectively, in highly conserved regions that are uniquely present in various sequenced Actinomycetales species. (C) Sequence alignment of the excision endonuclease UvrC showing a 1-aa deletion that is specific for the genus Mobiluncus. Information for another CSI that is specific for Actinomycetales is provided in File S26 in the supplemental material.
Molecular Signatures of the Micrococcales and Its Subclades
The order Micrococcales is the most diverse order within the phylum Actinobacteria, containing ecologically, morphologically, and chemotaxonomically divergent species (191, 283). The most updated taxonomic outline of this suborder encompasses 15 families and 86 genera, and information for some of these has been reviewed (38, 78, 157, 169, 191, 258, 282, 343, 343). In view of the importance of species of this order for bioremediation, industrial, and clinical purposes, large numbers of genomes of many species that are part of different genera and families of this order have been sequenced. The sequenced genera include Arthrobacter (202, 203), Beutenbergia, Brachybacterium (180), Cellulomonas (2), Clavibacter (105), Intrasporangium (66), Jonesia (233), Kocuria (296), Leifsonia (205), Renibacterium (328), Rothia, Sanguibacter (155), and Tropheryma (20, 239) (Table 1). In the phylogenetic tree based upon concatenated protein sequences (Fig. 2) or 16S rRNA (191, 343), species of the order Micrococcales are split into a number of clusters, with the orders Actinomycetales and Bifidobacteriales branching between them. Thus, the different families that are presently part of this order do not form a phylogenetically coherent group, and the taxonomy of this order needs to be emended. Hence, novel molecular markers that could serve to define and delimit different subclades of this order are of particular interest.
Our analyses of protein sequences from Actinobacteria have identified some CSIs that are specific for some of the subclades of Micrococcales. In the universally distributed and highly conserved β-subunit of the RNA polymerase (RpoB), a 2-aa insert is present in a conserved region that is specific for clade I Micrococcales (Fig. 15A). This clade, which is comprised of species of the families Micrococcaceae (Arthrobacter, Renibacterium, Micrococcus, Kocuria, and Rothia) and Brevibacteriaceae (Brevibacterium), is also strongly supported in the phylogenetic tree (Fig. 2). Another CSI that is specific for the Micrococcales has been identified in the ribose-5-phosphate isomerase (RPI) protein, which is a key enzyme of the pentose phosphate pathway that catalyzes the conversion of ribose-5-phosphate into ribulose-5-phosphate (340). In this highly conserved protein, a 4-aa insert in a conserved region is specifically present in all of the sequenced Micrococcales that are part of clusters I and III, but this insert is not found in cluster II Micrococcales or any other Actinobacteria (Fig. 15B). It should be noted that although clusters I and II branch in proximity of each other in the tree, they are phylogenetically quite distinct from each other. For cluster III Micrococcales species, although they branch deeply in the tree, their deep branching could be due to a long-branch length effect (82, 97), as Tropheryma, which is a part of this cluster, has a long branch length. Thus, although a clade consisting of cluster I and cluster III Micrococcales is not observed in the phylogenetic tree (Fig. 2), the CSI in the RPI protein suggests that these two subclades of Micrococcales might be more closely related to each other than the cluster II species. We have also identified one additional CSI consisting of a 4-aa insert in the pyruvate carboxylase protein that is uniquely present in various Micrococcales except Tropheryma (see File S27 in the supplemental material). Although homologs of this protein were not detected in all sequenced Micrococcales, this CSI suggests that despite their divergent branching in the phylogenetic trees, all of the Micrococcales might be derived from a common ancestor exclusive of other Actinobacteria.
Fig 15.
Partial sequence alignments of the RNA polymerase β-subunit showing a 2-aa insert in a conserved region that is specific for cluster I Micrococcales species (A) and a 4-aa insert in ribose-5-phosphate isomerase that is uniquely shared by both cluster I and cluster II Micrococcales species (B). Another CSI that is specific for Micrococcales can be found in File S27 in the supplemental material.
As noted above, in phylogenetic trees, species of the orders Actinomycetales and Bifidobacteriales branch between the different clusters of Micrococcales, indicating that these orders are closely related. One additional CSI that we have identified supports this inference. In the highly conserved DnaK or Hsp70 family of proteins, a 5-aa insert in a conserved region is present in all of the Bifidobacteriales, Actinomycetales, and Micrococcales (clusters I, II, and III), but with a few exceptions, this insert is not present in most other Actinobacteria (see File S28 in the supplemental material). The presence of this CSI in a few other Actinobacteria could be due to LGTs. The shared presence of this CSI in all species of these actinobacterial orders suggests that they likely shared a common ancestor exclusive of other Actinobacteria.
Molecular Signatures of the Propionibacteriales
The order Propionibacteriales contains the families Propionibacteriaceae and Nocardioidaceae (103, 343). Members of the Propionibacteriaceae thrive in diverse habitats, covering human epidermal surfaces, dairy products, silage, soil, water, Antarctic sandstone, and sewage treatment plants (279). They are either aerobic or facultative anaerobes, have different morphologies, and exhibit different peptidoglycan type variations (279, 281, 343). The genome sequences of several species of this order covering both families are now available. These include sequences of several strains of Propionibacterium acnes as well as those of Propionibacterium freudenreichii, Kribbella flavida, and Nocardioides sp. strain JS614 (34, 79, 235). Additionally, sequence information for large numbers of genes and proteins from Aeromicrobium marinum is also available in the NCBI database. Sequence alignments of actinobacteria have identified two CSIs that are specific for this order. In the helicase DinG, which is involved in DNA repair and replication, a 3-aa insert in a conserved region is specifically present in all available sequences from this order of bacteria but not in any other Actinobacteria or other bacteria (Fig. 16). Another CSI that is specific for this order is present in the cytochrome c oxidase subunit 1 (Cox1) protein (see File S29 in the supplemental material), which also contains a CSI that is specific for most Actinobacteria (97). In this case, all Propionibacteriales homologs contain a 1-aa deletion that is not present in other Actinobacteria (see File S29 in the supplemental material). Both these CSIs provide molecular markers that distinguish species of the order Propionibacteriales from all other Actinobacteria.
Fig 16.
Excerpts from sequence alignments for the helicase DinG showing a CSI consisting of a 3-aa insert that is uniquely present in various sequenced Propionibacteriales species. Sequence information for another CSI that is specific for Propionibacteriales can be found in File S29 in the supplemental material.
Molecular Signatures Identifying Larger Clades Consisting of the Orders Bifidobacteriales, Actinomycetales, Micrococcales, Kineosporiales, and Propionibacteriales
Due to the compact clustering of most actinobacterial orders in the 16S rRNA and protein trees, their branching orders are generally not resolved (Fig. 2). As indicated above, rare genetic changes such as CSIs, due to their rare and highly specific nature, are capable of resolving deep-branching relationships that are not resolved by phylogenetic trees (11, 119, 131, 243, 246). Our analyses have identified a number of CSIs that clarify the evolutionary relationships and branching orders of the actinobacterial orders Bifidobacteriales, Actinomycetales, Micrococcales, Kineosporiales, and Propionibacteriales.
In the phylogenetic tree shown in Fig. 2, the order Kineosporiales branches in the proximity of the order Micrococcales, which is interspersed by the orders Bifidobacteriales and Actinomycetales. Although a clade consisting of these orders is not supported by the 16S rRNA or protein trees (Fig. 2) (5, 343), a number of CSIs provide evidence that species of these orders are specifically related and that they shared a common ancestor exclusive of other Actinobacteria. In the highly conserved and universally distributed ribosomal protein S3, a 5-aa insert in a conserved region is uniquely present in all species of these 4 orders of actinobacteria, but this insert is not found in any other Actinobacteria (Fig. 17). The shared presence of this insert in this important protein involved in the information transfer process strongly suggests that the genetic change leading to this insert was introduced in a common ancestor of these 4 orders of Actinobacteria. Two other CSIs that also support that these orders are specifically related are found in the CgR_2975 and Cox1 proteins. In the CgR_2975 protein, whose cellular function is not known, a 3-aa insert in a conserved region is present in all species of the orders Bifidobacteriales, Micrococcales, and Kineosporiales but not in any other bacteria (see File S30 in the supplemental material). Because homologs of this protein were not detected in Actinomycetales, it is likely that this CSI was introduced in a common ancestor of the orders Bifidobacteriales, Actinomycetales, Micrococcales, and Kineosporiales, followed by the loss of this gene from the order Actinomycetales. Similarly, in the Cox1 protein, which contains two other CSIs, one specific for most Actinobacteria (97) and the other specific for Propionibacteriales (see File S29 in the supplemental material), one additional CSI that is uniquely shared by all species of the orders Micrococcales and Kineosporiales has been identified (see File S31 in the supplemental material). Because Cox1 homologs were not detected in the orders Actinomycetales and Bifidobacteriales, it is likely that this CSI was also introduced in a common ancestor of the above-described 4 orders, followed by the loss of this gene from the orders Bifidobacteriales and Actinomycetales. Recently, the existence of a clade consisting of these 4 actinobacterial orders based upon 16S rRNA trees was also suggested by Ludwig et al. (191). However, in the phylogenetic trees of 16S rRNA genes reported by Zhi et al. (343) and Adekambi et al. (3), this clade was not observed.
Fig 17.
Partial sequence alignment of ribosomal protein S3 showing a 5-aa conserved insert that is commonly shared by various sequenced species of the orders Bifidobacteriales, Actinomycetales, Micrococcales, and Kineosporiales but which is not found in any other Actinobacteria or in other phyla of bacteria. Information for two other CSIs showing similar specificities is provided in Files S30 and S31 in the supplemental material.
Lastly, the triosephosphate isomerase protein, which plays a key role in glycolysis, contains a 2-aa insert in a conserved region that is commonly shared by all species of the orders Bifidobacteriales, Actinomycetales, Micrococcales, Kineosporiales, and Propionibacteriales but which is not found in any other Actinobacteria or other phyla of bacteria (Fig. 18). The genetic changes responsible for this CSI were likely introduced in a common ancestor of these orders, providing evidence that they are specifically related. The evolutionary relationships among various taxa belonging to these actinobacterial orders that emerge based upon various identified CSIs and CSPs are summarized in Fig. 19.
Fig 18.
Partial sequence alignment of the triosephosphate isomerase protein showing a 2-aa conserved insert that is commonly shared by various sequenced species of the orders Bifidobacteriales, Actinomycetales, Micrococcales, Kineosporiales, and Propionibacteriales but which is not found in any other Actinobacteria.
Fig 19.
Summary diagram showing evolutionary relationships among the orders Bifidobacteriales, Actinomycetales, Micrococcales, Kineosporiales, and Propionibacteriales based upon phylogenetic trees (Fig. 2, and see File S25 in the supplemental material) and various identified CSIs and CSPs. Rib., ribosomal.
CONCLUSIONS AND FUTURE DIRECTIONS
The phylum Actinobacteria is very large and diverse in terms of its biology, ecology, and genetics, and it contains numerous organisms that are of great interest from medical, industrial, biotechnological, and environmental perspectives. The main focus of this review has been on the identification of molecular markers that are specific for either all Actinobacteria or their different constituent groups. Although this review describes a large number of signatures that are specific for Actinobacteria or their various subgroups, systematic studies to identify different CSIs or CSPs that are specific for various actinobacterial groups at different phylogenetic depths have not yet been carried out. As genome sequences of more actinobacteria become available, further studies in this regard should lead to the identification of many other signatures that are specific for various actinobacterial groups at different taxonomic levels. Nonetheless, the molecular markers thus far identified provide powerful new tools for a variety of studies that are briefly discussed below.
