Abstract
In 1977, Carl Woese and George Fox published a brief paper in PNAS that established, for the first time, that the overall phylogenetic structure of the living world is tripartite. We describe the way in which this monumental discovery was made, its context within the historical development of evolutionary thought, and how it has impacted our understanding of the emergence of life and the characterization of the evolutionary process in its most general form.
A fundamental breakthrough in biological science occurred in 1977, and most biologists did not notice. The paper by Woese and Fox (1) in 1977 was 2.5 pages in length and contained a single table of numbers that compared sequence snippets derived from small subunit rRNAs of different organisms. The table provided the first gene sequence-based quantitative assessment of phylogenetic (evolutionary) relationships between representatives of the major known kinds of organisms (1). The paper showed that all cellular life falls into one of three large relatedness groups: eukaryotes (our kind of cells, which contain a nuclear envelope), eubacteria [Woese and Fox (1) termed the group and this group is where classically studied bacteria fit], and archaebacteria [an unusual group of recently described organisms named by Woese and Fox (1) to distinguish the group from eubacteria]. In describing the phylogenetic relationships, the results also charted the first scientific view of deep evolutionary history. Both these fundamental aspects of biology, the phylogenetic structure of life and the course of early evolution, previously were only realms of speculation.
However, the methods and data used in the work by Woese and Fox (1) were unfamiliar to most biologists, even molecular biologists. Traditional biologists, students of plants and animals, paid little attention, because the results had little bearing on their interests. Because of a joint press release by the National Aeronautics and Space Administration and the National Science Foundation that supported Woese's research, the paper was heralded on the front page of The New York Times for discovery of “a third form of life” (2). However, the few biologists who noticed sometimes reacted negatively, and articles denouncing the claims were published. Subsequent developments showed that the methods and conclusions of the paper were sound.
Several aspects of the paper by Woese and Fox (1) sparked skepticism. One was the arcane nature of the molecular data, which few could appreciate. The reliance on a single gene to trace major trends in evolution was an equally alien concept. Some quibbled about the name archaebacteria; others objected to The New York Times publicity. Most importantly, however, the conclusions of the paper flew into the face of the common wisdom of the time regarding the basic divisions of biology and the nature of early evolution. It was generally believed—and still is taught in our textbooks—that life is of two kinds: on one hand, eukaryotes and on the other hand, prokaryotes, which lack nuclear membranes and as the name implies, were supposed to have preceded and evolved into eukaryotes. However, eubacteria and the newly discovered group of archaebacteria both lacked nuclear membranes. Eukaryotes seemed not derived from either bacteria or archaebacteria; all three kinds of organisms seemed to represent aboriginal lines of descent.
In this retrospective, we view the 1977 paper by Woese and Fox (1) from three standpoints. First, we discuss the specific accomplishments of this landmark paper and how the program of research initiated and led by Woese from the late 1960s to the present day has spawned a revolution in microbiology and other fields contingent on microbiology, including ecology and the health sciences. Second, we discuss the place of that paper in the history of evolutionary biology, where its unprecedented use of molecular sequences associated with rRNA provided the first window into the deep timeline of life, one independent of theoretical prejudices that had flawed earlier efforts to classify life. Finally, we describe how the understandings sparked by the paper are bringing a new face to the study of evolution by compelling biologists to address foundational issues related to the very concepts of species and organism and bringing to the fore the deep limitations of earlier accounts of the evolutionary process.
Lead Up to the Paper
The 1977 paper by Woese and Fox (1) was an early example of what we would today call molecular phylogenetics—the comparison of macromolecular sequences to infer genealogical and thereby, evolutionary relationships. The notion of comparing sequences to infer relationships was put forward in 1958 by Francis Crick (3) and more formally, by Emil Zuckerkandl and Linus Pauling in 1965 (4). This was a time when determination of protein sequences had become, to some extent, tractable with protocols developed by Fred Sanger (5), who received his first Nobel Prize for that development and the determination of the amino acid sequence of insulin in the 1950s (5). Protein biochemists began to develop phylogenetic relatedness maps, phylogenetic trees, based on amino acid sequences derived from various organisms, mainly animals. Russell Doolittle sketched out vertebrate evolution using blood-clotting fibrinopeptides in the work by Doolittle and Feng (6); the work by Fitch and Margoliash (7) used the mitochondrial protein cytochrome C to relate animals and some fungi. However, not all organisms possess cytochrome C, and for that reason alone, its amino acid sequence could not be used to infer the patterns of relationships among all of life.
Carl Woese came to the study of evolution from a background in biophysics and an interest in the genetic code and its origins. During the early 1960s, the nature of protein synthesis and the makeup of the genetic code were just being worked out (8). Woese was a contributor to early thought on the genetic code and had conducted experimental studies to try to understand the chemical basis of the canonical assignments of different amino acids to particular codons (the DNA or RNA base triplets that specify the amino acid sequence of a protein during protein synthesis) (9). His 1967 book The Genetic Code: The Molecular Basis for Genetic Expression focused prescient attention on the RNA elements of the protein synthesizing machinery (10). Woese, along with Francis Crick (11) and Leslie Orgel (12) after him, are considered founding champions of the idea that nucleic acids played more than template roles in the origin of biological systems, thus giving rise to the notion of a hypothetical prebiotic RNA world in which nucleic acids served as both catalytic entities and genetic templates (13). Woese was concerned that the emerging paradigm for the mechanism of protein synthesis was too static and had no evolutionary dimension. As early as 1969, as articulated in a letter to Francis Crick, he understood that the only way to reveal the essence of the process was to study its conservation and variation in different organisms—its evolution—in a phylogenetic framework. He set out to do that study by comparing rRNA sequences. This task was daunting at that time before the development of rapid sequencing protocols, but it was the only way to quantify evolutionary change.
All cells contain ribosomes, which carry out protein synthesis and typically are composed of ∼50% RNA and 50% protein. The ribosome consists of two subunits: a small subunit (SSU), which contains the 1,500- to 2,000-nt-long SSU rRNA (also called 16S or 18S rRNA to denote size), and a large subunit (LSU), which has two RNA molecules (a 120-nt 5S rRNA and a 3,000- to 5,000-nt LSU rRNA). [Most eukaryotes contain a fourth LSU rRNA (5.8S rRNA), but this RNA corresponds to one end of the bacterial, archaeal, and some eukaryotic LSU rRNAs.] Woese began with 5S rRNA, because its small size rendered it amenable to the sequencing technology of the time. He studied bacteria because of his background in working with Bacillus subtilis and the practical requirement to prepare highly radioactive RNA for sequence analysis.
