Abstract
A plant's best strategy for acquiring resources may often depend on the identity of neighbours. Here, I ask whether plants adjust their strategy to local relatedness: individuals may cooperate (reduce competitiveness) with kin but compete relatively intensely with non-kin. In a greenhouse experiment with Ipomoea hederacea, neighbouring siblings from the same inbred line were relatively uniform in height; groups of mixed lines, however, were increasingly variable as their mean height increased. The reproductive yield of mixed and sibling groups was similar overall, but when adjusted to a common mean height and height inequality, the yield of mixed groups was significantly less. Where this difference in yield was most pronounced (among groups that varied most in height), mixed groups tended to allocate more mass to roots than comparable sibling groups, and overall, mixed groups produced significantly fewer seeds per unit mass of roots. These results suggest that, from the group perspective, non-kin may have wasted resources in below-ground competition at the expense of reproduction; kin groups, on the other hand, displayed the relative efficiency that is expected of reduced competitiveness.
Keywords: Ipomoea, kin recognition, kin selection, root growth, size inequality, tragedy of the commons
1. Introduction
Most plants live in social groups, where neighbours interact in competition for above- and below-ground resources. Furthermore, because of limited dispersal, interacting plants are often close genealogical relatives [1]. The effect of relatedness on the outcome of intraspecific competition could inform two important perspectives in evolutionary ecology. One view is that a group of related (and thus phenotypically correlated) plants will inevitably suffer more competition and a lower reproductive yield than a more diverse group that is able to partition resources (e.g. [2]). This idea plays a key role in hypotheses for the evolution of sex [3,4] and outcrossing [5] and has received some support in plant ecology (e.g. [6–8] and references therein).
However, a plant's access to resources might also depend on its relative allocation to costly competitive traits, and from this perspective, local relatedness can have the opposite effect on group yield. When relatedness is low, self-interested individuals can outcompete their neighbours by taking a larger share of resources, often leading to an escalation of selfishness at the expense of group productivity (a ‘tragedy of the commons’; reviewed by Rankin et al. [9]). When relatedness is high, on the other hand, individuals share an interest in the group's productivity. In this case, cooperation (reduced selfishness) can be favoured because cooperators save the cost of competitive traits and also gain indirectly, by promoting the reproduction of relatives ([10]; see also [11]). Current examples of the ‘tragic’ nature of plant competition involve escalated allocation to roots and a correlated decline in reproduction among non-self competitors (e.g. [12–14]). Allocation to plant height in competition over light may be similarly costly (reviewed by Schmitt [15]), and in fact, artificial selection to minimize height in agricultural crops clearly demonstrates a collective benefit of reduced selfishness [16,17]. Similar examples of naturally selected cooperation among plants remain to be discovered.
Cooperation might be detected experimentally if plants have the ability to adjust their competitiveness to the relatedness of neighbours. At one extreme, individual organs should detect and avoid competition with other parts of the same individual, a type of self-recognition and coordination that appears common (reviewed by Novoplansky [18]). More uncertain is whether plants can recognize and adaptively coordinate their behaviour with physiologically separate kin. Some studies report higher reproductive yield in kin versus non-kin groups (e.g. [19–21]) without explicitly considering plasticity in resource allocation. Others find plasticity in below- and above-ground traits to the relatedness of neighbours (e.g. [22,23]) without measuring reproductive consequences.
Here, I study the effects of manipulating relatedness and light competition on root growth, plant height and lifetime reproductive yield in groups of the ivyleaf morning glory (Ipomoea hederacea). Morning glory, a climbing vine with gravity-dispersed seeds, is appropriate for a study of plasticity in competitive behaviour because its evolutionary history probably includes competition with siblings (when maternal plants grow vertically) and non-siblings (when maternal plants grow laterally). The resource-partitioning hypothesis may or may not involve plasticity in allocation, but it predicts that non-sibling groups will have higher yield than sibling groups. The recognition–cooperation hypothesis, on the other hand, predicts that: (i) the mean allocation to height and/or root growth in non-sibling groups will be greater than in sibling groups and (ii) as a consequence, sibling groups will have a higher yield than non-siblings. Because plant yield typically depends on measures of plant size [24–26], I also examine the reproductive allometry of groups, testing whether yield with respect to mean plant height (herein ‘reproductive allocation’) depends on relatedness. I focus on the yield of groups (specifically, the mean yield of plants in a group) because similar analyses at the level of individual plants become difficult to interpret if genotypes (families) differ in size and competitive ability (cf. [27]). For example, individuals from a competitively dominant genotype may yield the most seeds when dominating non-kin groups, but the yield of non-kin groups as a whole could nevertheless suffer (relative to kin groups) because of escalated allocation to competitive traits. In fact, the extent of size inequality within groups may act as an index of competitive dominance [28], which could also predict group yield [29]. I therefore use size inequality to refine the allometric approach described above: I test for an effect of relatedness on mean yield as a function of both the mean and the variability in the height of neighbouring plants.