Usefulness of CSIs and CSPs for an Understanding of the Phylogeny and Taxonomy of Actinobacteria
One of the immediate applications of these signatures is that they provide potentially more definitive means for understanding or clarifying actinobacterial phylogeny and taxonomy. Our understanding of the phylogeny and taxonomy of Actinobacteria currently relies solely on phylogenetic trees based on 16S rRNA (103, 191, 283, 343). Although such trees have been and will remain some of the primary means for understanding microbial phylogeny and taxonomy, some of the limitations of these trees should be recognized (50, 171, 225, 278). While phylogenetic trees in general are most effective in resolving evolutionary relationships at intermediate taxonomic levels (viz., genus, family, and order), their resolving power at either higher (among orders, classes, or phyla) or lower (i.e., among species or different strains of a species) phylogenetic depths is quite limited (192, 278, 286, 324, 343). Additionally, the phylogenetic trees are a continuum, with no fixed boundaries. Hence, on the basis of the branching in these trees, it is often difficult to delimit a phylum or any other taxa reliably, unless all members of the proposed clade or taxon share at least some unique and reliable molecular, biochemical, or physiological characteristics. The phylum Actinobacteria represented such a case, where no unique characteristic of any kind was known that was commonly shared by all species that have been assigned to this phylum. In this context, our identification of a number of CSIs and CSPs that are commonly and uniquely shared by most members of all other classes of the phylum Actinobacteria, except Coriobacteriia, argues strongly that the bacteria belonging to the class Coriobacteriia, which branches more deeply than all other Actinobacteria, should at present be excluded from the phylum Actinobacteria. If, in the future, some unique biochemical and/or molecular properties that are specifically shared by Coriobacteriia and Actinobacteria are discovered, the inclusion of Coriobacteriia in the phylum Actinobacteria could be reconsidered.
On the basis of the identified CSIs and CSPs, it is also now possible to identify and delimit most of the main orders within the phylum Actinobacteria in molecular terms (Fig. 11 and 19). For some orders that have been studied in detail (viz., Corynebacteriales, Bifidobacteriales, and Streptomycetales), individual families and genera as well as subclades of some genera (e.g., Corynebacterium and Mycobacterium) can now also be identified in clear molecular terms based upon multiple signatures. Additionally, based upon these molecular signatures, it is also possible to delineate the interrelationships among different orders of Actinobacteria, and several higher levels of clades can be identified (Fig. 11 and 19). These clades include those consisting of (i) the orders Corynebacteriales and Pseudonocardiales; (ii) the orders Corynebacteriales, Pseudonocardiales, Glycomycetales, and Micromonosporales; and (iii) the orders Corynebacteriales, Pseudonocardiales, Glycomycetales, and Micromonosporales and the genus Frankia (Fig. 11). Although the Frankiales species do not form a coherent clade, all sequenced species are part of this larger clade, indicating that they are related to this group of species. Phylogenetic studies also support a larger clade consisting of the orders Corynebacteriales, Pseudonocardiales, Glycomycetales, Micromonosporales, Frankiales, Streptosporangiales, and Streptomycetales, although no molecular signature that is specific for this large clade has thus far been identified. The other higher levels of clades within the phylum Actinobacteria that can be identified on the basis of identified molecular signatures include those consisting of the orders (iv) Bifidobacteriales, Actinomycetales, and Micrococcales; (v) Bifidobacteriales, Actinomycetales, Micrococcales, and Kineosporiales; and (vi) Bifidobacteriales, Actinomycetales, Micrococcales, Kineosporiales, and Propionibacteriales (Fig. 19). Several of these phylogenetic clades were also observed in a consensus phylogenetic tree for a limited number of actinobacteria constructed by using different approaches (5). Additionally, the phylogenetic analysis and molecular signatures reported here provide strong evidence that species of the order Streptomycetales are closely related to Catenulisporales, and a strong case can be made for the merger of Catenulisporales into the order Streptomycetales.
Recently, on the basis of 16S rRNA trees, Ludwig et al. (191) also indicated the identification of two large clades within the phylum Actinobacteria. One of these clades consists of the orders Actinopolysporales, Corynebacteriales, Glycomycetales, Jiangellales, Micromonosporales, Pseudonocardiales, and Propionibacteriales. This clade is similar to one of the large clades identified here, except that our results suggest that the genus Frankia and other species that are currently part of the order Frankiales are also affiliated with this clade, whereas those of the order Propionibacteriales are not part of this clade. There are no genome sequences available at present for the orders Actinopolysporales and Jiangellales. Hence, we are unable to determine the placement of these orders within this clade. The other large clade identified by Ludwig et al. (191), consisting of the orders Bifidobacteriales, Actinomycetales, Micrococcales, and Kineosporiales, is also supported by various identified signatures (Fig. 19). Although the identification of these large clades based upon different CSIs as well as the 16S rRNA trees strongly indicates that these clades are meaningful, it should be recognized that phylogenetic trees are dynamic constructs and that the branching of species within them is dependent upon large numbers of variables and assumptions, including the different species that are part of the data set and the models used to create the sequence alignment and phylogenetic trees (81, 83, 192, 206, 330). This is illustrated by the fact that the two large clades proposed by Ludwig et al. (191) were not observed in the phylogenetic trees for 16S rRNA reported by Zhi et al. (343) and Adekambi et al. (3). In contrast to the highly dynamic (and variable) nature of phylogenetic trees, the inferences derived from CSIs are based upon minimal assumptions, and their interpretation is generally straightforward (119, 126, 132). Based upon these CSIs, all of the identified clades are defined simply based upon the presence or absence of given indels in highly conserved regions of proteins (119, 126, 130, 132). Furthermore, these CSIs provide highly stable molecular markers with strong predictive abilities. This is evidenced by the fact that many of the Actinobacteria-specific CSIs and CSPs, which were identified when the number of sequenced genomes was very limited (97, 100), are still reliable characteristics of this phylum despite the nearly 10-fold increase in the number of sequenced genomes. Additionally, the investigated CSIs are also present in many other actinobacterial species whose genomes have not been sequenced, providing further strong evidence of their reliability and predictive power (97).
The specificity of various identified signatures for actinobacterial species or groups is presently based mainly upon the species and/or strains whose genomes have been sequenced (Table 1). Although these genomes represent only a small fraction of the actinobacterial species (52, 103), they cover most of the major orders and families of Actinobacteria. However, it is of much importance to obtain sequence information for these genes/proteins from other actinobacterial species to further validate and more precisely determine the boundaries of the clades that are defined by these signatures. These signatures are also very appealing for taxonomic studies, as the assignment of various species (or new isolates) to different clades can be readily done based upon the presence or absence of certain diagnostic signatures, without the need for the construction of detailed phylogenetic trees.
Interesting Cases of Lateral Gene Transfers Identified by CSIs and CSPs
Although most of the CSIs and CSPs described in this review are specific for particular clades of Actinobacteria, the shared presence of CSIs or CSPs in unrelated groups of bacteria also provides a novel means for the identification of lateral gene transfers. Two interesting cases of LGTs between Actinobacteria and Chlamydiae that have been identified by these means include those for the genes encoding the enzymes serine hydroxymethyltransferase (SHMT) (or the GlyA protein) and UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) (115, 117). In the enzyme SHMT, which links amino acid and nucleotide metabolisms by generating the key intermediate for one-carbon transfer reactions (238), two CSIs (3 and 31 aa long) are uniquely shared by all Chlamydiae species, Treponema species, as well as a subset of Actinobacteria (117). Interestingly, the actinobacterial species which contain these CSIs have multiple homologs of the glyA gene, and only one of them harbors the indicated CSIs (117). Similarly, in the MurA protein, which plays an important role in the synthesis of cell wall peptidoglycan, a 16-aa CSI was commonly shared by all Chlamydiae and a subset of Actinobacteria (115). In the phylogenetic trees based upon GlyA or MurA protein sequences, the Chlamydiae homologs branched with the various insert-containing Actinobacteria within a clade of other Actinobacteria. These results provide strong evidence that the shared presence of these CSIs in these two groups of bacteria is due to the lateral transfer of genes for these proteins from certain groups of Actinobacteria to a common ancestor of the Chlamydiae (117, 133). It is of much interest to understand the functional significances of the identified CSIs in these proteins and to determine why their genes were laterally transferred from Actinobacteria to the common ancestor of the Chlamydiae. Our work on actinobacterial CSPs has also revealed that homologs of some of them are also found in Magnetospirillum magnetotacticum (100), which is unrelated to the Actinobacteria. The CSI in the glycyl-tRNA synthetase, which is mainly a distinctive characteristic of Actinobacteria, is also found in this bacterium as well as in a few Planctomycetes (Table 2). The shared presence of these CSPs and CSI is again due to LGTs from Actinobacteria to M. magnetotacticum, and it is of much interest to determine what unique properties are shared by these two groups of bacteria.
Application of the Identified Molecular Signatures for Identification of Actinobacteria and Exploring Their Diversity
The phylum Actinobacteria is extremely diverse. In addition to containing many bacteria that are major human, animal, or plant pathogens (e.g., Mycobacterium, Actinomyces, Renibacterium, Atopobium, Gordonia, Gardnerella, Leifsonia, and Clavibacter), other actinobacterial taxa arguably provide the richest source for discovering diverse natural products that have proven to be of seminal importance in clinical and biotechnological applications (12, 21, 36, 45, 86, 87, 220, 249). Thus, it is of much interest and importance to discover novel means by which both known as well as novel species belonging to different actinobacterial groups can be readily and accurately identified in different settings (viz., clinical or environmental). Because some taxa of Actinobacteria (e.g., Streptomyces, Salinispora, Saccharopolyspora, Cellulomonas, Verrucosispora, Pseudonocardia, Micromonospora, Bifidobacterium, and Arthrobacter, etc.) have proven to be particularly important sources for the discovery of novel compounds such as antibiotics and probiotics and compounds useful in bioremediation (42, 110, 162, 197, 220, 221, 308, 325), there is enormous interest in the discovery of novel actinobacterial species belonging to these taxa, which could lead to the discovery of either novel antibiotics or other natural products that can be gainfully employed for various applications (35, 36, 84, 110, 197). As emphasized by Goodfellow and coworkers (110, 323), a sound knowledge of actinobacterial systematics is of particular importance in this regard. A reliable phylogenetic framework for Actinobacteria in conjunction with specific probes for identifying different groups of Actinobacteria can greatly facilitate the discovery of novel actinobacterial species in different environments. In this context, molecular markers (CSIs and CSPs) that are specific for different major clades of Actinobacteria are of particular importance, since probes based on them can serve as novel and specific tools for the identification and discovery of novel actinobacterial species belonging to these taxa. The primary sequences of many of the CSPs and most of the proteins that contain these CSIs are highly conserved. Based upon conserved regions in these genes/proteins, degenerate PCR primers for these genes/proteins can be readily designed, which should specifically amplify gene sequences from these clades (95, 97, 115) and should provide novel means for the identification of new as well as existing actinobacterial species belonging to these clades from different environments. Using these molecular signatures, it should also be possible to readily and more accurately determine the presence or absence of different families and orders of Actinobacteria in metagenomic samples obtained from various environments (35, 109, 110, 147, 201, 241, 276, 277). Likewise, CSIs and CSPs that are specific for the pathogenic Actinobacteria (viz., Mycobacterium, Corynebacterium, Propionibacterium, and Actinomyces) provide novel means for their diagnostics. Some of the CSPs and the proteins containing CSIs that are specific for these groups should also provide potential means for developing vaccines for these bacteria or potential targets for developing drugs that are specific for these bacteria.