RNA sequencing developed in the mid-1960s mainly through the efforts of Fred Sanger and his colleagues using the general approach used for Sanger's protein sequencing protocol (14). A 32P-labeled RNA was digested with base-specific RNases, and the sequences of the resulting oligonucleotides were determined by digestion with other nucleases. Next, fragments of incomplete digestion of the RNA were isolated and digested completely, and the digestion products were analyzed; eventually, the sequence could be inferred from the oligonucleotide contents of overlapping fragments. Mitchell Sogin, then a graduate student with Woese, learned the techniques from David H. L. Bishop, a postdoctoral student from Fred Sanger's laboratory who was then working in the Sol Spiegelman laboratory at the University of Illinois. Sogin set up the necessary facility for Woese's group. Woese and his students determined several bacterial 5S rRNA sequences. They showed that the rRNA sequences could be used as phylogenetic markers for bacteria (15). They also showed that evolutionary variation in sequences could be used to determine how the RNAs fold into secondary structure (so-called phylogenetic comparative RNA structure analysis) (16). However, it soon became clear that 5S rRNA, at only 120 nt in length, was too small in size and hence, information content to provide for accurate phylogenetic assessments.
The SSU rRNA, at 1,500–2,000 nt, was information-rich, but because of its relatively large size, it was practically impossible to determine the entire sequence using the Sanger method. Woese posited, however, that the full sequence of the RNA was not necessary for phylogenetic comparisons and that sufficient information was available in the collection of oligonucleotide fragments that result from specific nuclease digestions of SSU rRNA. Woese argued that any oligonucleotide six residues or longer had a low probability of random occurrence in a molecule the size of the SSU rRNA; therefore, they could be assumed to be homologous in different organisms and have common ancestry, and they could be used in phylogenetic assessments. He and his colleagues began to generate catalogs of sequences of oligonucleotides that resulted from digestion of the RNA with ribonuclease T1, which cleaves at G residues and therefore, produces oligonucleotides that are comprised of some collection of U, C, and A with a single G.
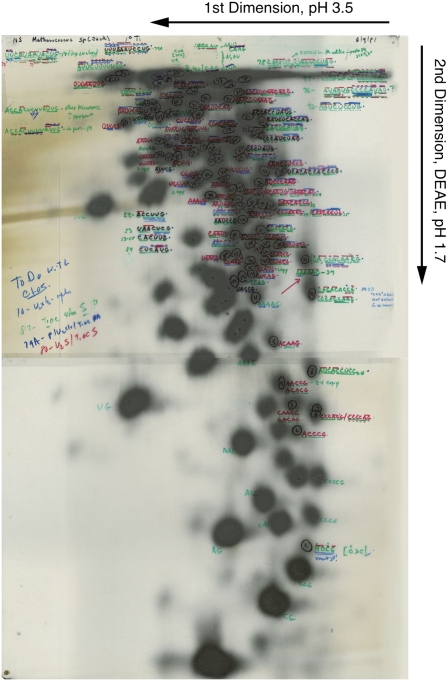
The resulting oligonucleotides were resolved by 2D electrophoresis, first on cellulose acetate at pH 3.5 and then on DEAE cellulose paper at pH 1.9 using Sanger's protocols. The autoradiogram shown in Fig. 1 is an example of such an RNase T1 fingerprint of a 32P-labeled SSU rRNA. The positions of the different oligonucleotides in the electropherogram reflect size, base composition, and sequence of the particular oligonucleotides. Longer oligonucleotides were excised from the paper for secondary and sometimes, tertiary digestions with other ribonucleases to determine the sequences. Preparation and analysis of any particular SSU rRNA were somewhat risky processes. They involved labeling cell cultures with many millicuries of 32P-orthophosphate, processing the highly labeled RNA, and conducting up to 5- to 8-kV electrophoresis in 100-L tanks filled with refined kerosene as a coolant. The analysis of RNA sequences in these ways probably could not be conducted today because of safety regulations.
Fig. 1.
Ribonuclease T1 oligonucleotide fingerprint. As outlined in the text, data reported in the paper by Woese and Fox (1) in 1977 consisted of catalogs of oligonucleotide sequences derived from RNase T1 digestion of small subunit (SSU) rRNAs. The first step of the analysis involved resolution of RNase T1 digests of 32P-labeled rRNA by 2D electrophoresis and locating labeled oligonucleotides on the 80 × 100-cm sheet of electrophoresis paper by autoradiography. The result, an RNase T1 fingerprint of the specific RNA, is shown; notations were made by Woese during the analysis of archaeal rRNAs. Larger phylogenetically informative oligonucleotides are in the upper one-half of the pattern.
Three Kinds of Life
George Fox joined Woese at the University of Illinois as a postdoctoral fellow. When the catalogs of SSU sequences proved informative, Fox worked with William Balch, then a graduate student in the laboratory of Ralph Wolfe at the University of Illinois, to prepare 32P-labeled SSU rRNAs from methanogens, organisms that produce methane (natural gas) as a metabolic product. Although environmentally important, methanogens were little studied because of their requirement for extremely anoxic conditions for growth, and as a result, there was no classification system of these organisms. Labeled RNAs went to technician Linda Magrum for fingerprinting and then to Woese for work up of oligonucleotide sequences and compilation of the catalogs. The SSU catalogs provided the first phylogeny and classification of those organisms, but more importantly, the methanogen oligonucleotide catalogs differed markedly from catalogs of any bacteria that had so far been examined (17).
Today, we would compare DNA or RNA sequences from different organisms explicitly in terms of percent identity, an intuitively meaningful comparison even to a nonspecialist. Comparison of oligonucleotide catalogs was not so straightforward, which possibly contributed to the lack of comprehension and resulting skepticism that greeted the paper by Woese and Fox (1). The single table in the work by Woese and Fox (1) was a matrix that compared association coefficients, SAB values, of different SSU oligonucleotide catalogs. (SAB values for catalogs of organisms A and B are calculated as SAB = 2NAB/(NA + NB), where NA and NB are the numbers of nucleotides in sequences of hexamers or larger in the catalogs of organisms A and B and NAB is the number in common to the two catalogs.) The higher the SAB, the more similar the sequences and the more closely related must be the organisms represented by the sequences. The single table in the paper by Woese and Fox (1), the matrix of SAB numbers, showed clearly that life—at least SSU rRNA sequences—fell into three distinct relatedness groups or urkingdoms as Woese and Fox termed them. Each urkingdom was further characterized by collections of signature oligonucleotides found only in that group and not the other groups. In addition to these idiosyncratic markers, there were universal signatures, oligonucleotide sequences found in all organisms examined. For the first time, it was actually shown that all life is related phylogenetically. This finding was a seminal finding not previously established but widely and implicitly assumed. However, more strikingly and wholly unexpectedly was their data indicating that there were at least three, not two, primary lineages of life.
The results of the paper showed that a large-scale map of life's diversity could be seen as three branches corresponding to eukaryotes, eubacteria, and archaebacteria. However, where was the root of the tree (assuming there was one)? Where was the origin of it all or some place in the tree that would correspond to a hypothetical last universal common ancestor? Were two of the urkingdoms more closely related to one another than to the third urkingdom, or did the three branches spring independently from some universal ancestor?