2. Methods
(a). Greenhouse experiment
The experiment was performed at the University of Toronto, where plants were grown from seed in September of 2008 until January of 2009. All I. hederacea seeds used in the experiment were the progeny of one of three inbred lines, herein referred to as black, blue and red. The lines were originally collected from adjacent populations in North Carolina, USA, and then self-fertilized and bred by single-seed descent for three generations in a common greenhouse environment (by J. R. Stinchcombe, University of Toronto). It follows that seeds from the same inbred line are siblings with a high level of genetic similarity. Seeds were sown at constant density in pots of either three siblings or three non-siblings (one seed from each of the three lines; herein ‘mixed’ pots). All pots were the same size (12.7 cm circumference) and contained the same soil medium (80% Promix BX, 20% Profile ceramic conditioner and Osmocote fertilizer at 4 ml l−1).
On each of seven tables in the greenhouse, 15 pots were uniformly arranged (at least 18 cm apart) and assigned to a relatedness treatment in a randomized block design (each table was a block). Pots were assigned so that on every table, each of the sibling pot types (black, blue and red) appeared at least twice. The second factor, stake ownership, was assigned in an alternating fashion: every second pot location on a table was assigned to the ‘shared-stake’ condition, and directly adjacent locations were assigned to the ‘own-stake’ condition. Shared-stake groups had a single wooden stake fixed next to the pot on which all three plants entwined; own-stake groups had three stakes, one for each plant, spaced 16 cm apart (stakes were outside of pots so that root interactions were left undisturbed). The purpose of this treatment was to give sharing plants a stronger signal of light competition (via shading) than plants with their own stake (after Weiner [30]).
Throughout the growing period, lighting and day length depended on the ambient conditions, although supplemental lighting (5 h mid-day) was provided during germination and fruiting stages. Temperature was held as close to room temperature as possible, and all pots were watered similarly, as a group. By the end of the experiment (accounting for the loss of some pots, missing at random), there were 52 mixed groups (24 low and 28 high shoot competition) and 48 sibling groups (24 low and 24 high shoot competition).
(b). Data collection and analysis
Plants flowered and autonomously self-fertilized in the greenhouse, and fruits were later collected (and seed yield counted) as the plants senesced. Each vine was then clipped at the soil level, and its (uncoiled) length was measured. In cases where plants had multiple vines, the sum of all vine lengths was used to measure the total plant height. I refer to this total as ‘plant height’.
Roots of the three plants per pot could not be untangled, but the combined root mass was measured as an index of the group's below-ground biomass allocation. Because of time limitations, I measured roots from a particular subset of pots: 21 mixed pots were paired with a sibling pot (10 blue, 11 red) that produced the most similar mean plant height (and pots were paired only if the match was within 5 cm). This sampling design was meant to match groups that were in similar overall condition. Pairs were always treated similarly during the harvesting process: they were washed of all soil at the same time and by the same observer, later dried together in a forced convection oven (30°C for 120 h) and then weighed. Although relatedness treatments were not randomized within each pair, I treat each pair as a block because they form natural groupings of experimental units.
I used analysis of variance to test for effects of relatedness and stake ownership on the mean height, height inequality, and mean yield of plants in a group. To measure the degree of height inequality, I used the coefficient of variation (CV; s.d./mean), which is commonly used for this purpose (e.g. [31,32]). When testing for treatment effects on the CV in height, I used the mean height of plants in a group as a covariate, and all models also included the blocking factor ‘table’. Residual analyses indicated that, for these data, linear models with a normally distributed error provided a satisfactory fit, and no transformations were necessary.
Allometric analyses of seed yield were performed at the level of individual plants (details in the electronic supplementary material) and, in the main text, at the level of plant groups (pots). The mean yield of plants in a group was modelled as a function of ‘table’ (block), relatedness, mean plant height and the CV in height (covariates), and both relatedness-by-covariate interactions. A scatterplot of mean seed yield against mean plant height showed a linear relationship but increasing variability as the number of seeds increased (indicating Poisson data). For all allometric analyses, I therefore used generalized linear models with a Poisson error distribution and an identity link function, fit by maximum likelihood. Inspection of residuals confirmed that such models produced a satisfactory fit to the data.
Finally, to test for an effect of relatedness on groups' allocation to roots, I estimated the mean difference in root mass among the matched pairs of sibling and mixed groups. A t-test was used to determine whether the mean difference was significantly different from zero, and I used multiple regression to test whether the size of the difference in root mass varied with the mean plant height and/or mean height inequality of pairs. I also tested for a difference in the seed yield per unit of root mass between matched sibling and mixed groups, in this case, using a paired t-test. Yield per unit of root mass was square-root transformed to stabilize variation in the distribution of residuals.
All statistical analyses in the main text were performed with JMP 8 (SAS Institute). Throughout the results, any significant treatment-by-covariate interactions were further studied with separate regression analyses at each treatment level and presented as either the predicted values from these whole models or as partial regressions on each predictor variable. Where partial regressions are plotted, I use leverage plots, which show the independent effect of a given predictor variable while holding all others fixed. Parameter estimates are reported as: estimate (lower 95% confidence value, upper value).
