Abstract
DNA ligases are required for DNA replication, repair, and recombination. In eukaryotes, there are three families of ATP-dependent DNA ligases. Members of the DNA ligase I and IV families are found in all eukaryotes, whereas DNA ligase III family members are restricted to vertebrates. These enzymes share a common catalytic region comprising a DNA-binding domain, a nucleotidyltransferase (NTase) domain, and an oligonucleotide/oligosaccharide binding (OB)-fold domain. The catalytic region encircles nicked DNA with each of the domains contacting the DNA duplex. The unique segments adjacent to the catalytic region of eukaryotic DNA ligases are involved in specific protein-protein interactions with a growing number of DNA replication and repair proteins. These interactions determine the specific cellular functions of the DNA ligase isozymes. In mammals, defects in DNA ligation have been linked with an increased incidence of cancer and neurodegeneration.
Keywords: cancer, genome stability, neurodegeneration, recombination, repair, replication
INTRODUCTION
Interruptions in the phosphodiester backbone of DNA that form as a consequence of discontinuous DNA synthesis on the lagging strand of the replication fork and during most recombination and repair pathways are repaired by DNA ligases. DNA-joining activity was initially described in extracts of Escherichia coli and bacteriophage T4-infected E. coli in 1967 and in the following year in extracts of mammalian cells (1, 2). DNA ligases are a universal feature of DNA-based life forms and have almost ubiquitous involvement in DNA transactions. These enzymes have played a critical role in the development of the recombinant DNA technologies that have transformed the investigation of biological processes. Their widespread but specialized roles in cell growth and DNA maintenance make ligases attractive targets for the development of antimicrobial agents and cancer therapeutics.
DNA ligases are nucleotidyltransferases (NTases) that utilize a high-energy cofactor, either NAD+ or ATP, to catalyze phosphodiester bond formation in a three-step reaction mechanism (1, 2). To date, all eukaryotic DNA ligases are ATP dependent, whereas both ATP- and NAD+-dependent DNA ligases have been identified in bacteria, archaea, and viruses. In the first part of this review, we describe how recent structures of NTases have provided insights into DNA substrate selection and the catalytic mechanisms of eukaryotic DNA ligases, comparing the conserved features and unique adaptations of specific enzymes.
On the basis of biochemical and immunological studies, the laboratory of Tomas Lindahl provided the first evidence that mammalian cells contain more than one species of DNA ligase (3). This was confirmed by the cloning of three mammalian LIG genes (4–6). More recently, genome sequencing has revealed that archaeal and prokaryotic organisms also possess multiple DNA ligases (7). In the latter part of this review, we describe the three families of eukaryotic DNA ligases, their cellular functions and protein partners, and how defects in DNA ligation contribute to human disease.
CATALYTIC ACTIVITIES OF DNA LIGASES AND RELATED NUCLEOTIDYLTRANSFERASES
DNA ligases catalyze an energetically favorable, multistep reaction in which an adenylate group, adenosine 5′-monophosphate (AMP), is sequentially transferred from ATP or NAD+ to an active-site lysine of the enzyme (step 1), then to the 5′ phosphate of the DNA substrate (step 2). Adenylation of the DNA activates the 5′ phosphate for the nucleophilic attack by a 3′ OH that displaces AMP and covalently joins together the ends of two DNA strands (step 3) (Figure 1). The highly favorable reaction equilibrium of each chemical step makes this reaction sequence effectively irreversible. Consequently, the adenylated form of a DNA ligase is relatively stable in the absence of a DNA substrate, and it is likely that most ligase molecules in a cell are adenylated and ready to react with DNA. Adenylation of the enzyme also enhances the specificity of binding to DNA substrates, an activity known as nick sensing (8). Nonetheless, ligases can react with DNA ends that are unsuitable for ligation, generating a chemically stable AMP adduct, which must be removed enzymatically from the DNA to enable further repair of these “dirty breaks” (9, 10). Aprataxin is a phosphodiesterase that specifically acts on dead-end DNA ligation products to hydrolyze the AMP-phosphate bond and enable repair (11). A deficiency of aprataxin results in early neurological defects and later neurodegeneration (12, 13).