These questions could not be answered from the necessarily limited 1977 data and indeed, would not be settled for more than another decade when Iwabe et al. (18) and Gogarten et al. (19) used paralogous rooting, a phylogenetic technique developed by Margaret Dayhoff, a pioneer in bioinformatics (20), to establish that the origin was deep on the eubacterial line. The eukaryotes and archaebacteria seemed to constitute a sister group to the exclusion of bacteria. The result came as no surprise, because by this time (1989), the phylogenetic work of Woese's group had stimulated a large-scale effort, particularly in Germany, to determine the molecular biology and biochemistry of archaebacteria. Many aspects of the fundamental molecular biology of eukaryotes and archaebacteria proved to resemble each other more than their bacterial counterparts. For instance, during transcription, eukaryotes and archaebacteria were both known to use TATA binding proteins in promoter selection, whereas bacteria use σ-factors, a different basic mechanism (21); the DNA replication enzymologies of eukaryotes and archaebacteria resemble one another far more so than either resembles the eubacterial version (22). The biochemical and molecular criteria generally supported the rooting of the tree and a deep, at least partial relationship of eukaryotes and archaebacteria. The rudiments of a scientifically grounded universal tree of life were in place.
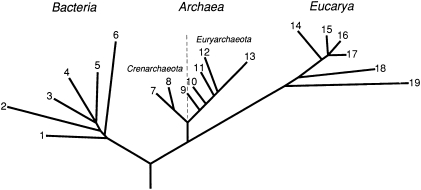
In 1990, Woese, with colleagues Otto Kandler and Mark Wheelis, proposed the formal designation domains to denote the three major phylogenetic groups, which they proposed be named Eucarya, Bacteria, and Archaea (formerly archaebacteria) (23). The diagrammatic phylogenetic tree that they used to support their classification is reproduced as Fig. 2. For the first time, a universal tree of life had been determined in a scientifically rigorous way. The field of molecular phylogeny is still a contentious one, but the large-scale organization shown in Fig. 2 is generally accepted.
Fig. 2.
Diagrammatic phylogenetic tree used to illustrate the proposal of three phylogenetic domains in 1990. Modified from Woese et al. (23).
Revolution in Microbiology
The development of a sequence-based phylogenetic framework for the identification of microbes would revolutionize microbial ecology (24) and more generally, bring together evolution and ecological studies under a single empirically based framework. It had long been appreciated that microbial ecology is the critical driver of the global biosphere. However, progress to achieving some understanding of environmental microbes had been hampered by the general necessity to culture microorganisms, even for identification. This need was a crippling constraint, because few environmental microbes, perhaps much less than 1%, can be cultured using standard methods (25). Consequently, the microbial ecologist had little access to environmental organisms and little knowledge of even the kinds of microbes that occur in the environment.
With the phylogenetic framework for classification in place and the subsequent development of recombinant DNA and sequencing technology, rRNA gene sequences began to be used to sidestep the requirement for culture to identify environmental organisms (26). rRNA genes could be isolated directly from environmental DNA as recombinant clones or the products of the PCR and sequenced to identify environmental organisms phylogenetically. Such sequences are incisive identifiers for microbes, and therefore, correlations between organisms and environments became possible. Moreover, the rRNA or other gene sequences also could be used for development of molecular tools, such as in situ hybridization probes, for the study of environmental organisms in their native environments. Surveys of rRNA gene sequences from different environments continue to expand knowledge of microbial life in all of the phylogenetic domains, and it is clear that, so far, we are only scratching the surface of a vast reservoir of microbial diversity. Currently, about 2 million rRNA sequences are held by the sequence databases, and more than 99% of these sequences are environmental sequences (27).
The 1977 paper (1) and its aftermath transformed microbiology by introducing a phylogenetic framework for exploring life's diversity and developing a universal tree of life, a natural system of classification based on genealogy. This phylogenetic framework spans most of evolutionary time on Earth as a microbial world, hitherto absent from classical evolutionary biology.
Evolution Without Microbes
The neo-Darwinian evolutionary synthesis that emerged in the 1930s and 1940s was a conception void of microbial foundation. It was formed in a world of two kingdoms: plants and animals. Microbiology largely remained a world apart from evolutionary biology until the development of new concepts and methods for determining the phylogenetic relationships based on universal characteristics at the core of the molecular genetic system of all organisms.
The concept of highly conserved characteristics, far removed from the vicissitudes of life, from which one could reconstruct the main course of evolution had been central to evolutionary biology from its beginnings. In Philosophie Zoologique, Jean Baptiste de Lamarck (28) argued that, to classify organisms correctly, one had to distinguish relatively trivial characteristics that were modified through the influence of environmental conditions from the essential system of organs. The former adaptive traits represented the branchings of the tree of life. The latter represented the course of increasing complexity of organization. Comparisons of the essential system of organs, he said, could be made only between the higher groupings of animals and not between species or genera; they were less conspicuous in plants and not conspicuous at all in the Infusoria (28).
The existence of highly preserved characteristics through which one could trace the tree of life was equally central to Charles Darwin's theorizing in regard to descent with modification. As Darwin wrote to his friend Thomas Henry Huxley in 1857, the “time will come I believe, though I shall not live to see it, when we shall have very fairly true genealogical trees of each great kingdom of nature” (29). Darwin (30) wrote in On the Origin of Species that all “true classification is genealogical; that community of descent is the hidden bond which naturalists have been unconsciously seeking, and not some unknown plan of creation, or … the mere putting together and separating objects more or less alike” (30).
For Darwin no less than for Lamarck, to have a natural classification that reflected the course of evolution, one needed to distinguish trivial characteristics from the “essential characteristics” or “organs of high vital or physiological importance” (30). As he explained in On the Origin of Species,
[i]t might have been thought … that those parts of the structure which determine the habits of life, and the general place of each being in the economy of nature, would be of very high importance in classification. Nothing can be more false … It may even be given as a general rule that the less any part of the organization is concerned with special habits, the more important it becomes for classification (30).
“Embryological characters,” he said, “are the most valuable of all” (30).
Comparative morphology, anatomy, embryology, and the fossil record were the ways to distinguish homology from analogy and thus, order organisms according to their evolutionary relatedness. However, bacteria lacked both morphological complexity and a fossil record. One could not tell which characteristics were ancient and of common ancestry, which characteristics were more recent adaptations, which characteristics were homologous, and which characteristics were analogous.
Microbiology's Scandal
In his great work Systema Naturae, Carl Linnaeus (31) had placed all Infusoria (microbes) in one species that he presciently baptized Chaos infusoria. Bacterial classification remained in chaos for the next 200 y. Bacteria were not put into groups based on principles of homology and evolution but based on as many characteristics as possible and principles of utility for industry and medicine. Bacteria were arranged into a nested hierarchy of orders, families, genera, and species based on cell shape, plane of cell division, ability to form spores and/or colonies, possession of flagella, whether cells were connected, whether they were branching, staining reactions, relation to temperature and oxygen, pigment production, pathogenicity, and a broad diversity of biochemical properties (32).