Functional Significance of Actinobacterial CSIs and CSPs
An important area for future research is to understand the functional significance of various CSIs and CSPs that are specific for either all Actinobacteria or their various clades. For the phylum Actinobacteria or most of its major clades, no biochemical or physiological characteristics that are unique to them are presently known. Hence, the identified CSIs and CSPs that are specific for different clades of Actinobacteria provide novel means for discovering biochemical and/or other characteristics that are unique to these groups. Most of the identified CSIs are located in widely distributed proteins (e.g., ribosomal proteins, RNA polymerase, gyrase, DNA polymerase, and various enzymes in key metabolic pathways) that are responsible for carrying out essential cellular functions. The primary functions of these proteins are vital for cell survival, and they are expected to remain the same in all organisms. Hence, the question arises, What is the functional significance of these evolutionarily conserved indels that are specific for different actinobacterial lineages?
Recent work on a number of conserved indels in the Hsp60 (GroEL) and Hsp70 (DnaK) proteins showed that the identified CSIs are essential for the groups of species where they are found and that deletions or most changes in them led to a failure of cell growth (269). Based upon this finding, we expect that the CSIs that are specific for Actinobacteria will also be essential for the particular lineages where they are found. An important observation in this regard is that most of these CSIs are generally present in the surface loops of various proteins (4, 118, 269). This is also true for most of the Actinobacteria-specific CSIs described in this work, and it is illustrated by the structures of two of the proteins, viz., S-adenosyl homocysteine hydrolase (240) and serine hydroxymethyltransferase (Fig. 20), which contain 5-aa and 9-aa CSIs, respectively, that are specific for most Actinobacteria (see Files S4 and S6 in the supplemental material for sequence alignments of these proteins). The structures shown in Fig. 20 are from M. tuberculosis, which contains these inserts, and the regions corresponding to the inserts are colored magenta. As shown in Fig. 20, the inserts in both proteins are present in surface loops, and they are seen as patches or knobs on the surfaces of these proteins. The surface loops in protein sequences are known to play an important role in mediating protein-protein interactions, and they can either facilitate or disrupt certain interactions (4, 152). In view of the predicted essential nature of these CSIs and their locations on protein surfaces (generally away from the active sites), we have postulated that these CSIs are involved in conferring novel functional capabilities (i.e., ancillary functions) on these essential proteins through protein-protein or other forms of interactions (269). These ancillary functions are expected to be important for the lineages in which these CSIs are found, and they could include the ability of the protein(s) to interact with other cellular proteins or ligands (with the CSI serving as a docking site) that either modulate the activity of these proteins or confer some new function(s) on them. Recent studies of two large CSIs in the gyrase B and RpoC proteins that are specific for a number of bacterial phyla support this hypothesis (46, 116, 255). Hence, further studies toward an understanding of the cellular functions of these Actinobacteria-specific CSIs should lead to the discovery of novel aspect of many important proteins that contain these CSIs.
Fig 20.
Structures of the S-adenosyl-l-homocysteine hydrolase (PDB accession number 3CE6) (240) (A and B) and serine hydroxymethyltransferase (PDB accession number 3H7F) (C and D) proteins from M. tuberculosis showing the locations in protein structures of the 9-aa and 5-aa actinobacterium-specific inserts that are found in these proteins (see Files S5 and S6 in the supplemental material). While panels A and C show ribbon representations, panels B and D depict the surface representations of these protein structures. The inserts in these proteins are shown in magenta.
Unlike CSIs, which are commonly found in essential proteins of known functions, the cellular functions of most of the CSPs that are limited to particular lineages of Actinobacteria are generally not known. The evolutionary conservation and retention of genes for these proteins by different lineages strongly suggest that they perform important functions (62, 96, 133, 244) that are specific for these lineages and which distinguish them from other Actinobacteria. Hence, an understanding of the cellular functions of these CSPs should provide valuable insights into the biochemical and physiological characteristics that are unique to different taxa of Actinobacteria. The significance of such proteins for particular lineages is illustrated by the examples of the well-studied EmbA, EmbB, EmbC, and AftA proteins, which are CSPs that are limited to either the order Corynebacteriales or the orders Corynebacteriales and Pseudonocardiales (Tables 3 and 9). The species of these two orders have cell wall chemotype IV, defined by the presence of meso-diaminopimelic acid, arabinose, and galactose in their cell walls, and these proteins play key roles in the biosynthesis of arabinan, which is a unique component of their cell walls (7, 24, 106, 259, 263, 300). Thus, the lineage specificity of these proteins correlates with a unique and essential biochemical property of these orders of Actinobacteria.
Of the four CSPs that are distinguishing characteristics of nearly all Actinobacteria, the structures of two of them, viz., SCO1997 and SCO1662 [gene identification from S. coelicolor A3(2), which corresponds to the ML1009 and ML1306 proteins from M. leprae TN], were recently solved (Protein Data Bank [PDB] accession number 3E35) (100). Although the structures of these two related proteins show limited structural similarity to purine nucleoside phosphorylase and the PAC2 family of proteins from the Archaea (PDB accession number 3GAA), at the sequence level, they exhibit no significant similarity to these proteins. Thus, the functions of these Actinobacteria-specific proteins are predicted to be novel. This inference is strongly supported by recent work showing that SCO1662 specifically interacts with the ParA protein, and it likely corresponds to the ParJ protein, which negatively regulates ParA polymerization in vitro, which is important for efficient chromosome segregation in sporulating aerial hyphae (71). However, further studies are needed to understand the roles of the two homologs of this protein (viz., SCO1662 and SCO1997) and why they are uniquely found in Actinobacteria. Similarly, the WhiB protein family, which has several gene copies in all Actinobacteria except the deepest-branching lineages, is indicated to play an essential role in controlling developmental transition in Streptomyces (43, 90). In nonsporulating actinobacterial species such as Mycobacterium species, WhiB proteins are differentially expressed, and they are important in regulating virulence, cell division, antibiotic resistance, and other stress responses (32, 208). These examples indicate that the CSPs that are specific for different actinobacterial lineages likely play important roles in different unique aspects of these bacteria, including their niche adaptation, pathogenic mechanisms, and other genetic, biochemical, and morphological characteristics that are unique to these bacteria. Hence, concerted efforts to understand their cellular functions should provide important insights into the unique biological aspects of these bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a research grant from the Natural Science and Engineering Research Council of Canada.
R.S.G. thanks Sanjan George, Balpreet Brar, Karen Kwofie, and Mobolaji Adeolu for their expert technical assistance in the identification and formatting of information for various CSIs and CSPs.
Biographies

Beile Gao completed her Ph.D. under the supervision of Radhey Gupta in December 2009. Her thesis work was on identifying molecular signatures that are specific for Actinobacteria, and a number of CSIs and CSPs specific for actinobacteria were identified. She also solved the structure for the first Actinobacteria-specific protein. After the completion of her Ph.D. thesis work, Dr. Gao worked for another 6 to 8 months on the identification of additional CSIs and CSPs that were specific for Actinobacteria. Since March 2010, Dr. Gao has been working as a postdoctoral fellow in the Section of Microbial Pathogenesis at Yale University School of Medicine.

Radhey S. Gupta is a Professor of Biochemistry at McMaster University in Canada. He joined McMaster in 1978, and his research interests have covered many areas, including studies on drug-resistant mutants of mammalian cells and heat shock proteins, structure-function studies on enzymes, and studies on novel aspects of mitochondria. Since the 1990s, his primary research interests have been in using molecular sequence data to understand the early evolutionary history of life. His earlier work provided evidence that the eukaryotic cell nucleus is a chimera formed by fusion between an archaeon and a bacterium. For the past 10 to 12 years, the main focus of his work has been on using genomic sequence data to understand microbial systematics and evolution. The aim of these studies is to discover novel molecular markers for identifying different groups of Bacteria in more definitive molecular terms and to understand their branching order from a common ancestor. His laboratory has pioneered the discovery of conserved signature indels and whole proteins that are specific for different phyla of bacteria at various phylogenetic depths. Large numbers of such signatures for different bacterial groups have been identified. In addition to providing more reliable means for identifying different groups of bacteria, these signatures also provide novel tools for microbial diagnostics, as potential targets for drug and vaccine development, and powerful means for genetic, biochemical, and evolutionary studies. Professor Gupta has published >260 articles in peer-reviewed articles, and the current “h” score of his publications is 51. Further information on his work can be found at his website, http://www.science.mcmaster.ca/biochem/faculty/gupta/index.htm.
Footnotes
Supplemental material for this article may be found at http://mmbr.asm.org/.