Figure 1.
Enzymatic ligation of DNA. The three-step reaction catalyzed by DNA ligases (Ligs) results in the serial transfer of AMP (adenosine 5′-monophosphate) to an active-site lysine (step 1) then to the 5′-PO4 end of DNA (step 2). During step 3, the 3′-OH end of a second DNA strand attacks the 5′-PO4 to release AMP and generate the ligated DNA product. Eukaryotic and archaeal DNA ligases use an ATP cofactor, whereas most bacterial ligases use NAD+.
The three-step reaction chemistry that is a universal feature of DNA ligases (Figure 1) is also shared by RNA ligases (14), whereas 5′ mRNA capping enzymes only catalyze steps 1 and 2 using GTP and RNA substrates (15). Together these three classes of enzymes comprise a large family of NTases with six conserved sequence motifs reflecting a common active-site structure (15). Capping enzymes catalyze the transfer of GMP (guanosine 5′-monophosphate) to the 5′-phosphorylated end of mRNA via a lysyl-GMP intermediate that is analogous to the AMP active-site modification of DNA ligases. The structures of DNA ligases, RNA ligases, and capping enzymes reveal a conserved domain architecture that supports closely related enzymatic activities toward different nucleic acid substrates.
The structural biology of DNA ligases has advanced significantly in recent years with reports of crystal structures of unliganded, AMP-modified, and DNA-bound forms of bacterial and eukaryotic ligases. Two themes have emerged. All DNA ligases, excepting the simplest viral ligases, completely encircle their DNA substrates using a multidomain architecture that imparts sufficient flexibility to open and close around DNA. Furthermore, local rearrangements within the active site accommodate progression through the three chemical steps of DNA end joining. The modular structure of DNA ligases may also be important for the nick-sensing activity that directs the enzyme to appropriate substrates. For this review, we use the pioneering studies of the Chlorella virus DNA ligase by the Shuman laboratory (8, 16–20) as the basis for discussing comparative aspects of other ligase structures and enzymatic functions.
STRUCTURES OF EUKARYOTIC DNA LIGASES
The ATP-dependent DNA ligase encoded by Chlorella virus PBCV-1 is among the smallest (298 amino acids) of eukaryotic ligases, consisting of only two domains and resembling bacteriophage DNA ligases (21) (Figure 2). The larger N-terminal NTase domain binds ATP and contains many of the active-site residues. The AMP moiety binds in an active-site pocket with the adenine ring remaining buried throughout the three-step ligation reaction as the AMP is transferred to lysine, the DNA 5′ phosphate, and ultimately to water. As mentioned above, AMP occupancy appears to also be important for DNA substrate selection (nick sensing) (8). Specificity for AMP binding is achieved through hydrogen bonding interactions by residues lining the pocket and the exocyclic groups of the adenine base (18). A corresponding pocket within the NTase domain of GTP-dependent capping enzymes makes guanine-specific hydrogen bonding interactions (22, 23).
Figure 2.