Although some microbiologists held out hope for a natural classification, one based on evolutionary relationships (33, 34), by the middle of the 20th century, they were forced to admit that their efforts were futile (35). Admitting defeat, Roger Stanier, Michael Doudoriff, and Edward Adelberg (36) declared in the first edition of their popular text The Microbial World that “it is a waste of time to attempt a natural system of classification for bacteria … bacteriologists should concentrate instead on the more humble practical task of devising determinative keys to provide the easiest possible identification of species and genera” (36).
Even distinguishing different microbes from one another was troublesome for microbiologists. For medical researchers, they were all simply germs as Joseph Lister called them in 1874; microbe was introduced 2 y later (37). Some thought they were little animals, and others thought that they were little plants. Louis Pasteur spoke sometimes of microscopic plants, sometimes as animalcules and sometimes as virus (the Latin word for poison). In the early 1870s, the term bacteria (Greek for little staff or rod) also began to be used for the smallest of germs: “for all those minute, rounded, ellipsoid, rod shaped, thread-like or spiral forms” (38, 39). Throughout most of the 20th century, bacteria were understood to be plants; their classification was the domain of botanists who referred to them as the fission fungi or Schizomycetes as Carl Nägeli had named them in 1857. This notion persists in our age today with common use of microflora to describe microbes. Breaking out of the plant–animal dualism would prove to be difficult.
In his speculative phylogenetic tree of 1866, Ernst Haeckel, who coined the terms ontogeny, phylogeny, and phylum, placed bacteria and blue-green algae (now cyanobacteria) in a division that he called Monera, and he christened another kingdom the Protista. The Monera were supposed to lack the fundamental division of labor of nucleus and cytoplasm exhibited in true cells and bridge the gap between the living and the nonliving. Whether the organization of the bacterium and the blue-green alga fit Haeckel's definition was hotly contested throughout the early 20th century (32, 40, 41). A few microbiologists proposed that bacteria and the blue-green algae should be granted their own kingdom, Monera (34, 42). However, others doubted that the grouping was monophyletic. Additionally, it was far from clear that the blue-green algae really lacked a nucleus and how smaller bacteria such as Rickettsia and Chlamydia could be distinguished from viruses (36).
In a famed paper titled “The Concept of a Virus,” André Lwoff (43) drew what he considered to be a unequivocal distinction between a virus and a bacterium on the basis of EM and chemistry. The virus contained only one kind of nucleic acid, either RNA or DNA, enclosed in a coat of protein; it possessed few, if any, enzymes, and it did not reproduce by division like a cell. In 1962, Stanier and Van Niel (44) wrote a sister paper, “The Concept of a Bacterium”, that aimed to resolve issues about the anatomy of the bacterium once and for all. “Any good biologist,” they wrote, “finds it intellectually distressing to devote his life to the study of a group that cannot be readily and satisfactorily defined in biological terms; and the abiding intellectual scandal of bacteriology has been the absence of a clear concept of a bacterium” (44).
Borrowing terms from Lwoff's former mentor Edouard Chatton (45, 46), Stanier and Van Niel (44) distinguished prokaryotic cells (Greek for before karyon or nucleus) from eukaryotic cells (Greek for true nucleus). The latter divided by mitosis and possessed a membrane-bound nucleus, an intricate cytoskeleton, mitochondria, and in the case of plants cells, chloroplasts. Prokaryotic cells were smaller and lacked those structures. Stanier and Van Niel (44) wrote that the
principle distinguishing features of the procaryotic cell are: 1 absence of internal membranes which separate the resting nucleus from the cytoplasm, and isolate the enzymatic machinery of photosynthesis and of respiration in specific organelles; 2 nuclear division by fission, not by mitosis, a character possibly related to the presence of a single structure which carries all the genetic information of the cell; and 3 the presence of a cell wall which contains a specific mucopeptide as its strengthening element (44).
The prokaryote was, thus, defined largely negatively. Although they had forsaken a natural classification for bacteria, they asserted nonetheless that the prokaryote was a monophyletic group of common origin (44). “All these organisms share the distinctive structural properties associated with the procaryotic cell … and we can therefore safely infer a common origin for the whole group in the remote evolutionary past” (47). “In fact,” they wrote, “this basic divergence in cellular structure, which separates the bacteria and blue-green algae from all other cellular organisms, probably represents the greatest single evolutionary discontinuity to be found in the present-day world” (47). The kingdom Monera, thus, rose like a phoenix from the ashes. It was to be one of five kingdoms, along with animals, plants, fungi, and protists (microbial eukaryotes) (48, 49).
The prokaryote–eukaryote dichotomy for the description of life's diversity was quickly instated among the canons of biology. Born in a time when the search for a natural system for microorganisms had been abandoned, the prokaryote concept contained just as many untested assumptions as the old taxonomic conjectures: the prokaryote was a monophyletic grouping that preceded and gave rise to eukaryotes. Indeed, nothing was known of prokaryote or eukaryote origins; they could have arisen one or many times.
Clash
The paper by Woese and Fox (1), “Phylogenetic Structure of the Prokaryotic Domain: The Primary Kingdoms,” clashed with the doctrines and methods of classical microbiology in several ways. First and foremost was the prokaryote–eukaryote dualism. Phylogenetically, Woese and Fox (1) wrote, life is not “structured in a bipartite way along the lines of organizationally dissimilar prokaryote and eukaryote. Rather it is (at least) tripartite” (ref. 1, p. 5090). Second, the eukaryotic cell organelles did not originate solely in the gradual neo-Darwinian manner by gene mutation and selection but rather, saltationally through symbiosis as long had been suggested. The SSU rRNA data left “no doubt that the chloroplast is of specific eubacterial origin” (1). The case was not yet as certain for mitochondria. The nature of the engulfing species (the pure urcaryote) that would have lacked eukaryotic organelles was unknown.
The manuscript received severe criticisms when it was submitted to PNAS in the summer of 1977 (1). One reviewer recommended that it not be published on methodological grounds that their claim for a tripartite division of the microbial world was as unfounded as their claims in regard to symbiosis and the origin of eukaryotic organelles. For Woese and Fox, rRNA was a highly conserved, nonadaptive structure at the core of all organisms. Nested deep in the center of essential cellular functions, the SSU rRNA was, as Woese (50) later put it, “the ultimate molecular chronometer.” However, for critics, an rRNA oligonucleotide catalog was merely a trait like any other, and classification on its basis held no more validity than classifying birds, bats, and insects on the basis of their possession of wings. That one molecule or many could be used to discern phylogenetic relationships was more than many classical microbiologists could accept; the molecular approach was pronounced by some as doomed to failure (51).
The belief that it was impossible to know the phylogenetic relationships among microbes because of a lack of fossil record was well-entrenched in the minds of microbiologists. As Stanier et al. (52) wrote in The Microbial World in 1970,
[r]eflection and experience have shown, however, that the goal of a phylogenetic classification can seldom be realized. The course that evolution has actually followed can be ascertained only from direct historical evidence contained in the fossil record. This record is at best fragmentary and becomes almost completely illegible in Precambrian rocks more than 400 million years old (52).