REFERENCES
- 1. Abdallah AM, et al. 2006. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol. Microbiol. 62:667–679 [DOI] [PubMed] [Google Scholar]
- 2. Abt B, et al. 2010. Complete genome sequence of Cellulomonas flavigena type strain (134). Stand. Genomic Sci. 3:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adekambi T, et al. 2011. Core gene set as the basis of multilocus sequence analysis of the subclass Actinobacteridae. PLoS One 6:e14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Akiva E, Itzhaki Z, Margalit H. 2008. Built-in loops allow versatility in domain-domain interactions: lessons from self-interacting domains. Proc. Natl. Acad. Sci. U. S. A. 105:13292–13297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alam MT, Merlo ME, Takano E, Breitling R. 2010. Genome-based phylogenetic analysis of Streptomyces and its relatives. Mol. Phylogenet. Evol. 54:763–772 [DOI] [PubMed] [Google Scholar]
- 6. Alderwick LJ, Seidel M, Sahm H, Besra GS, Eggeling L. 2006. Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 281:15653–15661 [DOI] [PubMed] [Google Scholar]
- 7. Amin AG, et al. 2008. EmbA is an essential arabinosyltransferase in Mycobacterium tuberculosis. Microbiology 154:240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Atlas RM. 1988. Microbiology: fundamentals and applications, p 1–807 Macmillan Publishing Co, New York, NY [Google Scholar]
- 9. Bagwell CE, et al. 2008. Survival in nuclear waste, extreme resistance, and potential applications gleaned from the genome sequence of Kineococcus radiotolerans SRS30216. PLoS One 3:e3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baldauf SL. 2003. Phylogeny for the faint of heart: a tutorial. Trends Genet. 19:345–351 [DOI] [PubMed] [Google Scholar]
- 11. Baldauf SL, Palmer JD. 1993. Animals and fungi are each other's closest relatives: congruent evidence from multiple proteins. Proc. Natl. Acad. Sci. U. S. A. 90:11558–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baltz RH. 2008. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 8:557–563 [DOI] [PubMed] [Google Scholar]
- 13. Bapteste E, Philippe H. 2002. The potential value of indels as phylogenetic markers: position of trichomonads as a case study. Mol. Biol. Evol. 19:972–977 [DOI] [PubMed] [Google Scholar]
- 14. Barabote RD, et al. 2009. Complete genome of the cellulolytic thermophile Acidothermus cellulolyticus 11B provides insights into its ecophysiological and evolutionary adaptations. Genome Res. 19:1033–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barrangou R, et al. 2009. Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J. Bacteriol. 191:4144–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Behal V. 2000. Bioactive products from Streptomyces. Adv. Appl. Microbiol. 47:113–156 [PubMed] [Google Scholar]
- 17. Bentley SD, Brosch R, Gordon SV, Hopwood DA, Cole ST. 2004. Genomics of Actinobacteria, the high G+C Gram-positive bacteria, p 333–359 In Fraser CM, Read TD, Nelson KE. (ed), Microbial genomes. Humana Press, Totowa, NJ [Google Scholar]
- 18. Bentley SD, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147 [DOI] [PubMed] [Google Scholar]
- 19. Bentley SD, et al. 2008. Genome of the actinomycete plant pathogen Clavibacter michiganensis subsp. sepedonicus suggests recent niche adaptation. J. Bacteriol. 190:2150–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bentley SD, et al. 2003. Sequencing and analysis of the genome of the Whipple's disease bacterium Tropheryma whipplei. Lancet 361:637–644 [DOI] [PubMed] [Google Scholar]
- 21. Berdy J. 2005. Bioactive microbial metabolites. J. Antibiot. (Tokyo) 58:1–26 [DOI] [PubMed] [Google Scholar]
- 22. Biavati B, Mattarelli P. 2006. The family Bifidobacteriaceae, p 322–382 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 23. Bignell DR, et al. 2010. Streptomyces scabies 87-22 contains a coronafacic acid-like biosynthetic cluster that contributes to plant-microbe interactions. Mol. Plant Microbe Interact. 23:161–175 [DOI] [PubMed] [Google Scholar]
- 24. Birch HL, et al. 2008. Biosynthesis of mycobacterial arabinogalactan: identification of a novel alpha(1→3) arabinofuranosyltransferase. Mol. Microbiol. 69:1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blackwood KS, et al. 2000. Evaluation of recA sequences for identification of Mycobacterium species. J. Clin. Microbiol. 38:2846–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blair C, Murphy RW. 2011. Recent trends in molecular phylogenetic analysis: where to next? J. Hered. 102:130–138 [DOI] [PubMed] [Google Scholar]
- 27. Blaise G, Nikkels AF, Hermanns-Le T, Nikkels-Tassoudji N, Pierard GE. 2008. Corynebacterium-associated skin infections. Int. J. Dermatol. 47:884–890 [DOI] [PubMed] [Google Scholar]
- 28. Bottacini F, et al. 2010. Comparative genomics of the genus Bifidobacterium. Microbiology 156:3243–3254 [DOI] [PubMed] [Google Scholar]
- 29. Boussau B, Daubin V. 2010. Genomes as documents of evolutionary history. Trends Ecol. Evol. 25:224–232 [DOI] [PubMed] [Google Scholar]
- 30. Bradshaw CS, et al. 2006. The association of Atopobium vaginae and Gardnerella vaginalis with bacterial vaginosis and recurrence after oral metronidazole therapy. J. Infect. Dis. 194:828–836 [DOI] [PubMed] [Google Scholar]
- 31. Brinkrolf K, Schroder J, Puhler A, Tauch A. 2010. The transcriptional regulatory repertoire of Corynebacterium glutamicum: reconstruction of the network controlling pathways involved in lysine and glutamate production. J. Biotechnol. 149:173–182 [DOI] [PubMed] [Google Scholar]
- 32. Brosch R, et al. 2007. Genome plasticity of BCG and impact on vaccine efficacy. Proc. Natl. Acad. Sci. U. S. A. 104:5596–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown JR, Douady CJ, Italia MJ, Marshall WE, Stanhope MJ. 2001. Universal trees based on large combined protein sequence data sets. Nat. Genet. 28:281–285 [DOI] [PubMed] [Google Scholar]
- 34. Bruggemann H, et al. 2004. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 305:671–673 [DOI] [PubMed] [Google Scholar]
- 35. Bull AT, Stach JE, Ward AC, Goodfellow M. 2005. Marine actinobacteria: perspectives, challenges, future directions. Antonie Van Leeuwenhoek 87:65–79 [PubMed] [Google Scholar]
- 36. Bull AT, Stach JEM. 2007. Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol. 15:491–499 [DOI] [PubMed] [Google Scholar]
- 37. Burkovski A. 2006. Proteomics of Corynebacterium glutamicum: essential industrial bacterium. Methods Biochem. Anal. 49:137–147 [PubMed] [Google Scholar]
- 38. Busse HJ. 2006. Renibacterium, p 972–974 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 39. Cerdeno-Tarraga AM, et al. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 31:6516–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Challis GL. 2008. Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology 154:1555–1569 [DOI] [PubMed] [Google Scholar]
- 41. Chater KF. 2006. Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chater KF, Biro S, Lee KJ, Palmer T, Schrempf H. 2010. The complex extracellular biology of Streptomyces. FEMS Microbiol. Rev. 34:171–198 [DOI] [PubMed] [Google Scholar]
- 43. Chater KF, Chandra G. 2006. The evolution of development in Streptomyces analysed by genome comparisons. FEMS Microbiol. Rev. 30:651–672 [DOI] [PubMed] [Google Scholar]
- 44. Chen CW, Huang CH, Lee HH, Tsai HH, Kirby R. 2002. Once the circle has been broken: dynamics and evolution of Streptomyces chromosomes. Trends Genet. 18:522–529 [DOI] [PubMed] [Google Scholar]
- 45. Chertkov O, et al. 2011. Complete genome sequence of Thermomonospora curvata type strain (B9). Stand. Genomic Sci. 4:13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chlenov M, et al. 2005. Structure and function of lineage-specific sequence insertions in the bacterial RNA polymerase betá subunit. J. Mol. Biol. 353:138–154 [DOI] [PubMed] [Google Scholar]
- 47. Choulet F, et al. 2006. Intraspecific variability of the terminal inverted repeats of the linear chromosome of Streptomyces ambofaciens. J. Bacteriol. 188:6599–6610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ciaramella M, Napoli A, Rossi M. 2005. Another extreme genome: how to live at pH 0. Trends Microbiol. 13:49–51 [DOI] [PubMed] [Google Scholar]
- 49. Ciccarelli FD, et al. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287 [DOI] [PubMed] [Google Scholar]
- 50. Coenye T, Gevers D, Van de Peer Y, Vandamme P, Swings J. 2005. Towards a prokaryotic genomic taxonomy. FEMS Microbiol. Rev. 29:147–167 [DOI] [PubMed] [Google Scholar]
- 51. Colbert CL, et al. 2010. Crystal structure of spot 14, a modulator of fatty acid synthesis. Proc. Natl. Acad. Sci. U. S. A. 107:18820–18825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cole JR, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cole ST. 1999. Learning from the genome sequence of Mycobacterium tuberculosis H37Rv. FEBS Lett. 452:7–10 [DOI] [PubMed] [Google Scholar]
- 54. Cole ST. 2002. Comparative and functional genomics of the Mycobacterium tuberculosis complex. Microbiology 148:2919–2928 [DOI] [PubMed] [Google Scholar]
- 55. Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–538 [DOI] [PubMed] [Google Scholar]
- 56. Cole ST, et al. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007–1011 [DOI] [PubMed] [Google Scholar]
- 57. Collins MD, Cummins CS. 1986. Genus Corynebacterium Lehmann and Neumann 1896, 350AL, p 1266–1276 In Sneath PHA, Mair NS, Sharpe ME, Holt JG. (ed), Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, MD [Google Scholar]
- 58. Collins MD, Goodfellow M, Minnikin DE. 1982. A survey of the structures of mycolic acids in Corynebacterium and related taxa. J. Gen. Microbiol. 128:129–149 [DOI] [PubMed] [Google Scholar]
- 59. Copeland A, et al. 2009. Complete genome sequence of Catenulispora acidiphila type strain (ID 139908). Stand. Genomic Sci. 1:119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Copeland A, et al. 2009. Complete genome sequence of Atopobium parvulum type strain (IPP 1246). Stand. Genomic Sci. 1:166–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cronin M, Ventura M, Fitzgerald GF, van Sinderen D. 2011. Progress in genomics, metabolism and biotechnology of bifidobacteria. Int. J. Food Microbiol. 149:4–18 [DOI] [PubMed] [Google Scholar]
- 62. Danchin A. 1999. From protein sequence to function. Curr. Opin. Struct. Biol. 9:363–367 [DOI] [PubMed] [Google Scholar]
- 63. Daubin V, Gouy M, Perriere G. 2002. A phylogenomic approach to bacterial phylogeny: evidence of a core of genes sharing a common history. Genome Res. 12:1080–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Daubin V, Ochman H. 2004. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 14:1036–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. D'Costa VM, McGrann KM, Hughes DW, Wright GD. 2006. Sampling the antibiotic resistome. Science 311:374–377 [DOI] [PubMed] [Google Scholar]
- 66. Del Rio TG, et al. 2010. Complete genome sequence of Intrasporangium calvum type strain (7 KIP). Stand. Genomic Sci. 3:294–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Delsuc F, Brinkmann H, Philippe H. 2005. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 6:361–375 [DOI] [PubMed] [Google Scholar]
- 68. Demain AL, Sanchez S. 2009. Microbial drug discovery: 80 years of progress. J. Antibiot. (Tokyo) 62:5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Demangel C, Stinear TP, Cole ST. 2009. Buruli ulcer: reductive evolution enhances pathogenicity of Mycobacterium ulcerans. Nat. Rev. Microbiol. 7:50–60 [DOI] [PubMed] [Google Scholar]
- 70. Devulder G, Perouse de Montclos M, Flandrois JP. 2005. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55:293–302 [DOI] [PubMed] [Google Scholar]
- 71. Ditkowski B, et al. 2010. The actinobacterial signature protein ParJ (SCO1662) regulates ParA polymerization and affects chromosome segregation and cell division during Streptomyces sporulation. Mol. Microbiol. 78:1403–1415 [DOI] [PubMed] [Google Scholar]