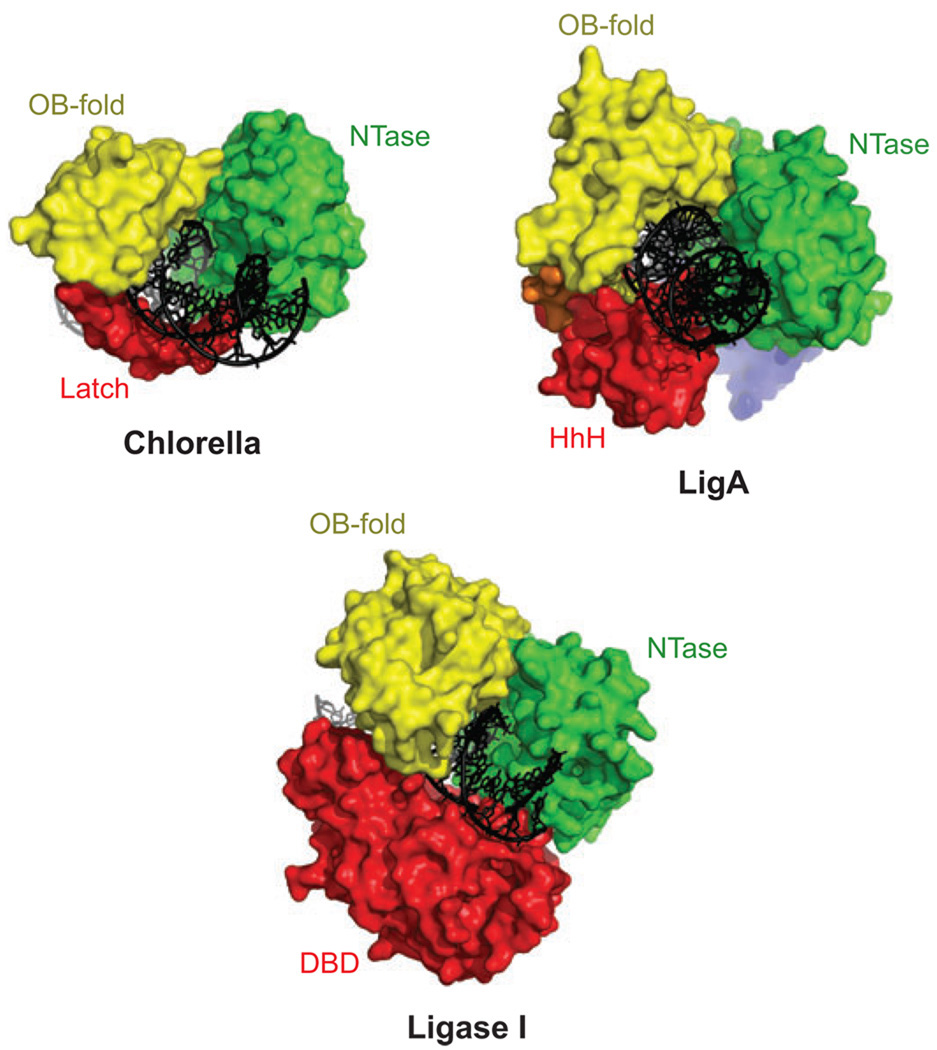
Comparative anatomy of DNA ligases. Examples of viral, bacterial, and mammalian DNA ligases are shown bound to DNA. The DNA ligases from Chlorella virus PCBV-1, Protein Data Bank (pdb) 2q2t, Escherichia coli (LigA protein; pdb 2owo), and human ligase I (pdb 1×9n) have a similar ring-shaped structures in complex with a nicked DNA substrate. A conserved catalytic core, consisting of the NTase domain (green) and OB-fold domain (yellow), contacts the 3′-OH and 5′-PO4 ends of DNA during steps 2 and 3 of the end-joining reaction (cf. Figure 1). An additional domain (red) in each enzyme interacts with DNA, completing the ring-shaped structure. The Chlorella ligase has an insertion in the OB-fold domain, termed the latch, which is disordered in the absence of DNA and bridges between the two domains of the catalytic core (16). E. coli LigA has a C-terminal helix-hairpin-helix (HhH) (25, 35) domain engaging the DNA that is functionally analogous to the N-terminal DNA-binding domain (DBD) (26) of human DNA ligase I. A small N-terminal motif in the LigA protein is important for sensing the NAD+ cofactor (25). Abbreviations: OB-fold, oligonucleotide/oligosaccharide binding fold; NTase, nucleotidyltransferase.