3. Results
(a). Plant height and height inequality
If related plants limited allocation to growing tall in competition for light, then sibling groups in the shared-stake condition may have had: (i) shorter average height than mixed groups on shared stakes and (ii) similar average height to groups in the own-stake condition. These predictions translate to an interaction between relatedness and stake ownership for measures of plant height. However, there was no evidence for such an interaction on the height of the tallest vine in a pot, and, overall, stake ownership had no detectable effects on plant height (table 1). It remains possible that treatment groups differed in stem elongation (i.e. plants of the same height may have differed in stem mass), but measures of above-ground biomass would be needed to test for these effects.
Table 1.
Results from models of above-ground growth responses to stake ownership and relatedness treatments. All models also include ‘table’ as a blocking factor (not shown). Effect sizes (with 95% CI) give the mean difference in least-squares means between treatment levels (estimated by a Student's t-test), or, for the covariate in c, a linear regression coefficient. (Notes: Total sample size was 100 groups. Stake ownership had no significant effect in c and was removed for simplicity. The interaction from c is plotted in figure 1. Statistically significant effects are in bold.)
| response variable | explanatory variables | F1,90 | p-value | effect size |
|---|---|---|---|---|
| (a) height of tallest vine | ||||
| stake ownership | 0.22 | 0.64 | own > shared; 2.5 cm (−8.1, 13.0) | |
| relatedness | 4.0 | 0.049 | mix > sibs; 10.6 cm (0.07, 21.2) | |
| stake ownership × relatedness | 0.039 | 0.84 | ||
| (b) mean height | ||||
| stake ownership | 0.64 | 0.43 | shared > own; 3.8 cm (−5.7, 13.3) | |
| relatedness | 1.7 | 0.19 | mix > sibs; 6.2 cm (−3.2, 15.7) | |
| stake ownership × relatedness | 0.0048 | 0.95 | ||
| (c) height inequality (CV) | ||||
| relatedness | 40.9 | <0.0001 | mix > sibs; 0.18 (0.12, 0.23) | |
| mean height | 10.2 | 0.0019 | 0.002 (0.0007, 0.003) | |
| mean height × relatedness | 10.6 | 0.0016 |
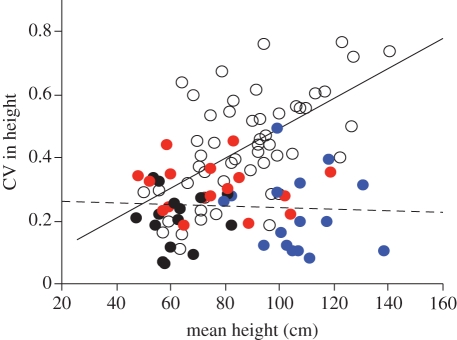
There was a large and unexpected difference among genotypes in the mean height of plants in sibling groups (least-squares (LS) means, after accounting for ‘table’: blue: 106.8 cm (92.8, 120.9); red: 75.2 cm (61.3, 89.2); black: 62.3 cm (48.1, 76.5)). These differences largely predicted plant height in mixtures, where a plant from the blue genotype was tallest in all but four cases. Overall, relatedness had no significant effect on the mean height of plants in a group (table 1b), but the LS mean height inequality (measured as the CV) was significantly greater in mixed groups (table 1c). In particular, the CV in height increased with mean height in mixed (but not sibling) groups (figure 1); hence, although the tallest mixed and sibling groups grew to similar mean heights, mixed groups were usually dominated by an individual from the blue genotype.
Figure 1.
Leverage plots showing the interacting effects of relatedness and mean height on the height inequality (coefficient of variation, CV) of plants in a group (corresponding to the model in table 1c). Filled circles are sibling groups (where colour corresponds to genotype), and open circles are mixed groups. Height inequality increased significantly in mixed groups (solid line; p < 0.0001; estimated slope: 0.0047 (0.003, 0.007)) but not in sibling groups (dashed line; p = 0.69; estimated slope: −0.0003 (−0.0002, 0.01)).
(b). Reproductive yield and allometry
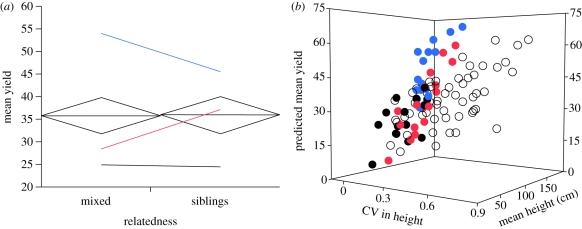
There was no evidence for effects of relatedness, stake ownership or their interaction on the mean yield of groups (all p-values >0.8). Figure 2a gives the overall LS mean yields of sibling and mixed groups from this analysis and also indicates the mean yield of individual plants from each relatedness–genotype combination. These data at the genotype level summarize the full allometric analysis of individual plant yield given in the electronic supplementary material.
Figure 2.