- 72. Domenech P, Barry CE, Cole ST. 2001. Mycobacterium tuberculosis in the post-genomic age. Curr. Opin. Microbiol. 4:28–34 [DOI] [PubMed] [Google Scholar]
- 73. Dong Z, Onrust R, Skangalis M, O'Donnell M. 1993. DNA polymerase III accessory proteins. I. holA and holB encoding delta and deltá. J. Biol. Chem. 268:11758–11765 [PubMed] [Google Scholar]
- 74. Dutilh BE, Snel B, Ettema TJ, Huynen MA. 2008. Signature genes as a phylogenomic tool. Mol. Biol. Evol. 25:1659–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dykhuizen EC, May JF, Tongpenyai A, Kiessling LL. 2008. Inhibitors of UDP-galactopyranose mutase thwart mycobacterial growth. J. Am. Chem. Soc. 130:6706–6707 [DOI] [PubMed] [Google Scholar]
- 76. Embley TM, Hirt RP, Williams DM. 1994. Biodiversity at the molecular level: the domains, kingdoms and phyla of life. Philos. Trans. R. Soc. Lond. B Biol. Sci. 345:21–33 [DOI] [PubMed] [Google Scholar]
- 77. Euzeby JP. 2011. List of prokaryotic names with standing in nomenclature. http://www.bacterio.cict.fr/a/actinobacteria
- 78. Evtushenko LI, Takeuchi M. 2006. The family Microbacteriaceae, p 1020–1098 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 79. Falentin H, et al. 2010. The complete genome of Propionibacterium freudenreichii CIRM-BIA1, a hardy actinobacterium with food and probiotic applications. PLoS One 5:e11748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fang G, Rocha EP, Danchin A. 2008. Persistence drives gene clustering in bacterial genomes. BMC Genomics 9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Felsenstein J. 1988. Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet. 22:521–565 [DOI] [PubMed] [Google Scholar]
- 82. Felsenstein J. 1996. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 266:418–427 [DOI] [PubMed] [Google Scholar]
- 83. Felsenstein J. 2004. Inferring phylogenies. Sinauer Associates, Inc, Sunderland, MA [Google Scholar]
- 84. Fenical W, Jensen PR. 2006. Developing a new resource for drug discovery: marine actinomycete bacteria. Nat. Chem. Biol. 2:666–673 [DOI] [PubMed] [Google Scholar]
- 85. Ferreira AC, et al. 1999. Characterization and radiation resistance of new isolates of Rubrobacter radiotolerans and Rubrobacter xylanophilus. Extremophiles 3:235–238 [DOI] [PubMed] [Google Scholar]
- 86. Fiedler HP, et al. 2005. Marine actinomycetes as a source of novel secondary metabolites. Antonie Van Leeuwenhoek 87:37–42 [DOI] [PubMed] [Google Scholar]
- 87. Fischbach MA, Walsh CT, Clardy J. 2008. The evolution of gene collectives: how natural selection drives chemical innovation. Proc. Natl. Acad. Sci. U. S. A. 105:4601–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Fleischmann RD, et al. 2002. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 184:5479–5490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Foster B, et al. 2010. Complete genome sequence of Xylanimonas cellulosilytica type strain (XIL07). Stand. Genomic Sci. 2:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fowler-Goldsworthy K, et al. 2011. The actinobacteria-specific gene wblA controls major developmental transitions in Streptomyces coelicolor A3(2). Microbiology 157:1312–1328 [DOI] [PubMed] [Google Scholar]
- 91. Freilich S, et al. 2009. Metabolic-network-driven analysis of bacterial ecological strategies. Genome Biol. 10:R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Froula JL, Francino MP. 2007. Selection against spurious promoter motifs correlates with translational efficiency across bacteria. PLoS One 2:e745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Fukuda S, et al. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469:543–547 [DOI] [PubMed] [Google Scholar]
- 94. Funke G, von Graevenitz A, Clarridge JE, III, Bernard KA. 1997. Clinical microbiology of coryneform bacteria. Clin. Microbiol. Rev. 10:125–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Galley KA, Singh B, Gupta RS. 1992. Cloning of HSP70 (dnaK) gene from Clostridium perfringens using a general polymerase chain reaction based approach. Biochem. Biophys. Acta 1130:203–208 [DOI] [PubMed] [Google Scholar]
- 96. Galperin MY, Koonin EV. 2004. ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res. 32:5452–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gao B, Gupta RS. 2005. Conserved indels in protein sequences that are characteristic of the phylum Actinobacteria. Int. J. Syst. Evol. Microbiol. 55:2401–2412 [DOI] [PubMed] [Google Scholar]
- 98. Gao B, Gupta RS. 2007. Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis. BMC Genomics 8:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gao B, Mohan R, Gupta RS. 2009. Phylogenomics and protein signatures elucidating the evolutionary relationships among the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 59:234–247 [DOI] [PubMed] [Google Scholar]
- 100. Gao B, Parmanathan R, Gupta RS. 2006. Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups. Antonie Van Leeuwenhoek 90:69–91 [DOI] [PubMed] [Google Scholar]
- 101. Garnier T, et al. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. U. S. A. 100:7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Garrigues C, Johansen E, Pedersen MB. 2010. Complete genome sequence of Bifidobacterium animalis subsp. lactis BB-12, a widely consumed probiotic strain. J. Bacteriol. 192:2467–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Garrity GM, Bell JA, Lilburn TG. 2005. The revised road map to the manual, p 159–220 In Brenner DJ, Krieg NR, Staley JT. (ed), Bergey's manual of systematic bacteriology, vol 2 Springer, New York, NY [Google Scholar]
- 104. Garrity GM, Holt JG. 2001. The road map to the manual, p 119–166 In Boone DR, Castenholz RW. (ed), Bergey's manual of systematic bacteriology, vol 2 Springer-Verlag, Berlin, Germany [Google Scholar]
- 105. Gartemann KH, et al. 2008. The genome sequence of the tomato-pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a large island involved in pathogenicity. J. Bacteriol. 190:2138–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Gibson KJ, et al. 2003. Identification of a novel mannose-capped lipoarabinomannan from Amycolatopsis sulphurea. Biochem. J. 372:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Glasby JS. 1979. Encyclopedia of antibiotics. John Wiley & Sons, New York, NY [Google Scholar]
- 108. Goker M, et al. 2010. Complete genome sequence of Olsenella uli type strain (VPI D76D-27C). Stand. Genomic Sci. 3:76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gontang EA, Fenical W, Jensen PR. 2007. Phylogenetic diversity of Gram-positive bacteria cultured from marine sediments. Appl. Environ. Microbiol. 73:3272–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Goodfellow M, Fiedler HP. 2010. A guide to successful bioprospecting: informed by actinobacterial systematics. Antonie Van Leeuwenhoek 98:119–142 [DOI] [PubMed] [Google Scholar]
- 111. Goodfellow M, Maldonado LA. 2006. The family Dietziaceae, Gordoniaceae, Nocardiaceae and Tsukamurellaceae, p 843–888 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 112. Goude R, Amin AG, Chatterjee D, Parish T. 2008. The critical role of embC in Mycobacterium tuberculosis. J. Bacteriol. 190:4335–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Graevenitz AV, Bernard K. 2006. The genus Corynebacterium—medical, p 819–842 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 114. Gribaldo S, Philippe H. 2002. Ancient phylogenetic relationships. Theor. Popul. Biol. 61:391–408 [DOI] [PubMed] [Google Scholar]
- 115. Griffiths E, Gupta RS. 2002. Protein signatures distinctive of chlamydial species: horizontal transfer of cell wall biosynthesis genes glmU from Archaebacteria to Chlamydiae, and murA between Chlamydiae and Streptomyces. Microbiology 148:2541–2549 [DOI] [PubMed] [Google Scholar]
- 116. Griffiths E, Gupta RS. 2004. Signature sequences in diverse proteins provide evidence for the late divergence of the order Aquificales. Int. Microbiol. 7:41–52 [PubMed] [Google Scholar]
- 117. Griffiths E, Gupta RS. 2006. Lateral transfers of serine hydroxymethyl transferase (glyA) and UDP-N-acetylglucosamine enolpyruvyl transferase (murA) genes from free-living Actinobacteria to the parasitic chlamydiae. J. Mol. Evol. 63:283–296 [DOI] [PubMed] [Google Scholar]
- 118. Gupta RS, Mathews DW. 2010. Signature proteins for the major clades of Cyanobacteria. BMC Evol. Biol. 10:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Gupta RS. 1998. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62:1435–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Gupta RS. 1998. What are archaebacteria: life's third domain or monoderm prokaryotes related to Gram-positive bacteria? A new proposal for the classification of prokaryotic organisms. Mol. Microbiol. 29:695–708 [DOI] [PubMed] [Google Scholar]
- 121. Gupta RS. 2000. The phylogeny of Proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol. Rev. 24:367–402 [DOI] [PubMed] [Google Scholar]
- 122. Gupta RS. 2001. The branching order and phylogenetic placement of species from completed bacterial genomes, based on conserved indels found in various proteins. Int. Microbiol. 4:187–202 [DOI] [PubMed] [Google Scholar]
- 123. Gupta RS. 2003. Evolutionary relationships among photosynthetic bacteria. Photosynth. Res. 76:173–183 [DOI] [PubMed] [Google Scholar]
- 124. Gupta RS. 2005. Molecular sequences and the early history of life, p 160–183 In Sapp J. (ed), Microbial phylogeny and evolution: concepts and controversies. Oxford University Press, New York, NY [Google Scholar]
- 125. Gupta RS. 2009. Protein signatures (molecular synapomorphies) that are distinctive characteristics of the major cyanobacterial clades. Int. J. Syst. Evol. Microbiol. 59:2510–2526 [DOI] [PubMed] [Google Scholar]
- 126. Gupta RS. 2010. Applications of conserved indels for understanding microbial phylogeny, p 135–150 In Oren A, Papke RT. (ed), Molecular phylogeny of microorganisms. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 127. Gupta RS. 2010. Microbial phylogeny and evolution based on protein sequences (the change from targeted genes to proteins), p 35–53 In Shah HN, Gharbia SE. (ed), Mass spectrometry for microbial proteomics. Wiley, Chichester, United Kingdom [Google Scholar]
- 128. Gupta RS. 2010. Molecular signatures for the main phyla of photosynthetic bacteria and their subgroups. Photosynth. Res. 104:357–372 [DOI] [PubMed] [Google Scholar]
- 129. Gupta RS. 2011. Origin of diderm (Gram-negative) bacteria: antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes. Antonie Van Leeuwenhoek 100:171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Gupta RS, Gao B. 2010. Recent advances in understanding microbial systematics, p 1–14 In Xu J. (ed), Microbial population genetics. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 131. Gupta RS, Golding GB. 1993. Evolution of HSP70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J. Mol. Evol. 37:573–582 [DOI] [PubMed] [Google Scholar]
- 132. Gupta RS, Griffiths E. 2002. Critical issues in bacterial phylogenies. Theor. Popul. Biol. 61:423–434 [DOI] [PubMed] [Google Scholar]
- 133. Gupta RS, Griffiths E. 2006. Chlamydiae-specific proteins and indels: novel tools for studies. Trends Microbiol. 14:527–535 [DOI] [PubMed] [Google Scholar]
- 134. Gupta RS, Mok A. 2007. Phylogenomics and signature proteins for the alpha proteobacteria and its main groups. BMC Microbiol. 7:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Gupta RS, Sneath PHA. 2007. Application of the character compatibility approach to generalized molecular sequence data: branching order of the proteobacterial subdivisions. J. Mol. Evol. 64:90–100 [DOI] [PubMed] [Google Scholar]
- 136. Gurtler V, Mayall BC, Seviour R. 2004. Can whole genome analysis refine the taxonomy of the genus Rhodococcus? FEMS Microbiol. Rev. 28:377–403 [DOI] [PubMed] [Google Scholar]
- 137. Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Gust B, Kieser T, Chater KF. 2002. REDIRECT technology: PCR-targeting system in Streptomyces coelicolor. John Innes Centre, Norwich, England [Google Scholar]