The C-terminal oligonucleotide/oligosaccharide binding (OB)-fold domain of the Chlorella ligase assists in the step 1 transfer of AMP to the active-site lysine of the NTase domain and then functions subsequently as a DNA-binding domain during steps 2 and 3. Although the OB-fold protein architecture is typically associated with single-strand DNA-binding proteins (24), the OB-fold domain of DNA ligases binds to double-stranded DNA in the minor groove, primarily interacting with the DNA strand adjacent to the 5′ phosphate end of the nick (16). The OB-fold domain is flexibly tethered to the NTase domain, and it undergoes a large rotation between the step 1 adenylation of the enzyme and the DNA-dependent steps 2 and 3. During step 1, residues of one of the six conserved motifs of NTases (15), motif VI, reside within the OB-fold domain and face the active site (18), where they are required for AMP transfer to the active-site lysine. For steps 2 and 3, the OB-fold domain turns to orient DNA-binding residues for interactions with the DNA substrate (16), and the motif VI residues are exposed on the protein surface away from the active site. This conformational flexibility of the ligase protein is accommodated by an extended polypeptide linker joining the NTase and OB-fold domains, which presumably also assists in loading the enzyme onto DNA.
The crystal structure of the Chlorella ligase-AMP complex (18) presaged another important conformational change that accompanies the progression from step 1 to step 2. The deoxyribose moiety of AMP swings between the active-site lysine and the 5′ DNA phosphate during the transition from step 1 to step 2, whereas the adenine base of AMP remains fixed in the binding pocket of the NTase domain. A rotation about the glycosylic bond between adenine and 2′ deoxyribose realigns the AMP phosphate for bonding with the DNA 5′ phosphate (step 2). This change in the conformation of the bound AMP appears to be triggered by DNA binding to the enzyme and was originally intuited from the Chlorella ligase-AMP complex structure (18), which showed a bound SO42− ion that might mimic the 5′ phosphate of a DNA substrate. The authors correctly reasoned that the AMP group would have to isomerize to react with phosphate at the proposed binding site. This prediction was subsequently borne out by the crystal structure of the Chlorella ligase-DNA complex (16). The handoff of AMP from a lysine side chain to a DNA phosphate may be accompanied by a reconfiguration of bound metals in the active site. Divalent metals are required for normal enzymatic activity during all three steps of end joining. The metals presumably orient the phosphate atoms of AMP and the DNA as well as neutralize the charge during phosphoryl transfer reactions. However, the identification of discrete metal-binding sites within DNA ligase structures has been elusive. Crystal structures of viral, bacterial, and mammalian DNA ligases include extra electron density within the active site that has been interpreted as bound metal ion(s) or a bound water standing in for a metal that is coordinated by protein oxygen ligands (16, 18, 25–27). There is uncertainty about the number of metals and their locations during each step of end joining because of the limited quality of electron density, particularly when using anomalous X-ray scattering to locate metal-binding sites.
STRUCTURAL SPECIALIZATION OF MAMMALIAN DNA LIGASES
The NTase and OB-fold domains together comprise a minimal catalytic unit that is conserved in all known DNA and RNA ligases and capping enzymes. These domains harbor the active site as well as many DNA-binding residues. The catalytic core of mammalian DNA ligases and NAD+-dependent bacterial ligases is embellished with additional N- and C-terminal domains (Figure 2), which are essential for biological activity and participate in protein-protein and/or DNA-binding interactions. For example, the three mammalian DNA ligases all require an additional N-terminal DNA-binding domain (DBD) for efficient ligation activity (26).
The DBD interacts in the minor groove, contacting the DNA upstream and downstream of the nick. The α-helical DBD includes several helix-turn-helix motifs that insert into the minor groove surface of the DNA opposite from the 3′-OH and 5′-PO4 ends engaged by the NTase domain. The DBD appears to stabilize the DNA in an under-wound conformation with a widened minor groove that exposes the ligatable ends of the DNA to conserved residues that are important for catalytic activity. The DBD also makes protein-protein interactions with the NTase domain and the OB-fold domain, completing a ring-shaped protein structure that encircles the DNA. The addition of the DBD domain in trans greatly stimulates DNA nick joining by the catalytic core of human DNA ligase I (26) and DNA ligase III (157). Stimulation of catalytic activity may result from the DBD shaping the conformation of the DNA substrate and/or positioning the catalytic core on the DNA.