The average yield of mixed and sibling groups as overall means (horizontal lines in (a)) and as a function of mean height and the height inequality (CV) of plants in a group ((b), corresponding to the model in table 2a). In (a), the vertical span of each diamond is a 95% CI, and coloured lines (one for each of the three genotypes) illustrate the average change in yield at the level of individual plants. In (b), predicted values come from separate multiple regression models (one for siblings, one for mixed groups) that included ‘table’ (block), CV in height and mean height. The points shown (sibling groups as filled circles, mixed groups as open circles) fall upon the least-squares regression plane predicted by the corresponding model. Partial regression coefficients and associated p-values: sibling groups: 19.0 (p = 0.021) for CV in height, 0.50 (p < 0.0001) for mean height; mixed groups: −2.8 (p = 0.64) for CV in height, 0.36 (p < 0.0001) for mean height.
In contrast to the overall means, sibling groups had significantly higher LS mean yield than mixed groups when considering yield as a function of mean height and the CV in height (where LS means are adjusted to the mean value of the covariates; [33]), and there was evidence for both relatedness-by-covariate interactions (table 2a). These interactions were plotted by fitting two regression planes, one for each level of relatedness, from a multiple regression of yield on both covariates (figure 2b). The figure reveals that the difference in yield between mixed and sibling groups was most pronounced among tall and variable sibling groups and tall and variable mixed groups (see also the partial regression coefficients, given in the caption). To reconcile the results of figure 2a,b, I show in the electronic supplementary material that the distribution of mean height in both mixed and sibling groups was skewed towards groups with a low mean height. This helps to explain why a difference in the overall yield could not be detected, given that the effect of relatedness emerged as mean height increased.
Table 2.
Results from Poisson regression models of the mean yield of plants in a group as a function relatedness, mean height and height inequality (CV). Both models also include the blocking factor ‘table’ (not shown). Effect sizes (with 95% CI) give the mean difference in least-squares means between levels of relatedness or, for covariates, a linear regression coefficient. (Notes: Total sample size was 100 groups. A non-significant interaction between relatedness and the mean height of subordinates was removed from b. Interactions from a are plotted in figure 2b; the interaction from b is plotted in figure 3b. p-Values were not adjusted for multiple testing of the same dataset.)
| response variable | explanatory variables | 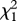 |
p-value | effect size |
|---|---|---|---|---|
| (a) mean yield | ||||
| relatedness | 9.16 | 0.0025 | sibs > mix; 4.52 seeds (1.52, 7.53) | |
| mean height | 209.5 | <0.0001 | 0.43 (0.38, 0.48) | |
| relatedness × mean height | 3.80 | 0.050 | ||
| CV in height | 1.88 | 0.17 | 6.50 (−2.8, 15.8) | |
| relatedness × CV | 5.91 | 0.015 | ||
| (b) mean yield of subordinates | ||||
| relatedness | 10.7 | 0.0011 | sibs > mix; 4.10 seeds (1.51, 6.70) | |
| mean height of subordinates | 242.8 | <0.0001 | 0.48 (0.42, 0.54) | |
| CV in height (whole group) | 4.90 | 0.027 | 9.31 (1.07, 17.5) | |
| relatedness × CV | 7.08 | 0.0078 |
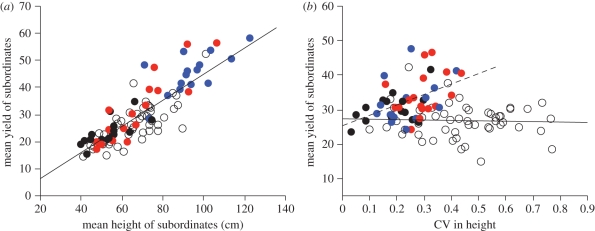
The interacting effects of relatedness and height inequality were further studied by focusing on the reproductive allocation of subordinate plants (the two shortest plants) in sibling and mixed groups (table 2b). The marginal increase in the yield of subordinates with an increase in their mean height was similar for all groups (figure 3a). But while holding their mean height fixed, the yield of subordinates in sibling groups (but not mixed groups) increased significantly with the group's height inequality (figure 3b). (Note that the same conclusion holds when using the height of the dominant plant in place of the CV in height). Given that subordinates in mixed groups were almost always from the black and red genotypes, also note that after excluding blue sibling groups and the few exceptional mixed groups (leaving only black and red genotypes in the analysis), all conclusions from figure 3 remain unchanged. Similar analyses (not shown) revealed no effect of relatedness on the reproductive allocation of dominant plants from the blue genotype.
Figure 3.
Leverage plots from a multiple regression of the mean yield of subordinate plants on their mean height and the height inequality (CV) of plants in their group (corresponding to the model in table 2b). While accounting for a common positive relationship between the mean height of subordinates and their mean yield (a), a marginal increase in height inequality (b) had a significantly positive effect on the mean yield of subordinates in sibling groups (p = 0.005; estimated slope: 21.9 (6.6, 37.2) but not in mixed groups (p = 0.85; estimated slope: −0.80 (−8.9, 7.3)).