- 139. Hansmann S, Martin W. 2000. Phylogeny of 33 ribosomal and six other proteins encoded in an ancient gene cluster that is conserved across prokaryotic genomes: influence of excluding poorly alignable sites from analysis. Int. J. Syst. Evol. Microbiol. 50(Pt 4):1655–1663 [DOI] [PubMed] [Google Scholar]
- 140. Hao B, Qi J. 2004. Prokaryote phylogeny without sequence alignment: from avoidance signature to composition distance. J. Bioinform. Comput. Biol. 2:1–19 [DOI] [PubMed] [Google Scholar]
- 141. Hao Y, et al. 2011. Complete genome sequence of Bifidobacterium longum subsp. longum BBMN68, a new strain from a healthy Chinese centenarian. J. Bacteriol. 193:787–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Hartmans S, Debont JAM, Stackebrandt E. 2006. The genus Mycobacterium—nonmedical, p 889–918 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 143. Hershfield MS, Krodich NM. 1978. S-Adenosylhomocysteine hydrolase is an adenosine-binding protein: a target for adenosine toxicity. Science 202:757–760 [DOI] [PubMed] [Google Scholar]
- 144. Hirano S, Tanaka K, Ohnishi Y, Horinouchi S. 2008. Conditionally positive effect of the TetR-family transcriptional regulator AtrA on streptomycin production by Streptomyces griseus. Microbiology 154:905–914 [DOI] [PubMed] [Google Scholar]
- 145. Hopwood DA. 2006. Soil to genomics: the Streptomyces chromosome. Annu. Rev. Genet. 40:1–23 [DOI] [PubMed] [Google Scholar]
- 146. Hopwood DA. 2007. Streptomyces in nature and medicine. Oxford University Press, New York, NY [Google Scholar]
- 147. Hugenholtz P, Tyson GW. 2008. Microbiology: metagenomics. Nature 455:481–483 [DOI] [PubMed] [Google Scholar]
- 148. Ikeda H, et al. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526–531 [DOI] [PubMed] [Google Scholar]
- 149. Ikeda M, Nakagawa S. 2003. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 62:99–109 [DOI] [PubMed] [Google Scholar]
- 150. Inoue K, Habe H, Yamane H, Nojiri M. 2006. Characterization of novel carbazole catabolism genes from gram-positive carbazole degrader Nocardioides aromaticivorans IC177. Appl. Environ. Microbiol. 72:3321–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ishikawa J, et al. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. U. S. A. 101:14925–14930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Itzhaki Z, Akiva E, Altuvia Y, Margalit H. 2006. Evolutionary conservation of domain-domain interactions. Genome Biol. 7:R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Ivanova N, et al. 2010. Complete genome sequence of Gordonia bronchialis type strain (3410). Stand. Genomic Sci. 2:19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Ivanova N, et al. 2010. Complete genome sequence of Geodermatophilus obscurus type strain (G-20). Stand. Genomic Sci. 2:158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Ivanova N, et al. 2009. Complete genome sequence of Sanguibacter keddieii type strain (ST-74). Stand. Genomic Sci. 1:110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Jakimowicz D, et al. 2007. Characterization of the mycobacterial chromosome segregation protein ParB and identification of its target in Mycobacterium smegmatis. Microbiology 153:4050–4060 [DOI] [PubMed] [Google Scholar]
- 157. Jones D, Keddie RM. 2006. The genus Arthrobacter, p 945–960 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 158. Joshi SM, et al. 2006. Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc. Natl. Acad. Sci. U. S. A. 103:11760–11765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kalinowski J, et al. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5–25 [DOI] [PubMed] [Google Scholar]
- 160. Kampfer P. 2006. The family Streptomycetaceae, part I: taxonomy, p 538–604 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 161. Kasai H, Ezaki T, Harayama S. 2000. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38:301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Keller S, et al. 2007. Abyssomicins G and H and atrop-abyssomicin C from the marine Verrucosispora strain AB-18-032. J. Antibiot. (Tokyo) 60:391–394 [DOI] [PubMed] [Google Scholar]
- 163. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, London, United Kingdom [Google Scholar]
- 164. Kim JF, et al. 2009. Genome sequence of the probiotic bacterium Bifidobacterium animalis subsp. lactis AD011. J. Bacteriol. 191:678–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Kirby R. 2011. Chromosome diversity and similarity within the Actinomycetales. FEMS Microbiol. Lett. 319:1–10 [DOI] [PubMed] [Google Scholar]
- 166. Kirchner O, Tauch A. 2003. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 104:287–299 [DOI] [PubMed] [Google Scholar]
- 167. Klijn A, Mercenier A, Arigoni F. 2005. Lessons from the genomes of bifidobacteria. FEMS Microbiol. Rev. 29:491–509 [DOI] [PubMed] [Google Scholar]
- 168. Koch AL. 2003. Were Gram-positive rods the first bacteria? Trends Microbiol. 11:166–170 [DOI] [PubMed] [Google Scholar]
- 169. Kochur M, Kloos WE, Schleifer KH. 2006. The genus Micrococcus, p 961–971 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 170. Konstantinidis KT, Tiedje JM. 2004. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl. Acad. Sci. U. S. A. 101:3160–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Konstantinidis KT, Tiedje JM. 2007. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr. Opin. Microbiol. 10:504–509 [DOI] [PubMed] [Google Scholar]
- 172. Koonin EV. 2003. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 1:127–136 [DOI] [PubMed] [Google Scholar]
- 173. Kunisawa T. 2003. Gene arrangements and branching orders of Gram-positive bacteria. J. Theor. Biol. 222:495–503 [DOI] [PubMed] [Google Scholar]
- 174. Kunisawa T. 2007. Gene arrangements characteristic of the phylum Actinobacteria. Antonie Van Leeuwenhoek 92:359–365 [DOI] [PubMed] [Google Scholar]
- 175. Kuo CH, Ochman H. 2009. The fate of new bacterial genes. FEMS Microbiol. Rev. 33:38–43 [DOI] [PubMed] [Google Scholar]
- 176. Kurahashi M, Fukunaga Y, Sakiyama Y, Harayama S, Yokota A. 2010. Euzebya tangerina gen. nov., sp. nov., a deeply branching marine actinobacterium isolated from the sea cucumber Holothuria edulis, and proposal of Euzebyaceae fam. nov., Euzebyales ord. nov. and Nitriliruptoridae subclassis nov. Int. J. Syst. Evol. Microbiol. 60:2314–2319 [DOI] [PubMed] [Google Scholar]
- 177. Kyrpides NC, Woese CR. 1998. Universally conserved translation initiation factors. Proc. Natl. Acad. Sci. U. S. A. 95:224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Labeda DP, Goodfellow M, Chun J, Zhi XY, Li WJ. 2011. Reassessment of the systematics of the suborder Pseudonocardineae: transfer of the genera within the family Actinosynnemataceae Labeda and Kroppenstedt 2000 emend. Zhi et al. 2009 into an emended family Pseudonocardiaceae Embley et al. 1989 emend. Zhi et al. 2009. Int. J. Syst. Evol. Microbiol. 61:1259–1264 [DOI] [PubMed] [Google Scholar]
- 179. Lake JA, Herbold CW, Rivera MC, Servin JA, Skophammer RG. 2007. Rooting the tree of life using nonubiquitous genes. Mol. Biol. Evol. 24:130–136 [DOI] [PubMed] [Google Scholar]
- 180. Lapidus A, et al. 2009. Complete genome sequence of Brachybacterium faecium type strain (Schefferle 6-10). Stand. Genomic Sci. 1:3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Lawrence JG, Hendrickson H. 2005. Genome evolution in bacteria: order beneath chaos. Curr. Opin. Microbiol. 8:572–578 [DOI] [PubMed] [Google Scholar]
- 182. Lechevalier MP. 1977. Lipids in bacterial taxonomy—a taxonomist's view. Crit. Rev. Microbiol. 5:109–210 [DOI] [PubMed] [Google Scholar]
- 183. Lee JH, et al. 2008. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Letek M, et al. 2008. Evolution of the Rhodococcus equi vap pathogenicity island seen through comparison of host-associated vapA and vapB virulence plasmids. J. Bacteriol. 190:5797–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Levine C, Hiasa H, Marians KJ. 1998. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta 1400:29–43 [DOI] [PubMed] [Google Scholar]
- 186. Li L, et al. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. U. S. A. 102:12344–12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Liebl W. 2006. Corynebacterium—nonmedical, p 796–818 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 188. Liolios K, et al. 2010. Complete genome sequence of Thermobispora bispora type strain (R51). Stand. Genomic Sci. 2:318–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Little AEF, Currie CR. 2007. Symbiotic complexity: discovery of a fifth symbiont in the attine ant-microbe symbiosis. Biol. Lett. 3:501–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Louie GV, et al. 1992. Structure of porphobilinogen deaminase reveals a flexible multidomain polymerase with a single catalytic site. Nature 359:33–39 [DOI] [PubMed] [Google Scholar]
- 191. Ludwig W, et al. Road map of the Actinobacteria. In Goodfellow M, et al. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 5, in press Springer-Verlag, Berlin, Germany: http://www.bergeys.org/outlines/bergeys_vol_5_roadmap_outline.pdf [Google Scholar]
- 192. Ludwig W, Klenk H-P. 2005. Overview: a phylogenetic backbone and taxonomic framework for prokaryotic systamatics, p 49–65 In Brenner DJ, Krieg NR, Staley JT, Garrity GM. (ed), Bergey's manual of systematic bacteriology. Springer-Verlag, Berlin, Germany [Google Scholar]
- 193. Ludwig W, Schleifer KH. 1999. Phylogeny of Bacteria beyond the 16S rRNA standard. ASM News 65:752–757 [Google Scholar]
- 194. Lykidis A, et al. 2007. Genome sequence and analysis of the soil cellulolytic actinomycete Thermobifida fusca YX. J. Bacteriol. 189:2477–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Mavrommatis K, et al. 2009. Complete genome sequence of Cryptobacterium curtum type strain (12-3). Stand. Genomic Sci. 1:93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. McLeod MP, et al. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. U. S. A. 103:15582–15587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Miao VP, Davies J. 2010. Actinobacteria: the good, the bad, and the ugly. Antonie Van Leeuwenhoek 98:143–150 [DOI] [PubMed] [Google Scholar]
- 198. Mikusova K, et al. 2005. Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. J. Bacteriol. 187:8020–8025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Miller BG, Wolfenden R. 2002. Catalytic proficiency: the unusual case of OMP decarboxylase. Annu. Rev. Biochem. 71:847–885 [DOI] [PubMed] [Google Scholar]
- 200. Miyatake K, Nakano Y, Kitaoka S. 1979. Pantothenate synthetase from Escherichia coli [D-pantoate: beta-alanine ligase (AMP-forming), EC 6.3.2.1]. Methods Enzymol. 62:215–219 [DOI] [PubMed] [Google Scholar]
- 201. Monciardini P, Sosio M, Cavaletti L, Chiocchini C, Donadio S. 2002. New PCR primers for the selective amplification of 16S rDNA from different groups of actinomycetes. FEMS Microbiol. Ecol. 42:419–429 [DOI] [PubMed] [Google Scholar]
- 202. Mongodin EF, et al. 2006. Secrets of soil survival revealed by the genome sequence of Arthrobacter aurescens TC1. PLoS Genet. 2:e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Monnet C, et al. 2010. The Arthrobacter arilaitensis Re117 genome sequence reveals its genetic adaptation to the surface of cheese. PLoS One 5:e15489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Monot M, et al. 2009. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat. Genet. 41:1282–1289 [DOI] [PubMed] [Google Scholar]
- 205. Monteiro-Vitorello CB, et al. 2004. The genome sequence of the Gram-positive sugarcane pathogen Leifsonia xyli subsp. xyli. Mol. Plant Microbe Interact. 17:827–836 [DOI] [PubMed] [Google Scholar]
- 206. Moreira D, Philippe H. 2000. Molecular phylogeny: pitfalls and progress. Int. Microbiol. 3:9–16 [PubMed] [Google Scholar]
- 207. Morohoshi T, Wang WZ, Someya N, Ikeda T. 2011. Genome sequence of Microbacterium testaceum StLB037, an N-acylhomoserine lactone-degrading bacterium isolated from potato leaves. J. Bacteriol. 193:2072–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Morris RP, et al. 2005. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 102:12200–12205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Na KS, et al. 2005. Isolation and characterization of benzene-tolerant Rhodococcus opacus strains. J. Biosci. Bioeng. 99:378–382 [DOI] [PubMed] [Google Scholar]