However, this publication was well after Zuckerkandl and Pauling (4) had written “Molecules as Documents of Evolutionary History,” and Francis Crick (3) had written as far back as 1958 that the amino acid sequences of proteins “are the most delicate expression possible of the phenotype of an organism and vast amounts of evolutionary information may be hidden away within them” (3). There was, indeed, a strange disconnect between microbiology and molecular biology, one that was finally resolved through the SSU rRNA sequencing method reported in the paper by Woese and Fox (1) in 1977.
The SSU rRNA sequencing method had remarkable predictive success. Most of the higher taxa above the genus would have to be reclassified on the basis of SSU rRNA phylogenies. When that technology surprised microbiologists by predicting unexpected relationships, it was corroborated by other data. Nothing was more striking than the phenotypically diverse organisms that came to be included in the archaebacteria urkingdom by 1980 (domain archaea). It comprised methanogens, found in the guts of rumens, extreme halophiles known for rotting salted fish, and extreme thermoacidophiles that live in conditions that would cook other organisms (53). The group was found to have many highly conserved characteristics in common: their walls lacked murein peptidoglycan (the only positive characteristic of the prokaryotes), their tRNAs were unique, the lipids in their membranes were ether- and not ester-linked, their transcription enzymes were unlike the enzymes of the classic bacteria, and their viruses were unlike the viruses of the classic bacteria.
The SSU rRNA phylogenies also definitively resolved the venerable question of whether chloroplasts and mitochondria originated as symbionts (1, 16, 54). Conjectures in regard to the symbiotic origin of cell organelles had been discussed throughout the 20th century, but that question remained far outside the realm of empirical inquiry (55). As Wilson (56) commented in his famed book The Cell in Development and Heredity, “[t]o many, no doubt, such speculations [symbiotic origin of organelles] may appear too fantastic for present mention in polite biological society; nevertheless it is within the range of possibility that they may someday call for more serious consideration” (56).
Such speculation did, indeed, call for more serious consideration in the early 1960s with evidence that mitochondria and chloroplasts each possessed their own DNA and translation apparatus. That these organelles might have originated as symbionts was the conclusion of virtually every paper, showing that they possessed their own DNA (57–59). However, proof of origin was lacking, and the debates were sterile. As Woolhouse (60) remarked, “the time has come to bury this kind of speculation with, by way of an epitaph, a parody of Wittgenstein's well-known remark, ‘Whereof one cannot know, thereof one should not speak’” (60). Stanier (61) decreed that the problem of eukaryotic cell origins would always remain in the realm of metascience: “[i]t might have happened thus; but we shall surely never know with certainty,” Stanier (61) quipped.
Evolutionary speculation constitutes a kind of metascience, which has the same intellectual fascination for some biologists that metaphysical speculation possessed for some medieval scholastics. It can be considered a relatively harmless habit, like eating peanuts, unless it assumes the form of an obsession; then it becomes a vice (61).
SSU rRNA phylogenetics belied that statement. Molecular evolution revitalized evolutionary biology and provided a new basis for empirical investigations of profound new questions concerning the evolution of the cell and its components. Molecular phylogenetic methods and concepts constituted a paradigm apart from classical evolutionary biology. Molecular methods for classification based on GC content, DNA–RNA hybridization, and amino acid sequencing of proteins had begun to revitalize the aim for a phylogenetic classification of bacteria in the 1960s and 1970s. However, those approaches could not offer universal and quantitative methods for a universal tree of life.
Influence on Evolutionary Biology
The work by Woese and Fox (1) and its continuation within the Woese group have had an enormous impact (e.g., roughly one-quarter of the 40,000 total citations to Woese's work originate just from references) (1, 23, 50, 62), and its methods are revolutionizing disciplines as varied as marine science and the study of the human microbiome for medicine. This success has perhaps overshadowed the original motivations and may even have stimulated so much activity in genomics per se that Woese's overarching interests in fundamental evolutionary questions have been relatively neglected. We close out this retrospective with a brief account of how the work by Woese and Fox (1) began, the extent to which their work has influenced evolutionary thought, and the way in which their ideas are still unfolding.
Woese and Fox's discovery that all living systems are representatives of one of three “aboriginal lines of descent” did not emerge in a vacuum (1). It was a watershed event (but not the culmination) in an iconoclastic program of research that Woese had been pursuing since the mid-1960s, one with the goal of understanding the evolution of the complex structure of the modern cell from the origin of life (32). In this goal, he was arguably more ambitious than Darwin's statement of the problem, which was reflected in the title of Darwin's great work, On the Origin of Species (30). Indeed, Woese's concept of the evolution problem greatly surpassed prevailing views on what at that time passed for the deepest thinking on the question. Woese would not find it necessary to cite On the Origin of Species until 1992 (63), reflecting neither a stubbornness of character nor a contrived striving for originality. The fact of the matter is that Woese had not even read Darwin's original work until around 2000, because its relevance to Woese's program had seemed remote until that time.
So what was Woese's conception of the evolution problem? First and foremost, it explicitly drew a connection between the origin of the genetic code, the translation mechanism, and the emergence of cells and their organization. For Woese, speciation was an epiphenomenon of the evolutionary process; the more important question was that of degree of organization. Woese and Fox (1) wrote that “[e]volution seems to progress in a ‘quantized’ fashion. One level or domain of organization gives rise ultimately to a higher (more complex) one … Ideally one would like to know whether this is a frequent or a rare (unique) evolutionary event” (1). Even earlier, in 1972, Woese (64) had argued that
there are no such things as ‘special’ evolutionary problems. Evolution is not a mixed bag of various ‘historical accidents’ … All evolutionary problems appear to have important common aspects—whether they are intracellular problems or problems on higher levels of biological organization. Thus in ostensibly addressing ourselves to evolutions of genetic codes, translating mechanisms, etc., we are actually discovering and defining the basic principles for all evolution … The nature of the ‘elements’ involved changes from one example to another, but the patterning of events in time, the ‘principles of evolutionary construction,’ seem to remain invariant (64).
Woese understood that the goal of exploring evolution on the longest possible time scales was not to elucidate the idiosyncratic history of genes. “It is too easy to think of evolution solely in terms of gene mutations, flow in gene pools, and, of course, some vague ‘selection’ parameters. Too much emphasis is placed on the micro-changes at the expense of the macro-ordering” (64). He wanted to understand universal aspects of the process of evolution, not the particular sample path represented by the history of life on Earth, whose dynamics were imperfectly captured by the slogan natural selection. Woese and Fox (1), thus, approached their work in an open-ended way, one not constrained by theoretical prejudices about the particular dynamics of the evolutionary process. This strategy was brilliant; not only did they uncover an unanticipated new domain of life, but also, they pointed out that there were many possible modes and tempos of evolution, of which only one—long time-scale, core cellular mechanisms—could be properly probed by molecular phylogeny.