(c). Allocation to roots
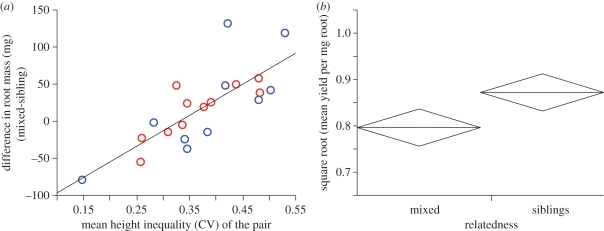
Overall, the mean difference in root mass between mixed and sibling groups (paired to match in mean height) was positive but not significantly greater than zero (t-test; t20 = 1.60; p = 0.12). However, it might be expected that mixed groups had greater allocation to roots where the difference in yield between sibling and mixed groups was most pronounced (among tall and variable sibling groups and tall and variable mixed groups). The size of the difference in root mass (mixed-sibling pots) did not vary with the average height of the pair (F1,18 = 0.83; p = 0.52), but it did increase significantly with the average height inequality of the pair (figure 4a). Hence, variable mixed groups tended to yield fewer seeds than variable sibling groups of similar height (e.g. figures 2b and 3b), and within matched pairs of highly variable groups, the mixed groups tended to allocate more to roots. Accordingly, there was strong evidence that mixed groups yielded fewer seeds per unit of root mass than their matched sibling groups (figure 4b).
Figure 4.
(a) Leverage plot of the difference in root mass between paired mixed and sibling groups (paired to match in mean height; colours indicate the genotype of the sibling group) regressed on the mean height inequality (CV) of the pair, showing a significant positive slope (F1,18 = 27.4; p < 0.0001). (b) Mixed groups also yielded significantly fewer seeds per unit of root mass, on average, than their paired sibling group (paired t-test; t20 = 2.74; p = 0.013). Mean values are block-centred (where ‘pair’ is the blocking factor), and the vertical span of each diamond is a 95% CI.
4. Discussion
Based only on the overall mean yield of kin and non-kin groups, which showed no influence of relatedness, it would appear that the current study cannot distinguish between the resource partitioning and recognition–cooperation hypotheses. Allometric analyses of group yield, however, revealed a major effect of relatedness: as the mean height and height inequality of groups increased, kin groups tended to yield more seeds than non-kin. I expected that increasing height inequality might negatively affect the reproductive allocation of mixed groups in particular, where one genotype consistently dominated in height. Yet surprisingly, it was the effect of height inequality in sibling groups that accounted for much of the difference in yield between kin and non-kin groups. Increasing height inequality in sibling (but not mixed) groups was associated with an increase in the reproductive allocation of subordinate plants, suggesting that siblings were relatively tolerant of competition with their taller kin, or perhaps that taller plants somehow facilitated the reproduction of shorter siblings. Regardless of the mechanisms involved, it is critical that among kin and non-kin groups with greatest height inequality (where the reproductive advantage of kin groups was pronounced), it was the non-kin groups that tended to allocate more mass to roots. Together, these results are consistent with the recognition–cooperation hypothesis, which predicts higher mean yield in kin groups as a result of avoiding ‘wasteful’ allocation to competitive traits.
The current study builds on evidence that below-ground competition among non-self plants can lead to over-proliferation of roots at the expense of reproduction (e.g. [12–14]) and evidence that kin groups allocate less biomass to roots than comparable non-kin groups [22]. The latter result provides evidence of kin recognition, but without measuring the reproductive consequences of root allocation, it does not necessarily imply cooperation [34–36]. Hence, a main advance of the current study is to link a difference in root allocation between kin and non-kin groups with a correlated difference in group yield. Before discussing further work that is required to understand apparent kin recognition and cooperation among plants, I first consider some alternative interpretations of the data presented here.
(a). Alternative interpretations
There was strong evidence that the genotypes used in the current study differed in size (height) and competitive ability. One consequence is that, when considering the yield of individuals (details given in the electronic supplementary material), plants from subordinate genotypes (red and black) can seem to fit the predictions of cooperation (enhanced growth and reproduction with siblings), whereas plants from the dominant genotype (blue) seem to fit the predictions of resource partitioning (enhanced growth and reproduction when dominating mixed groups). Masclaux et al. [27] interpreted similar data from Arabidopsis thaliana in this way, concluding that the outcome of competition depends on the relative competitive abilities of interacting genotypes—not the relatedness of neighbours per se. Certainly, whenever genotypes differ in competitive ability (perhaps owing to slight differences in average growth rates; [21]) and the outcome of competition is measured by individual yield, it is inevitable that a focal genotype can perform better or worse with kin, depending on its competitive ability in mixtures. But nevertheless, it is also possible that plants adjust their competitiveness to the relatedness of neighbours, with consequences for the productivity of the group as a whole. This is the key mechanism of the recognition–cooperation hypothesis, and it is supported by evidence from the current study.
It might appear that mixed groups suffered in productivity simply because they included ‘weak’ genotypes that were dominated by a superior competitor. However, I found that while holding the mean height of subordinate plants from the black and red genotypes fixed, they tended to produce more seeds when growing with a dominant sibling than with a dominant non-sibling of the same height. Given that competitive dominance seems at least partly owing to plant height, this result suggests that subordinate plants did indeed respond to the relatedness of competing neighbours. To fully untangle the effects of plant genotype and relatedness, however, it may be useful to compare the reproductive allometry of plants from a number of genotypes in kin competition, non-kin competition and no-competition environments.