- 210. Narra HP, Cordes MH, Ochman H. 2008. Structural features and the persistence of acquired proteins. Proteomics 8:4772–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. NCBI 2011. NCBI completed microbial genomes. http://www.ncbi.nlm.nih.gov/PMGifs/Genomes/micr.html
- 212. Nguyen HT, Wolff KA, Cartabuke RH, Ogwang S, Nguyen L. 2010. A lipoprotein modulates activity of the MtrAB two-component system to provide intrinsic multidrug resistance, cytokinetic control and cell wall homeostasis in Mycobacterium. Mol. Microbiol. 76:348–364 [DOI] [PubMed] [Google Scholar]
- 213. Nishio Y, et al. 2003. Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res. 13:1572–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Nolan M, et al. 2010. Complete genome sequence of Streptosporangium roseum type strain (NI 9100). Stand. Genomic Sci. 2:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Normand P. 2006. The families Frankiaceae, Geodtermatophilaceae, Acidothermaceae and Sproicchtthyaceae, p 669–681 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 216. Normand P, et al. 2007. Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Res. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Nouioui I, Ghodhbane-Gtari F, Beauchemin NJ, Tisa LS, Gtari M. 2011. Phylogeny of members of the Frankia genus based on gyrB, nifH and glnII sequences. Antonie Van Leeuwenhoek 100:579–587 [DOI] [PubMed] [Google Scholar]
- 218. Ochman H. 2005. Genomes on the shrink. Proc. Natl. Acad. Sci. U. S. A. 102:11959–11960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Ohnishi Y, et al. 2008. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190:4050–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Olano C, Mendez C, Salas JA. 2009. Antitumor compounds from actinomycetes: from gene clusters to new derivatives by combinatorial biosynthesis. Nat. Prod. Rep. 26:628–660 [DOI] [PubMed] [Google Scholar]
- 221. Olano C, Mendez C, Salas JA. 2009. Antitumor compounds from marine actinomycetes. Mar. Drugs 7:210–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Oliynyk M, et al. 2007. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat. Biotechnol. 25:447–453 [DOI] [PubMed] [Google Scholar]
- 223. Olsen GJ, Woese CR. 1993. Ribosomal RNA: a key to phylogeny. FASEB J. 7:113–123 [DOI] [PubMed] [Google Scholar]
- 224. Olson JM, Nguyen VQ, Yoo J, Kuechle MK. 2009. Cutaneous manifestations of Corynebacterium jeikeium sepsis. Int. J. Dermatol. 48:886–888 [DOI] [PubMed] [Google Scholar]
- 225. Oren A. 2004. Prokaryote diversity and taxonomy: current status and future challenges. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359:623–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Paradkar A, Trefzer A, Chakraburtty R, Stassi D. 2003. Streptomyces genetics: a genomic perspective. Crit. Rev. Biotechnol. 23:1–27 [DOI] [PubMed] [Google Scholar]
- 227. Pascual C, Lawson PA, Farrow JA, Gimenez MN, Collins MD. 1995. Phylogenetic analysis of the genus Corynebacterium based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 45:724–728 [DOI] [PubMed] [Google Scholar]
- 228. Pati A, et al. 2009. Complete genome sequence of Saccharomonospora viridis type strain (P101). Stand. Genomic Sci. 1:141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Penn K, et al. 2009. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J. 3:1193–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Persson BC, Bylund GO, Berg DE, Wikstrom PM. 1995. Functional analysis of the ffh-trmD region of the Escherichia coli chromosome by using reverse genetics. J. Bacteriol. 177:5554–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Philippot L, et al. 2010. The ecological coherence of high bacterial taxonomic ranks. Nat. Rev. Microbiol. 8:523–529 [DOI] [PubMed] [Google Scholar]
- 232. Pitulle C, Dorsch M, Kazda J, Wolters J, Stackebrandt E. 1992. Phylogeny of rapidly growing members of the genus Mycobacterium. Int. J. Syst. Bacteriol. 42:337–343 [DOI] [PubMed] [Google Scholar]
- 233. Pukall R, et al. 2009. Complete genome sequence of Jonesia denitrificans type strain (Prevot 55134). Stand. Genomic Sci. 1:262–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Pukall R, et al. 2010. Complete genome sequence of Conexibacter woesei type strain (ID131577). Stand. Genomic Sci. 2:212–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Pukall R, et al. 2010. Complete genome sequence of Kribbella flavida type strain (IFO 14399). Stand. Genomic Sci. 2:186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Qin Y, et al. 2006. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 127:721–733 [DOI] [PubMed] [Google Scholar]
- 237. Ranea JAG. 2006. Micro(be)-economics. Heredity 96:337–338 [DOI] [PubMed] [Google Scholar]
- 238. Rao NA, Talwar R, Savithri HS. 2000. Molecular organization, catalytic mechanism and function of serine hydroxymethyltransferase—a potential target for cancer chemotherapy. Int. J. Biochem. Cell Biol. 32:405–416 [DOI] [PubMed] [Google Scholar]
- 239. Raoult D, et al. 2003. Tropheryma whipplei twist: a human pathogenic Actinobacteria with a reduced genome. Genome Res. 13:1800–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Reddy MC, et al. 2008. Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors. Protein Sci. 17:2134–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Richert K, Brambilla E, Stackebrandt E. 2005. Development of PCR primers specific for the amplification and direct sequencing of gyrB genes from microbacteria, order Actinomycetales. J. Microbiol. Methods 60:115–123 [DOI] [PubMed] [Google Scholar]
- 242. Ripoll F, et al. 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Rivera MC, Lake JA. 1992. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science 257:74–76 [DOI] [PubMed] [Google Scholar]
- 244. Roberts RJ. 2004. Identifying protein function—a call for community action. PLoS Biol. 2:E42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Rohmer M. 1999. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 16:565–574 [DOI] [PubMed] [Google Scholar]
- 246. Rokas A, Holland PW. 2000. Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 15:454–459 [DOI] [PubMed] [Google Scholar]
- 247. Rokas A, Williams BL, King N, Carroll SB. 2003. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425:798–804 [DOI] [PubMed] [Google Scholar]
- 248. Roller C, Ludwig W, Schleifer KH. 1992. Gram-positive bacteria with a high DNA G+C content are characterized by a common insertion within their 23S rRNA genes. J. Gen. Microbiol. 138:167–175 [DOI] [PubMed] [Google Scholar]
- 249. Rosamond J, Allsop A. 2000. Harnessing the power of the genome in the search for new antibiotics. Science 287:1973–1976 [DOI] [PubMed] [Google Scholar]
- 250. Saiki K, Mogi T, Anraku Y. 1992. Heme O biosynthesis in Escherichia coli: the cyoE gene in the cytochrome bo operon encodes a protoheme IX farnesyltransferase. Biochem. Biophys. Res. Commun. 189:1491–1497 [DOI] [PubMed] [Google Scholar]
- 251. Saunders E, et al. 2009. Complete genome sequence of Eggerthella lenta type strain (IPP VPI 0255). Stand. Genomic Sci. 1:174–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Saviola B, Bishai W. 2006. The genus Mycobacterium—medical, p 919–933 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 253. Schaal KP, Yassin AF, Stackebrandt E. 2006. The family Actinomycetaceae: the genera Actinomyces, Actinobaculum, Arcanobacterium, Varibaculum and Mobiluncus, p 430–537 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 254. Schell MA, et al. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U. S. A. 99:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Schoeffler AJ, May AP, Berger JM. 2010. A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function. Nucleic Acids Res. 38:7830–7844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Schrempf H. 2001. Recognition and degradation of chitin by streptomycetes. Antonie Van Leeuwenhoek 79:285–289 [DOI] [PubMed] [Google Scholar]
- 257. Schrempf H. 2006. The family Streptomycetaceae, part II: molecular biology, p 605–622 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 258. Schumann P, Kampfer P, Busse HJ, Evtushenko LI. 2009. Proposed minimal standards for describing new genera and species of the suborder Micrococcineae. Int. J. Syst. Evol. Microbiol. 59:1823–1849 [DOI] [PubMed] [Google Scholar]
- 259. Seidel M, et al. 2007. Identification of a novel arabinofuranosyltransferase AftB involved in a terminal step of cell wall arabinan biosynthesis in corynebacterianeae, such as Corynebacterium glutamicum and Mycobacterium tuberculosis. J. Biol. Chem. 282:14729–14740 [DOI] [PubMed] [Google Scholar]
- 260. Seki M, et al. 2009. Whole genome sequence analysis of Mycobacterium bovis bacillus Calmette-Guerin (BCG) Tokyo 172: a comparative study of BCG vaccine substrains. Vaccine 27:1710–1716 [DOI] [PubMed] [Google Scholar]
- 261. Sekine M, et al. 2006. Sequence analysis of three plasmids harboured in Rhodococcus erythropolis strain PR4. Environ. Microbiol. 8:334–346 [DOI] [PubMed] [Google Scholar]
- 262. Sela DA, et al. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U. S. A. 105:18964–18969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Shi L, et al. 2006. The carboxy terminus of EmbC from Mycobacterium smegmatis mediates chain length extension of the arabinan in lipoarabinomannan. J. Biol. Chem. 281:19512–19526 [DOI] [PubMed] [Google Scholar]
- 264. Shinnick TM. 2006. Mycobacterium leprae, p 934–944 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 265. Siew N, Fischer D. 2003. Twenty thousand ORFan microbial protein families for the biologist? Structure (Camb.) 11:7–9 [DOI] [PubMed] [Google Scholar]
- 266. Sikorski J, et al. 2010. Complete genome sequence of Segniliparus rotundus type strain (CDC 1076). Stand. Genomic Sci. 2:203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Silva A, et al. 2011. Complete genome sequence of Corynebacterium pseudotuberculosis I19, a strain isolated from a cow in Israel with bovine mastitis. J. Bacteriol. 193:323–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Sims D, et al. 2009. Complete genome sequence of Kytococcus sedentarius type strain (541). Stand. Genomic Sci. 1:12–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Singh B, Gupta RS. 2009. Conserved inserts in the Hsp60 (GroEL) and Hsp70 (DnaK) proteins are essential for cellular growth. Mol. Genet. Genomics 281:361–373 [DOI] [PubMed] [Google Scholar]
- 270. Smith JJ, Tow LA, Stafford W, Cary C, Cowan DA. 2006. Bacterial diversity in three different Antarctic cold desert mineral soils. Microb. Ecol. 51:413–421 [DOI] [PubMed] [Google Scholar]
- 271. Sneath PHA. 2001. Numerical taxonomy, p 39–42 In Boone DR, Castenholz RW. (ed), Bergey's manual of systematic bacteriology. Springer-Verlag, Berlin, Germany [Google Scholar]
- 272. Sneath PHA, Sokal RR. 1973. Numerical taxonomy—the principles and practice of numerical classification, p 1–573 WH Freeman, San Francisco, CA [Google Scholar]
- 273. Snel B, Bork P, Huynen MA. 2002. Genomes in flux: the evolution of archaeal and proteobacterial gene content. Genome Res. 12:17–25 [DOI] [PubMed] [Google Scholar]
- 274. Snel B, Huynen MA, Dutilh BE. 2005. Genome trees and the nature of genome evolution. Annu. Rev. Microbiol. 59:191–209 [DOI] [PubMed] [Google Scholar]
- 275. Soltero-Higgin M, Carlson EE, Gruber TD, Kiessling LL. 2004. A unique catalytic mechanism for UDP-galactopyranose mutase. Nat. Struct. Mol. Biol. 11:539–543 [DOI] [PubMed] [Google Scholar]
- 276. Stach JE, Maldonado LA, Ward AC, Goodfellow M, Bull AT. 2003. New primers for the class Actinobacteria: application to marine and terrestrial environments. Environ. Microbiol. 5:828–841 [DOI] [PubMed] [Google Scholar]