Woese and Fox's approach yielded a major surprise that could be interpreted directly and plainly without contamination from theory. The phylogenetic tree implied by table 1 in ref. 1 clearly showed three fundamental groupings, results that were subsequently confirmed and elaborated in compelling detail by Woese and his collaborators (63, 65). At a stroke, those data demolished the conception of the prokaryote as a monophyletic group that preceded and gave rise to the eukaryotic cell. The draft of the phylogenetic tree that became available during the 1990s provided evidence for a last universal common ancestor, which is summarized above.
Progenote
Woese's conception of the evolution question had already led him to surmise by at least 1971 that life, as we know it today, descended from an earlier, radically different protocell, which lacked the translation mechanisms of cells (64). His rationale for this conclusion is important to appreciate, because the paper by Woese and Fox (1) in 1977 bore directly on the properties of that hypothetical primitive entity.
Woese and Fox (66) had, in parallel to their experimental work on molecular phylogeny, published a conceptual paper that introduced the concept of “a primitive entity … called [the] progenote, to recognize the possibility that it had not yet completed evolving the link between genotype and phenotype” (66). The progenotes were hypothetical ancient life forms in the throes of developing the relationship between nucleic acid and protein. Woese had suggested the existence of the progenote from rather detailed arguments based on his unrivalled knowledge of the translation apparatus, which was articulated in his book The Genetic Code: The Molecular Basis for Genetic Expression in 1967 (10); he considered these arguments to indicate that a general principle of evolution was that complexity emerged through (i) a process of refinement and (ii) a two-step cyclical dynamic process that he proposed to explain the quaternary structure of proteins and that turned out to have remarkable predictive power (ref. 67, p. 388). These same arguments suggested a resolution of the old “chicken-and-egg” paradox of whether translation preceded the gene or vice versa. The resolution is that both emerged from what we would today call a co-evolutionary process of refinement of translation, in which the earliest proteins would have been statistical in character. In other words, early translation produced not a single protein but a family of similar proteins, any one of which was adequate for the task at hand. Thus, early life was a rickety, free-wheeling construct whose lack of complexity could tolerate imprecision at the level of amino acids. The complex modern translation apparatus would have evolved in a progenote evolutionary era, with a tempo and mode based on a high mutation rate of the primitive error-prone translation system and a pervasive horizontal exchange of genes within the population. During this era, there were no lineages as such.
Woese and Fox (1) noticed that their work already implied something fundamentally interesting about the progenote state. Table 1 in ref. 1 established that the three urkingdoms had to have been at least 3 billion y old each and therefore, that the
time available to form each phenotype (from their common ancestor) is then short by comparison … we think that this implies that the common ancestor … was not a prokaryote. It was a far simpler entity; it probably did not evolve at the ‘slow’ rate characteristic of prokaryotes (1).
In short, Woese and Fox's early data already suggested to them that life evolved from at least a two-step process: a late stage that they characterized as having a slow rate of evolution, one which could be measured using the slowly changing sequence of rRNA, and an earlier stage refractory to molecular phylogenetic analysis, where complexity evolved much more rapidly, leading to the emergence of genotype and phenotype as separate cellular features along with a fully functional translation mechanism.
Woese would return to the progenote speculations in the succeeding years, emphasizing especially the characteristics of high gene mutation and horizontal gene transfer (HGT) (68), but by 1998 (69), the context had become genomics and the ever-increasing evidence for the widespread occurrence of HGT. Its occurrence was interpreted by some as being antithetical to the entire program of building a tree of life—all which he had addressed so many years earlier.
Is There Really a Tree of Life?
HGT is the capability that organisms possess to transfer genes to a nongenealogically related recipient (70). Once thought to be an exclusive aspect of microbial life, HGT has now been documented to occur between bacteria and multicellular eukaryotes (71) as well as between eukaryotes (72). With genomic evidence for the widespread occurrence of lateral gene transfer, the notion of a universal phylogenetic tree came under severe scrutiny and criticism (73). However, among Bacteria and Archaea, the genes that are horizontally transferred are typically metabolic genes or genes that confer such traits as antibiotic resistance and not the nonadaptive informational genes that are involved in transcription and translation.
It is now widely recognized that there can be wide compositional variations in the genomes of organisms that are nominally classified together on the basis of their SSU rRNA phylogeny. That within-group variation reflects the presence of cosmopolitan genes associated more closely with a particular environment than with any one particular organism (74, 75). Woese's original motivation for constructing a deep universal phylogenetic tree based on SSU rRNA sequences to understand the evolution of the molecular genetic system remains largely untouched by HGT. Even those elements of the translational apparatus that are most susceptible to HGT (the aminoacyl-tRNA synthetases) show that the canonical pattern of the rRNA tree is largely preserved (76). Moreover, the frequency of HGT is known to depend on the evolutionary distance between the lineages concerned, and thus, even if there is significant HGT, the impact on the rRNA phylogenetic tree would be expected to be rather small: the tree can reflect both the evolutionary history of the lineage as well as the predominant patterns of gene transfer into and out of the lineage (77).
Molecular Phylogeny vs. Morphological Taxonomy
When Woese, Otto Kandler, and Mark Wheelis (23) formally proposed the three domains as the natural classification of taxa in 1990, a direct confrontation with the status quo ensued. The issues were well-revealed in Woese's debates with his most distinguished opponent, Ernst Mayr (78, 79). First, there was the question of the great molecular and biochemical diversity of the microbial world compared with the great morphological diversity exhibited by multicellular eukaryotes. For Mayr (78, 79), the degree of differences exhibited within the prokaryotic world simply could not compare with the morphological diversity so readily observed in the eukaryotic world, and the measurement of that difference using molecular sequences was anathema compared with the more descriptive phenotypic distinctions that he favored.
A second issue concerned the purpose of a classification system. Mayr (78, 79) privileged the facility of information retrieval over a system based on evolutionary relationships. In his view, the classification system should enable a biologist to quickly identify an organism with “a minimum of effort and loss of time” (79) rather than reflect the genealogy of the organism. In his response, Woese (80) broadened the discussion to include the foundational evolutionary questions that we have presented above and that, by that time, had occupied his thoughts for nearly three decades.
Interestingly enough, the Mayr–Woese debate did not address one of the most contentious issues arising from the paper by Woese and Fox (1): the definition of species in the microbial world. Mayr (78, 79) had famously defined the Biological Species Concept in 1942 as groups of interbreeding natural populations that are reproductively isolated from each other, but clearly, this concept did not apply to Bacteria, Archaea, or even microbial eukaryotes. With the advent of SSU rRNA phylogeny, it became common practice to define operational taxonomic units by the requirement of close sequence similarity (typically 97%) (24). However, as more and more sequencing capability became available, it became clear that the spectrum of microbial life was continuous rather than discrete because of cosmopolitan genes and HGT. Bacteria seem to form a radiation, with no specific species boundaries (27, 81). According to some estimates, only ∼40% of the genes in a particular bacterium typically occur in all of the genomes of that particular named species; the other ∼60% of genes in the typical bacterial genome occur only sporadically in other representatives of the species or not at all (81). More than 30 y after the paper by Woese and Fox (1) was published, the fundamental biological concept of species remains unresolved.