The structure of a human DNA ligase III fragment (157) closely resembles the structure of DNA ligase I in complex with DNA (26). Structural information about DNA ligase IV is currently limited to a short peptide from the C terminus of ligase IV bound to its partner protein XRCC4 (28), although the amino acid sequence of DNA ligase IV includes the conserved three-domain catalytic segment (DBD-NTase-OB-fold domains) found in DNA ligases I and III. These observations make it likely that all three mammalian DNA ligases encircle DNA in a similar manner. However, these enzymes have different efficiencies of ligating DNA single-strand breaks versus double-strand breaks—functional differences that may be explained by subtle adaptations of their conserved three-domain core structures and auxiliary flanking domains that contribute to end-joining activity (29).
Mammalian DNA ligase III has an additional functionally important domain flanking the conserved three-domain core. An N-terminal zinc-binding domain termed a zinc finger binds to nicks and gaps in DNA, serving as a nick sensor (30, 31). The structure of the DNA ligase III zinc finger resembles that of a zinc finger in the GATA-1 transcription factor but uses a different interface to bind to DNA (32). Residues located at the tip of a β-hairpin motif within the zinc finger are strongly perturbed by binding to nicked DNA, suggesting that the hairpin may insert into nicks or gaps in DNA (32). The zinc finger is not required for DNA nick joining by DNA ligase III, but it enhances ligation of RNA-containing substrates and the ligation of DNA with an unpaired flap (30, 33), suggestive of a broader substrate range when the zinc finger is present. The nick-sensing activity of the zinc finger stimulates the catalytic efficiency of single-strand break ligation, and it dramatically improves intermolecular ligation of blunt-ended DNA molecules (157). DNA ligase III is substantially better than DNA ligase I at DNA blunt-end joining, and the zinc finger may provide additional DNA contacts that help juxtapose two duplexes for ligation. A “jackknife model” has been proposed to explain the role of DNA binding by the zinc finger during ligation of single-strand breaks and blunt-end joining by DNA ligase III (Figure 3).
Figure 3.
The “jackknife model” of nick sensing by ligase III. (a) DNA ligase III has an additional zinc finger (ZnF) that binds to nicks and gaps in DNA (b), stimulating DNA nick-joining by ligase III (c). The ZnF domain added in trans inhibits ligation by the catalytic core of ligase III, consistent with a competitive binding interaction. The ZnF and adjacent DNA-binding domain (DBD) constitute one DNA-binding surface of ligase III, and the catalytic core consisting of the nucleotidyltransferase (NTase) and OB-fold domains constitutes a second DNA-binding moiety. The jackknife model posits a handoff of the nicked DNA substrate from the ZnF-DBD to the catalytic core through protein motions resembling the opening and closing of blades of a jackknife. In this model, the ZnF-DBD initially recruits ligase III to an interruption in the DNA backbone, and then the DNA ends are engaged by the NTase-OB-fold catalytic core for the ligation reaction. (d) The ZnF also contributes strongly to the intermolecular ligation of linear DNA molecules. In this case, the two independent DNA-binding surfaces of ligase III (ZnF-DBD versus NTase-OB-fold) could align two DNAs for ligation (157). Abbreviations: AdD, adenylation domain; OB-fold, oligonucleotide/oligosaccharide binding fold.
Bacterial DNA ligases provide an interesting example of a permuted domain organization in comparison to eukaryotic ligases. E. coli LigA protein and Thermus filiformis DNA ligase are NAD+-dependent enzymes with the highly conserved NTase and OB-fold domains but lack the N-terminal DBD found in mammalian ligases. Instead, a C-terminal helix-hairpin-helix (HhH) domain (35) binds DNA in the minor groove (25), serving as a functional mimic of the mammalian DBD (Figure 2). Thus, LigA encircles its DNA substrates in a manner similar to the mammalian DNA ligases, but using domains that are arrayed differently in the primary sequence of the protein. In another variation on this theme, the Chlorella ligase has a C-terminal latch, which is disordered in the absence of DNA but completes the ring-shaped structure of the enzyme bound to DNA (16). This small β-hairpin structure bridges between the NTase and OB-fold domains and penetrates the major groove of the DNA (Figure 2). Mutations affecting residues in the latch cause variable defects in DNA nick-joining activity (17). Thus, DNA ligases of bacteria, viruses, and eukaryotes have evolved different auxilliary domains for extending the DNA-binding interface of the enzyme to completely encircle the DNA substrate. A ring-shaped protein may have a longer residence time on DNA, enhancing processivity and the search for ligatable DNA nicks.