- 277. Stach JEM, Bull AT. 2005. Estimating and comparing the diversity of marine actinobacteria. Antonie Van Leeuwenhoek 87:3–9 [DOI] [PubMed] [Google Scholar]
- 278. Stackebrandt E. 2006. Defining taxonomic ranks, p 29–57 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 279. Stackebrandt E, Cummins CS, Johnson JL. 2006. Family Propionibacteriaceae: the genus Propionibacterium, p 400–418 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 280. Stackebrandt E, Rainey FA, Ward-Rainey NL. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Evol. Microbiol. 47:479–491 [Google Scholar]
- 281. Stackebrandt E, Schaal KP. 2006. The family Propionibacteriaceae: the genera Friedmanniella, Leuteococcus, Microlunatus, Microprunia, Propioniferax, Propionimicrobium and Tessarococcus, p 383–399 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 282. Stackebrandt E, Schumann D, Prauser H. 2006. The family Cellulomonadaceae, p 983–1001 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 283. Stackebrandt E, Schumann P. 2006. Introduction to the taxonomy of Actinobacteria. Prokaryotes 3:297–321 [Google Scholar]
- 284. Stackebrandt E, et al. 1997. Phylogenetic analysis of the genus Desulfotomaculum: evidence for the misclassification of Desulfotomaculum guttoideum and description of Desulfotomaculum orientis as Desulfosporosinus orientis gen. nov., comb. nov. Int. J. Syst. Bacteriol. 47:1134–1139 [DOI] [PubMed] [Google Scholar]
- 285. Stahl DA, Urbance JW. 1990. The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria. J. Bacteriol. 172:116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Staley JT. 2006. The bacterial species dilemma and the genomic-phylogenetic species concept. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1899–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Stinear TP, et al. 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 18:729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Stinear TP, et al. 2007. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 17:192–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Strong M, et al. 2006. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 103:8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Stryer L. 1995. Biochemistry. WH Freeman and Co, New York, NY [Google Scholar]
- 291. Sun H, et al. 2010. Complete genome sequence of Nocardiopsis dassonvillei type strain (IMRU 509). Stand. Genomic Sci. 3:325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Sun Z, et al. 2010. Complete genome sequence of probiotic Bifidobacterium animalis subsp. lactis strain V9. J. Bacteriol. 192:4080–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Sutcliffe IC. 2011. Cell envelope architecture in the Chloroflexi: a shifting frontline in a phylogenetic turf war. Environ. Microbiol. 13:279–282 [DOI] [PubMed] [Google Scholar]
- 294. Sutcliffe IC, Harrington DJ. 2004. Lipoproteins of Mycobacterium tuberculosis: an abundant and functionally diverse class of cell envelope components. FEMS Microbiol. Rev. 28:645–659 [DOI] [PubMed] [Google Scholar]
- 295. Suzuki K, Goodfellow M, O'Donnell AG. 1993. Cell envelopes and classification, p 195–250 In Goodfellow M, O'Donnell AG. (ed), Handbook of new bacterial systematics. Academic Press, New York, NY [Google Scholar]
- 296. Takarada H, et al. 2008. Complete genome sequence of the soil actinomycete Kocuria rhizophila. J. Bacteriol. 190:4139–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Tauch A, et al. 2005. Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. J. Bacteriol. 187:4671–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Tauch A, et al. 2008. Ultrafast pyrosequencing of Corynebacterium kroppenstedtii DSM44385 revealed insights into the physiology of a lipophilic corynebacterium that lacks mycolic acids. J. Biotechnol. 136:22–30 [DOI] [PubMed] [Google Scholar]
- 299. Tauch A, et al. 2008. The lifestyle of Corynebacterium urealyticum derived from its complete genome sequence established by pyrosequencing. J. Biotechnol. 136:11–21 [DOI] [PubMed] [Google Scholar]
- 300. Telenti A, et al. 1997. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat. Med. 3:567–570 [DOI] [PubMed] [Google Scholar]
- 301. Tian J, Bryk R, Itoh M, Suematsu M, Nathan C. 2005. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate decarboxylase. Proc. Natl. Acad. Sci. U. S. A. 102:10670–10675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Tice H, et al. 2010. Complete genome sequence of Nakamurella multipartita type strain (Y-104). Stand. Genomic Sci. 2:168–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Trejo AG, Chittenden GJ, Buchanan JG, Baddiley J. 1970. Uridine diphosphate alpha-D-galactofuranose, an intermediate in the biosynthesis of galactofuranosyl residues. Biochem. J. 117:637–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Trost E, et al. 2010. Complete genome sequence and lifestyle of black-pigmented Corynebacterium aurimucosum ATCC 700975 (formerly C. nigricans CN-1) isolated from a vaginal swab of a woman with spontaneous abortion. BMC Genomics 11:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Tsolaki AG, et al. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. U. S. A. 101:4865–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Turroni F, et al. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. U. S. A. 107:19514–19519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Turroni F, van Sinderen D, Ventura M. 2009. Bifidobacteria: from ecology to genomics. Front. Biosci. 14:4673–4684 [DOI] [PubMed] [Google Scholar]
- 308. Turroni F, van Sinderen D, Ventura M. 2011. Genomics and ecological overview of the genus Bifidobacterium. Int. J. Food Microbiol. 149:37–44 [DOI] [PubMed] [Google Scholar]
- 309. Udwary DW, et al. 2007. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. U. S. A. 104:10376–10381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Ueda K, et al. 2004. Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism. Nucleic Acids Res. 32:4937–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Valas RE, Bourne PE. 2011. The origin of a derived superkingdom: how a Gram-positive bacterium crossed the desert to become an archaeon. Biol. Direct 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Ventura M, et al. 2006. Analysis of bifidobacterial evolution using a multilocus approach. Int. J. Syst. Evol. Microbiol. 56:2783–2792 [DOI] [PubMed] [Google Scholar]
- 313. Ventura M, Canchaya C, Fitzgerald GF, Gupta RS, van Sinderen D. 2007. Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie Van Leeuwenhoek 91:351–372 [DOI] [PubMed] [Google Scholar]
- 314. Ventura M, et al. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71:495–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Ventura M, Canchaya C, Zink R, Fitzgerald GF, van Sinderen D. 2004. Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: genetic, transcriptional, and phylogenetic analyses. Appl. Environ. Microbiol. 70:6197–6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Ventura M, et al. 2007. From bacterial genome to functionality; case bifidobacteria. Int. J. Food Microbiol. 120:2–12 [DOI] [PubMed] [Google Scholar]
- 317. Ventura M, et al. 2009. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat. Rev. Microbiol. 7:61–71 [DOI] [PubMed] [Google Scholar]
- 318. Ventura M, et al. 2009. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 5:e1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Ventura M, van Sinderen D, Fitzgerald GF, Zink R. 2004. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Van Leeuwenhoek 86:205–223 [DOI] [PubMed] [Google Scholar]
- 320. Ventura M, Zink R. 2003. Comparative sequence analysis of the tuf and recA genes and restriction fragment length polymorphism of the internal transcribed spacer region sequences supply additional tools for discriminating Bifidobacterium lactis from Bifidobacterium animalis. Appl. Environ. Microbiol. 69:7517–7522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Volff JN, Altenbuchner J. 2000. A new beginning with new ends: linearisation of circular chromosomes during bacterial evolution. FEMS Microbiol. Lett. 186:143–150 [DOI] [PubMed] [Google Scholar]
- 322. Wang XJ, et al. 2010. Genome sequence of the milbemycin-producing bacterium Streptomyces bingchenggensis. J. Bacteriol. 192:4526–4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Ward AC, Goodfellow M. 2004. Phylogeny and functionality: taxonomy as a roadmap to genes, p 288–313 In Bull AT. (ed), Microbial diversity and bioprospecting. ASM Press, Washington, DC [Google Scholar]
- 324. Ward N, Fraser CM. 2005. How genomics has affected the concept of microbiology. Curr. Opin. Microbiol. 8:564–571 [DOI] [PubMed] [Google Scholar]
- 325. Watve MG, Tickoo R, Jog MM, Bhole BD. 2001. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 176:386–390 [DOI] [PubMed] [Google Scholar]
- 326. Wei YX, et al. 2010. Complete genome sequence of Bifidobacterium longum JDM301. J. Bacteriol. 192:4076–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Wendisch VF, Bott M, Eikmanns BJ. 2006. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr. Opin. Microbiol. 9:268–274 [DOI] [PubMed] [Google Scholar]
- 328. Wiens GD, et al. 2008. Genome sequence of the fish pathogen Renibacterium salmoninarum suggests reductive evolution away from an environmental Arthrobacter ancestor. J. Bacteriol. 190:6970–6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Woese CR. 1987. Bacterial evolution. Microbiol. Rev. 51:221–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Woese CR. 1992. Prokaryote systematics: the evolution of a science, p 3–18 In Balows A, Tr̈uper HG, Dworkin M, Harder W, Schleifer KH. (ed), The prokaryotes. Springer-Verlag, New York, NY [Google Scholar]
- 331. Woese CR. 2003. How we do, don't and should look at bacteria and bacteriology, p 3–23 In Dworkin M, et al. (ed), The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag, New York, NY [Google Scholar]
- 332. Wright GD. 2007. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5:175–186 [DOI] [PubMed] [Google Scholar]
- 333. Wu D, et al. 2009. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462:1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Xu Z, Hao B. 2009. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 37:W174–W178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Yang S, Doolittle RF, Bourne PE. 2005. Phylogeny determined by protein domain content. Proc. Natl. Acad. Sci. U. S. A. 102:373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336. Yasawong M, et al. 2010. Complete genome sequence of Arcanobacterium haemolyticum type strain (11018). Stand. Genomic Sci. 3:126–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Yeoman CJ, et al. 2010. Comparative genomics of Gardnerella vaginalis strains reveals substantial differences in metabolic and virulence potential. PLoS One 5:e12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Yip MJ, et al. 2007. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J. Bacteriol. 189:2021–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Yukawa H, et al. 2007. Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153:1042–1058 [DOI] [PubMed] [Google Scholar]
- 340. Zhang R, et al. 2003. Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle. Structure 11:31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341. Zhao W, et al. 2010. Complete genome sequence of the rifamycin SV-producing Amycolatopsis mediterranei U32 revealed its genetic characteristics in phylogeny and metabolism. Cell Res. 20:1096–1108 [DOI] [PubMed] [Google Scholar]
- 342. Zheng H, et al. 2008. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS One 3:e2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343. Zhi XY, Li WJ, Stackebrandt E. 2009. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int. J. Syst. Evol. Microbiol. 59:589–608 [DOI] [PubMed] [Google Scholar]
- 344. Zhurina D, et al. 2011. Complete genome sequence of Bifidobacterium bifidum S17. J. Bacteriol. 193:301–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.