Legacy
Woese and Fox set out to determine the degrees of relatedness between all living organisms using rRNA sequences as a marker of cellular evolution on slow time scales subsequent to the emergence of the modern lines of descent. They discovered that there are three domains of life, not two domains as had been previously believed. Their work also strongly constrained the nature of life for times shorter than about 1 billion y, indicating that before the emergence of a strong phylogenetic signal of vertical descent, early life had to evolve rapidly; we now suspect that its evolution was reticulate in nature. Modern versions of the techniques used by Woese and Fox (1) are now routinely used to sample environments as varied as geothermal hot springs and gastrointestinal microbiomes, providing unprecedented insight into community structure and dynamics. The results challenged the foundations of classical evolutionary theory, requiring new modes of evolution to be considered, indicating the presence of an unexpectedly large microbial pangenome (“a field of genes” to use Woese's favorite phrase) (1), and forcing us to reconsider basic concepts such as the nature of species. Perhaps no other paper in evolutionary biology has left a richer legacy of accomplishments and promise for the future.
Acknowledgments
We thank Professors Carl Woese and George Fox for reviewing the manuscript for accuracy. N.R.P. is supported by the Alfred P. Sloan Foundation, J.S. is supported by the Social Sciences and Humanities Research Council of Canada, and N.G. is supported by National Science Foundation Grant EF-0526747.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Classic Article “Phylogenetic structure of the prokaryotic domain: The primary kingdoms” on page 5088 in issue 11 of volume 74.
See Classic Profile on page 1019.
References
- 1.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyons RD. NY Times. November 3, 1977. Scientists discover a form of life that predates higher organisms. Section A, p 1. [Google Scholar]
- 3.Crick FHC. The biological replication of macromolecules. Symp Soc Exp Biol. 1958;12:138–163. [PubMed] [Google Scholar]
- 4.Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965;8:357–366. doi: 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]
- 5.Sanger F. Chemistry of insulin; determination of the structure of insulin opens the way to greater understanding of life processes. Science. 1959;129:1340–1344. doi: 10.1126/science.129.3359.1340. [DOI] [PubMed] [Google Scholar]
- 6.Doolittle RF, Feng DF. Reconstructing the evolution of vertebrate blood coagulation from a consideration of the amino acid sequences of clotting proteins. Cold Spring Harb Symp Quant Biol. 1987;52:869–874. doi: 10.1101/sqb.1987.052.01.095. [DOI] [PubMed] [Google Scholar]
- 7.Fitch WM, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 8.Judson HF. The Eighth Day of Creation: Makers of the Revolution in Biology: Expanded Edition. Plainview, NY: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 9.Woese CR, Dugre DH, Saxinger WC, Dugre SA. The molecular basis for the genetic code. Proc Natl Acad Sci USA. 1966;55:966–974. doi: 10.1073/pnas.55.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woese CR. The Genetic Code: The Molecular Basis for Genetic Expression. New York: Harper and Row; 1967. [Google Scholar]
- 11.Crick FHC. The origin of the genetic code. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 12.Orgel LE. Evolution of the genetic apparatus. J Mol Biol. 1968;38:381–393. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert W. Origin of life: The RNA world. Nature. 1986;319:618. [Google Scholar]
- 14.Sanger F, Brownlee GG, Barrell BG. A two-dimensional fractionation procedure for radioactive nucleotides. J Mol Biol. 1965;13:373–398. doi: 10.1016/s0022-2836(65)80104-8. [DOI] [PubMed] [Google Scholar]
- 15.Rogers MJ, et al. Construction of the mycoplasma evolutionary tree from 5S rRNA sequence data. Proc Natl Acad Sci USA. 1985;82:1160–1164. doi: 10.1073/pnas.82.4.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox GW, Woese CR. 5S RNA secondary structure. Nature. 1975;256:505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- 17.Fox GE, Magrum LJ, Balch WE, Wolfe RS, Woese CR. Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc Natl Acad Sci USA. 1977;74:4537–4541. doi: 10.1073/pnas.74.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwabe N, Kuma KI, Hasegawa M, Osawa S, Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci USA. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gogarten JP, et al. Evolution of the vacuolar H+-ATPase: Implications for the origin of eukaryotes. Proc Natl Acad Sci USA. 1989;86:6661–6665. doi: 10.1073/pnas.86.17.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz RM, Dayhoff MO. Origins of prokaryotes, eukaryotes, mitochondria, and chloroplasts. Science. 1978;199:395–403. doi: 10.1126/science.202030. [DOI] [PubMed] [Google Scholar]
- 21.Thomm M. Transcription: Mechanism and regulation. In: Cavicchioli R, editor. Archaea: Molecular and Cellular Biology. New York: Wiley-Blackwell; 2007. pp. 139–157. [Google Scholar]
- 22.Lao-Sirieix S-B, Marsh VL, Bell SD. DNA replication and cell cycle. In: Cavicchioli R, editor. Archaea: Molecular and Cellular Biology. New York: Wiley-Blackwell; 2007. pp. 93–109. [Google Scholar]
- 23.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 25.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA. Microbial ecology and evolution: A ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 27.Pace NR. Mapping the tree of life: Progress and prospects. Microbiol Mol Biol Rev. 2009;73:565–576. doi: 10.1128/MMBR.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Lamarck JBM. In: Zoological Philosophy: An Exposition with Regard to the Natural History of Animals. Elliot H, translator. Chicago: University of Chicago Press; 1984. [Google Scholar]
- 29.Darwin C. Darwin to Thomas Henry Huxley: Darwin Correspondence Project. 1857. Letter 2143. Available at http://www.darwinproject.ac.uk/entry-2143.