EUKARYOTIC DNA LIGASE FAMILIES
The DNA ligases encoded by the human genes LIG1, LIG3, and LIG4 serve as the prototypic members of the three families of eukaryotic DNA ligases. These enzymes share a related catalytic region, described above, that is flanked by distinct N- and C-terminal regions (Figures 4, 5, 6). Homologs of the LIG1 and LIG4 genes appear to be present in all eukaryotes. In contrast, the LIG3 gene appears to be restricted to vertebrates. Below, we describe the characteristic features of the DNA ligase polypeptides in each of the three families and the cellular phenotypes caused by DNA ligase deficiency.
Figure 4.
DNA ligase I family. Alignment of the DNA ligases encoded by the Saccharomyces cerevisiae CDC9 and human LIG1 genes. The DNA-binding domain (DBD, red) and the catalytic core composed of the adenylation and OB-fold domains (gray) and the nuclear localization signals (NLS) (yellow) are indicated. The position of the active-site lysine residues (K419, K442, K568), within the adenylation domain that form the covalent bond with AMP are shown. Two polypeptides are generated from the CDC9 gene by alternative translation initiation. The longer polypeptide has an N-terminal mitochondrial (mito) leader sequence (MLS, turquoise). The N-terminal region of human DNA ligase I (hLigI) contains a replication factory-targeting sequence (RFTS, purple) and four serine (Ser) residues, which are phosphorylated, in addition to the NLS (40–44). Abbreviations: aa, amino acids.
Figure 5.
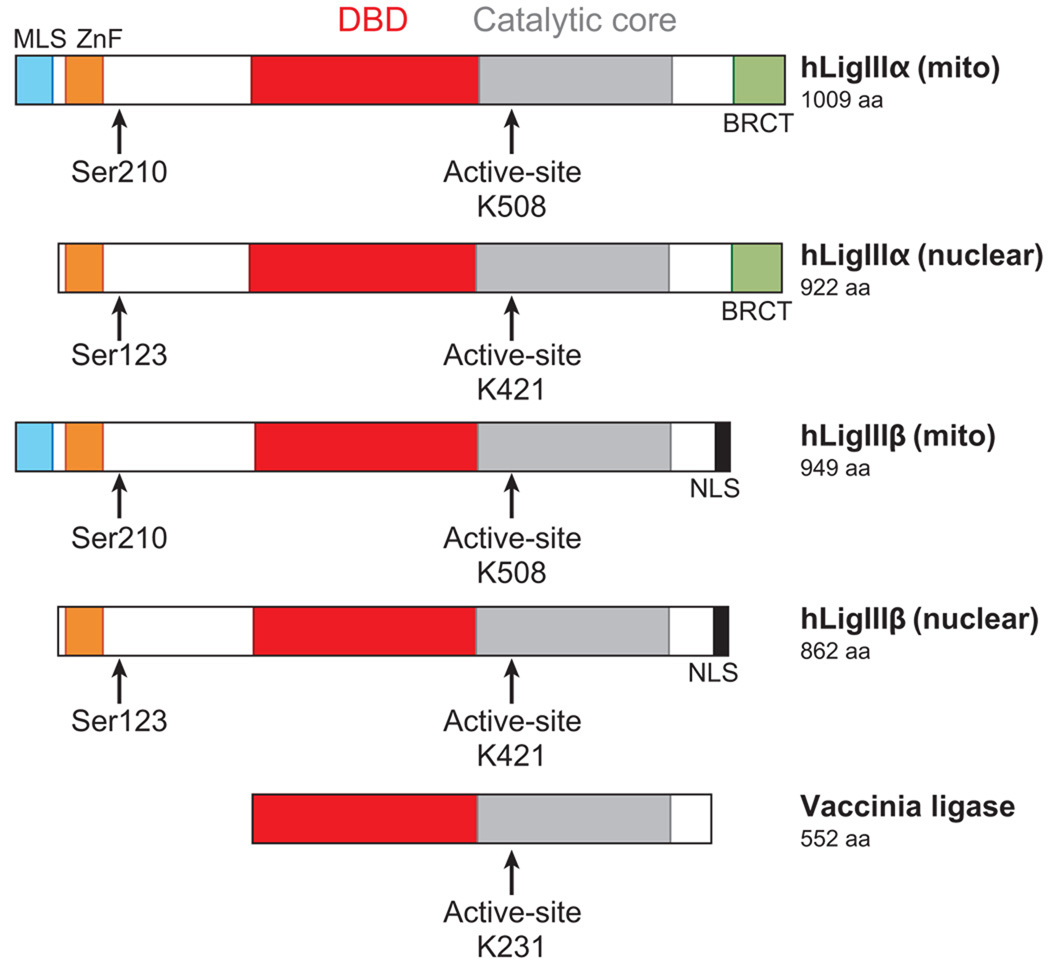
DNA ligase III family. Alignment of the DNA ligases encoded by vaccinia virus and human LIG3. The DNA-binding domain (DBD, red), catalytic core composed of the adenylation and OB-fold domains (gray), and nuclear localization signals (NLS, black) are indicated. The position of the active-site lysine residues (K508, K421, K231) within the adenylation domain that form the covalent bond with AMP are shown. Multiple DNA ligase polypeptides are encoded by the LIG3 gene. The N-terminal region of each of the polypeptides contains a zinc finger (ZnF, orange) and a serine residue (Ser123 or Ser210) that is phosphorylated (79). Two transcripts encoding hLigIIIα and hLigIIIβ are generated by alternative splicing. DNA ligase IIIα polypeptides have a breast and ovarian cancer susceptibility protein 1 C-terminal (BRCT) domain (green), whereas DNA ligase IIIβ polypeptides have a C-terminal NLS (gray) (62). Two polypeptides are generated from each of the transcripts by alternative translation initiation. The longer polypeptides have an N-terminal mitochondrial leader sequence (MLS, turquoise) (60). Abbreviations: aa, amino acids; mito, mitochondria.