Finally, the difference in yield among kin and non-kin groups could be explained as an inevitable consequence of the relatively large size inequality among non-kin. This inevitable consequence can arise whenever the smallest or largest plants produce a disproportionately small number of seeds for their size [21,37]. For example, in the context of the current study, if the relationship between individual plant height and yield were diminishing, then mixed groups that varied in height would necessarily have lower mean yield than uniform sibling groups of the same mean height (a consequence of Jensen's inequality; [37]). I confirm in the electronic supplementary material, however, that yield increased nearly linearly with plant height in mixed groups. This suggests that the relatively low mean yield of highly variable non-kin groups was not a simple consequence of their height inequality.
(b). Future directions
In addition to those already discussed, the current study suggests a number of avenues for future work. First, because roots could be measured only at the group level (see also [12,22]), the allocation strategies of individual plants remain obscure. It would be useful to characterize the phenotypes of dominant and subordinate plants in kin and non-kin groups, especially focusing on potential confrontation/avoidance behaviours of roots (cf. [38]) and on specific allocation to root types (e.g. fine roots versus main roots). Such individual-level allocation traits would be usefully compared with the biomass allocation of plants grown in the absence of competition; in this way, normal ontogenetic changes in resource allocation can be distinguished from true plasticity to the social environment [39]. Second, it is not yet clear how plants might distinguish kin from non-kin, although recent work implies chemical signalling among neighbouring roots [40]. A strong test of adaptive kin discrimination would involve eliminating such putative signalling mechanisms to induce the expression of ‘inappropriate’ phenotypes (cf. [41]). Interacting kin would then be expected to express a relatively selfish allocation strategy and to produce a lower mean yield than a more cooperative alternative strategy.
Finally, it is important to clarify that kin recognition is not a prerequisite for cooperation among plants. Local relatedness will usually result from limited dispersal [10], and in such contexts, selection can favour indiscriminant cooperation among neighbours. Studies of multilevel selection in plant populations (e.g. [42,43]) suggest that kin groups should indeed be selected to reduce their competitiveness in this way. Still, the appeal of kin recognition is that some species (those that experience variation in local relatedness) may have the ability to acquire additional information about the relatedness of neighbours, and if so, cooperation could be identified by experiment. The data here emphasize that such experiments should involve measures of plants' allocation to competitive traits along with measures of reproductive success or resource availability. By doing so at the group level, the current study takes a critical step towards identifying strategies that adjust a plant's competitiveness to its social environment.
Acknowledgements
I thank Peter Abrams, Aneil Agrawal, Joel Brown, Jen Perry, Locke Rowe, John Stinchcombe and Stuart West for comments on the manuscript. Ruben Milla and two anonymous referees provided insightful reviews. Brandon Campitelli, Anna Simonsen, and J. Stinchcombe gave advice on the experimental design and donated the seeds, and Jim Dix, Bruce Hall, Trung Luu and Andrew Petrie offered technical support. Daniel Lunn produced the R model in the electronic supplementary material. I was funded by a scholarship and a Discovery grant (to P.A.) from the Natural Sciences and Engineering Council of Canada and a grant (to S.W.) from the European Research Council.
References
- 1.Vekemans X., Hardy O. J. 2004. New insights from fine-scale spatial genetic structure analyses in plant populations. Mol. Ecol. 13, 921–935 10.1046/j.1365-294X.2004.02076.x (doi:10.1046/j.1365-294X.2004.02076.x) [DOI] [PubMed] [Google Scholar]
- 2.Ellstrand N., Antonovics J. 1985. Experimental studies of the evolutionary significance of sexual reproduction II. A test of the density-dependent selection hypothesis. Evolution 39, 657–666 10.2307/2408660 (doi:10.2307/2408660) [DOI] [PubMed] [Google Scholar]