- 30.Darwin C. On the Origin of Species: Facsimile Edition. Cambridge, MA: Harvard University Press; 1969. [Google Scholar]
- 31.Linnaeus C. Systema Naturae. 10th Ed. Stockholm: Laurentius Salvius; 1774. [Google Scholar]
- 32.Sapp J. The New Foundations of Evolution: On the Tree of Life. New York: Oxford University Press; 2009. [Google Scholar]
- 33.Kluyver AJ, Van Niel CB. Prospects for a natural system of classification of bacteria. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt II. 1936;94:369–403. [Google Scholar]
- 34.Stanier RY, Van Niel CB. The main outlines of bacterial classification. J Bacteriol. 1941;42:437–466. doi: 10.1128/jb.42.4.437-466.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Niel CB. Natural selection in the microbial world. J Gen Microbiol. 1955;13:201–217. doi: 10.1099/00221287-13-2-201. [DOI] [PubMed] [Google Scholar]
- 36.Stanier RY, Doudoriff M, Adelberg EA. The Microbial World. Englewood Cliffs, NJ: Prentice Hall; 1957. [Google Scholar]
- 37.Carter KC. The development of Pasteur's concept of disease causation and the emergence of specific causes in nineteenth-century medicine. Bull Hist Med. 1991;65:528–548. [PubMed] [Google Scholar]
- 38.Cohn F. Uber die Bacterien, die Kleinstein Lebenden Wesen. Samsung Gemeinverständlicher Wissenschaftlicher Vorträge. 1872;7:1–35. [Google Scholar]
- 39.Woodhead GS. Bacteria and Their Products. London: Walter Scott; 1892. [Google Scholar]
- 40.Sapp J. Transcending Darwinism thinking laterally on the tree of life. Hist Philos Life Sci. 2009;31:161–181. [PubMed] [Google Scholar]
- 41.Sapp J. The prokaryote-eukaryote dichotomy: Meanings and mythology. Microbiol Mol Biol Rev. 2005;69:292–305. doi: 10.1128/MMBR.69.2.292-305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Copeland HF. The kingdoms of organisms. Q Rev Biol. 1938;13:383–420. [Google Scholar]
- 43.Lwoff A. The concept of virus. J Gen Microbiol. 1957;17:239–253. doi: 10.1099/00221287-17-2-239. [DOI] [PubMed] [Google Scholar]
- 44.Stanier RY, Van Niel CB. The concept of a bacterium. Arch Mikrobiol. 1962;42:17–35. doi: 10.1007/BF00425185. [DOI] [PubMed] [Google Scholar]
- 45.Chatton E. Titres et travaux scientifiques (1906–1937) Italy: Sottano; 1938. [Google Scholar]
- 46.Chatton E. Pansporella perplexa, Amoebien á spores protégées parasite des Daphnies. Reflexions sur la biologie et la phylogenie des Protozoairies. Ann Sci Nat (Zool) 1925;8:5–84. [Google Scholar]
- 47.Stanier RY, Doudoroff M, Adelberg EA. The Microbial World. 2nd Ed. Englewood Cliffs, NJ: Prentice Hall; 1963. [Google Scholar]
- 48.Whittaker RH. New concepts of kingdoms or organisms. Evolutionary relations are better represented by new classifications than by the traditional two kingdoms. Science. 1969;163:150–160. doi: 10.1126/science.163.3863.150. [DOI] [PubMed] [Google Scholar]
- 49.Whittaker RH, Margulis L. Protist classification and the kingdoms of organisms. Biosystems. 1978;10:3–18. doi: 10.1016/0303-2647(78)90023-0. [DOI] [PubMed] [Google Scholar]
- 50.Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Margulis L, Guerrero R. Kingdoms in turmoil. New Sci. 1991;1761:46–50. [PubMed] [Google Scholar]
- 52.Stanier RY, Doudoroff M, Adelberg EA. The Microbial World. 3rd Ed. Englewood Cliffs, NJ: Prentice Hall; 1970. [Google Scholar]
- 53.Fox GE, et al. The phylogeny of prokaryotes. Science. 1980;209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 54.Gray MW, Doolittle WF. Has the endosymbiont hypothesis been proven? Microbiol Rev. 1982;46:1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sapp J. Evolution by Association: A History of Symbiosis. New York: Oxford University Press; 1994. [Google Scholar]
- 56.Wilson EB. The Cell in Development and Heredity. New York: Macmillan; 1925. [Google Scholar]
- 57.Ris H, Plaut W. Ultrastructure of DNA-containing areas in the chloroplast of Chlamydomonas. J Cell Biol. 1962;13:383–391. doi: 10.1083/jcb.13.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nass MMK, Nass S. Intramitochondrial fibers with DNA characteristics. I. Fixation and electron staining reactions. J Cell Biol. 1963;19:593–611. doi: 10.1083/jcb.19.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nass S. The significance of the structural and functional similarities of bacteria and mitochondria. Int Rev Cytol. 1969;25:55–129. doi: 10.1016/s0074-7696(08)60201-6. [DOI] [PubMed] [Google Scholar]
- 60.Woolhouse WH. A review of the plastids by JTO Kirk and RAE Tilney-Bassett. New Phytol. 1967;66:832–833. [Google Scholar]
- 61.Stanier RY. Some aspects of the biology of cells and their possible evolutionary significance. In: Charles HP, Knight BCJG, editors. Organization and Control in Prokaryotic and Eukaryotic Cells. Vol 20. Cambridge, UK: Cambridge University Press; 1970. pp. 1–38. [Google Scholar]
- 62.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: Reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wheelis ML, Kandler O, Woese CR. On the nature of global classification. Proc Natl Acad Sci USA. 1992;89:2930–2934. doi: 10.1073/pnas.89.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Woese CR. The emergence of genetic organization. In: Ponnamperuma C, editor. Exobiology. Amsterdam: North-Holland; 1972. pp. 301–341. [Google Scholar]
- 65.Cavicchioli R. Archaea: Molecular and Cellular Biology. New York: Wiley-Blackwell; 2007. [Google Scholar]
- 66.Woese CR, Fox GE. The concept of cellular evolution. J Mol Evol. 1977;10:1–6. doi: 10.1007/BF01796132. [DOI] [PubMed] [Google Scholar]
- 67.Goldenfeld N, Woese C. Life is physics: Evolution as a collective phenomenon far from equilibrium. Ann Rev Condensed Matter Physics. 2011;2:375–399. [Google Scholar]
- 68.Woese CR. Archaebacteria and cellular origins: An overview. Zbl Bakt Hyg I Abt Orig C. 1982;3:1–17. [Google Scholar]
- 69.Woese C. The universal ancestor. Proc Natl Acad Sci USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chia N, Goldenfeld N. Statistical mechanics of horizontal gene transfer in evolutionary ecology. J Stat Phys. 2011;142:1287–1301. [Google Scholar]
- 71.Dunning Hotopp JC. Horizontal gene transfer between bacteria and animals. Trends Genet. 2011;27:157–163. doi: 10.1016/j.tig.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- 73.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 74.Woese CR. A new biology for a new century. Microbiol Mol Biol Rev. 2004;68:173–186. doi: 10.1128/MMBR.68.2.173-186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frigaard NU, Martinez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- 76.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andam CP, Williams D, Gogarten JP. Natural taxonomy in light of horizontal gene transfer. Biol Philos. 2010;25:589–602. [Google Scholar]
- 78.Mayr E. A natural system of organisms. Nature. 1990;348:491. [Google Scholar]
- 79.Mayr E. Two empires or three? Proc Natl Acad Sci USA. 1998;95:9720–9723. doi: 10.1073/pnas.95.17.9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Woese CR. Default taxonomy: Ernst Mayr's view of the microbial world. Proc Natl Acad Sci USA. 1998;95:11043–11046. doi: 10.1073/pnas.95.19.11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. The bacterial species challenge: Making sense of genetic and ecological diversity. Science. 2009;323:741–746. doi: 10.1126/science.1159388. [DOI] [PubMed] [Google Scholar]