- 3.Maynard Smith J. 1978. The evolution of sex. Cambridge, UK: Cambridge University Press [Google Scholar]
- 4.Young J. 1981. Sib competition can favour sex in two ways. J. Theor. Biol. 88, 755–756 10.1016/0022-5193(81)90249-6 (doi:10.1016/0022-5193(81)90249-6) [DOI] [PubMed] [Google Scholar]
- 5.Schmitt J., Ehrhardt D. W. 1987. A test of the sib-competition hypothesis for outcrossing advantage in Impatiens capensis. Evolution 41, 579–590 10.2307/2409259 (doi:10.2307/2409259) [DOI] [PubMed] [Google Scholar]
- 6.Cheplick G., Kane K. 2004. Genetic relatedness and competition in Triplasis purpurea (Poaceae): resource partitioning or kin selection? Int. J. Plant Sci. 165, 623–630 10.1086/386556 (doi:10.1086/386556) [DOI] [Google Scholar]
- 7.Milla R., Forero D. M., Escudero A., Iriondo J. M. 2009. Growing with siblings: a common ground for cooperation or for fiercer competition among plants? Proc. R. Soc. B 276, 2531–2540 10.1098/rspb.2009.0369 (doi:10.1098/rspb.2009.0369) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotowska A. M., Cahill J. F., Jr, Keddie B. A. 2010. Plant genetic diversity yields increased plant productivity and herbivore performance. J. Ecol. 98, 237–245 10.1111/j.1365-2745.2009.01606.x (doi:10.1111/j.1365-2745.2009.01606.x) [DOI] [Google Scholar]
- 9.Rankin D. J., Bargum K., Kokko H. 2007. The tragedy of the commons in evolutionary biology. Trends Ecol. Evol. 22, 643–651 10.1016/j.tree.2007.07.009 (doi:10.1016/j.tree.2007.07.009) [DOI] [PubMed] [Google Scholar]
- 10.Hamilton W. D. 1964. The genetical evolution of social behaviour I and II. J. Theor. Biol. 7, 1–52 10.1016/0022-5193(64)90038-4 (doi:10.1016/0022-5193(64)90038-4) [DOI] [PubMed] [Google Scholar]
- 11.Frank S. A. 1995. Mutual policing and repression of competition in the evolution of cooperative groups. Nature 377, 520–522 10.1038/377520a0 (doi:10.1038/377520a0) [DOI] [PubMed] [Google Scholar]
- 12.Gersani M., Brown J. S., O'Brien E., Maina G., Abramsky Z. 2001. Tragedy of the commons as a result of root competition. J. Ecol. 89, 660–669 10.1046/j.0022-0477.2001.00609.x (doi:10.1046/j.0022-0477.2001.00609.x) [DOI] [Google Scholar]
- 13.O'Brien E., Gersani M., Brown J. S. 2005. Root proliferation and seed yield in response to spatial heterogeneity of below-ground competition. New Phytol. 168, 401–412 10.1111/j.1469-8137.2005.01520.x (doi:10.1111/j.1469-8137.2005.01520.x) [DOI] [PubMed] [Google Scholar]
- 14.Maina G. G., Brown J. S., Gersani M. 2002. Intra-plant versus inter-plant root competition in beans: avoidance, resource matching or tragedy of the commons. Plant Ecol. 160, 235–247 10.1023/A:1015822003011 (doi:10.1023/A:1015822003011) [DOI] [Google Scholar]
- 15.Schmitt J. 1997. Is photomorphogenic shade avoidance adaptive? Perspectives from population biology. Plant Cell Environ. 20, 826–830 10.1046/j.1365-3040.1997.d01-96.x (doi:10.1046/j.1365-3040.1997.d01-96.x) [DOI] [Google Scholar]
- 16.Denison R. F., Kiers E. T., West S. A. 2003. Darwinian agriculture: when can humans find solutions beyond the reach of natural selection? Q. Rev. Biol. 78, 145–168 10.1086/374951 (doi:10.1086/374951) [DOI] [PubMed] [Google Scholar]
- 17.Jennings P., de Jesus J. 1968. Studies on competition in rice I. Competition in mixtures of varieties. Evolution 22, 119–124 10.2307/2406656 (doi:10.2307/2406656) [DOI] [PubMed] [Google Scholar]
- 18.Novoplansky A. 2009. Picking battles wisely: plant behaviour under competition. Plant Cell Environ. 32, 726–741 10.1111/j.1365-3040.2009.01979.x (doi:10.1111/j.1365-3040.2009.01979.x) [DOI] [PubMed] [Google Scholar]
- 19.Andalo C., Goldringer I., Godelle B. 2001. Inter- and intragenotypic competition under elevated carbon dioxide in Arabidopsis thaliana. Ecology 82, 157–164 [Google Scholar]
- 20.Donohue K. 2003. The influence of neighbour relatedness on multilevel selection in the Great Lakes sea rocket. Am. Nat. 162, 77–92 10.1086/375299 (doi:10.1086/375299) [DOI] [PubMed] [Google Scholar]
- 21.Tonsor S. 1989. Relatedness and intraspecific competition in Plantago lanceolata. Am. Nat. 134, 897–906 10.1086/285020 (doi:10.1086/285020) [DOI] [Google Scholar]
- 22.Dudley S. A., File A. L. 2007. Kin recognition in an annual plant. Biol. Lett. 3, 435–438 10.1098/rsbl.2007.0232 (doi:10.1098/rsbl.2007.0232) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy G. P., Dudley S. A. 2009. Kin recognition: competition and cooperation in Impatiens (Balsaminaceae). Am. J. Bot. 96, 1990–1996 10.3732/ajb.0900006 (doi:10.3732/ajb.0900006) [DOI] [PubMed] [Google Scholar]
- 24.Aarssen L. W., Taylor D. R. 1992. Fecundity allocation in herbaceous plants. Oikos 65, 225–232 [Google Scholar]
- 25.Samson D. A., Werk K. S. 1986. Size-dependent effects in the analysis of reproductive effort in plants. Am. Nat. 127, 667–680 10.1086/284512 (doi:10.1086/284512) [DOI] [Google Scholar]
- 26.Weiner J., Campbell L. G., Pino J., Echarte L. 2009. The allometry of reproduction within plant populations. J. Ecol. 97, 1220–1233 10.1111/j.1365-2745.2009.01559.x (doi:10.1111/j.1365-2745.2009.01559.x) [DOI] [Google Scholar]
- 27.Masclaux F., Hammond R. L., Meunier J., Gouhier-Darimont C., Keller L., Reymond P. 2009. Competitive ability not kinship affects growth of Arabidopsis thaliana accessions. New Phytol. 185, 322–331 10.1111/j.1469-8137.2009.03057.x (doi:10.1111/j.1469-8137.2009.03057.x) [DOI] [PubMed] [Google Scholar]
- 28.Weiner J. 1990. Asymmetric competition in plant populations. Trends Ecol. Evol. 5, 360–364 10.1016/0169-5347(90)90095-U (doi:10.1016/0169-5347(90)90095-U) [DOI] [PubMed] [Google Scholar]
- 29.Ballare C. L., Scopel A. L. 1997. Phytochrome signalling in plant canopies: testing its population-level implications with photoreceptor mutants of Arabidopsis. Funct. Ecol. 11, 441–450 10.1046/j.1365-2435.1997.00108.x (doi:10.1046/j.1365-2435.1997.00108.x) [DOI] [Google Scholar]
- 30.Weiner J. 1986. How competition for light and nutrients affects size variability in Ipomoea tricolor populations. Ecology 67, 1425–1427 10.2307/1938699 (doi:10.2307/1938699) [DOI] [Google Scholar]
- 31.Ballare C. L., Scopel A. L., Jordan E. T., Vierstra R. D. 1994. Signaling among neighbours and the development of size inequalities in plant populations. Proc. Natl Acad. Sci. USA 91, 10 094–10 098 10.1073/pnas.91.21.10094 (doi:10.1073/pnas.91.21.10094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiner J., Thomas S. C. 1986. Size variability and competition in plant monocultures. Oikos 47, 211–222 10.2307/3566048 (doi:10.2307/3566048) [DOI] [Google Scholar]
- 33.Milliken G. A., Johnson D. E. 2002. Analysis of messy data: analysis of covariance. London, UK: Chapman and Hall [Google Scholar]
- 34.Callaway R., Mahall B. 2007. Plant ecology: family roots. Nature 448, 145–147 10.1038/448145a (doi:10.1038/448145a) [DOI] [PubMed] [Google Scholar]
- 35.de Kroon H. 2007. How do roots interact? Science 318, 1562–1563 10.1126/science.1150726 (doi:10.1126/science.1150726) [DOI] [PubMed] [Google Scholar]
- 36.Klemens J. A. 2008. Kin recognition in plants? Biol. Lett. 4, 67–68 10.1098/rsbl.2007.0518 (doi:10.1098/rsbl.2007.0518) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laird R., Aarssen L. 2005. Size inequality and the tragedy of the commons phenomenon in plant competition. Plant Ecol. 179, 127–131 10.1007/s11258-004-6737-4 (doi:10.1007/s11258-004-6737-4) [DOI] [Google Scholar]
- 38.Holzapfel C., Alpert P. 2003. Root cooperation in a clonal plant: connected strawberries segregate roots. Oecologia 134, 72–77 10.1007/s00442-002-1062-x (doi:10.1007/s00442-002-1062-x) [DOI] [PubMed] [Google Scholar]
- 39.McConnaughay K. D. M., Coleman J. S. 1999. Biomass allocation in plants: ontogeny or optimality? Ecology 80, 2581–2593 10.1890/0012-9658(1999)080[2581:BAIPOO]2.0.CO;2 (doi:10.1890/0012-9658(1999)080[2581:BAIPOO]2.0.CO;2) [DOI] [Google Scholar]
- 40.Biedrzycki M. L., Jilany T. A., Dudley S. A., Bais H. P. 2010. Root exudates mediate kin recognition in plants. Commun. Integr. Biol. 3, 28–35 10.4161/cib.3.1.10118 (doi:10.4161/cib.3.1.10118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitt J., Dudley S. A., Pigliucci M. 1999. Manipulative approaches to testing adaptive plasticity: phytochrome-mediated shade-avoidance responses in plants. Am. Nat. 154, S43–S54 10.1086/303282 (doi:10.1086/303282) [DOI] [PubMed] [Google Scholar]
- 42.Stevens L., Goodnight C. J., Kalisz S. 1995. Multilevel selection in natural populations of Impatiens capensis. Am. Nat. 145, 513–526 10.1086/285753 (doi:10.1086/285753) [DOI] [Google Scholar]
- 43.Weinig C., Johnston J. A., Willis C. G., Maloof J. N. 2007. Antagonistic multilevel selection on size and architecture in variable density settings. Evolution 61, 58–67 10.1111/j.1558-5646.2007.00005.x (doi:10.1111/j.1558-5646.2007.00005.x) [DOI] [PubMed] [Google Scholar]