Figure 6.
DNA ligase IV family. Alignment of the DNA ligases encoded by the S. cerevisiae DNL4 and human LIG4 genes. The DNA-binding domain (DBD, red) and the catalytic core composed of the adenylation and OB-fold domains (gray) are indicated. The position of the active-site lysine residues (K282, K213) within the adenylation domain that form the covalent bond with AMP are shown. The positions of the tandem BRCT domains (green) in the C-terminal regions of yeast Dnl4 and human DNA ligase IV (human LigIV) and a phosphorylated serine (Ser650) residue in human DNA ligase IV are shown (89). Abbreviation: aa, amino acids.
DNA Ligase I Family
The CDC9 gene, which encodes the Saccharomyces cerevisiae homolog of human LIG1, was first identified in the screen for cell division cycle mutants (36). The CDC9 gene is essential for viability, and temperature-sensitive alleles are hypersensitive to DNA damage, implicating the Cdc9 DNA ligase in DNA replication and repair (37, 38). Interestingly, an alternative translation initiation mechanism generates two forms of Cdc9 DNA ligase (Figure 4) that are directed to either the nuclear or the mitochondrial compartment and are essential for genome maintenance in these organelles (39). The conditional lethal phenotype of a cdc9 mutant strain was utilized to identify cDNAs, encoding human DNA ligase I, that permitted growth at the nonpermissive temperature (4). Unlike the yeast CDC9 gene, the human LIG1 gene does not encode a polypeptide that is targeted to mitochondria. In vertebrates, the LIG3 gene encodes the mitochondrial DNA ligase (see below).
As shown schematically in Figure 4, the polypeptides encoded by the human LIG1 and the yeast CDC9 genes contain a highly related C-terminal catalytic core. The adjacent N-terminal regions of these enzymes share very little overall amino acid sequence homology and are not required for catalytic activity (4, 26). This N-terminal region of human DNA ligase I was not present in the catalytic fragment that was crystallized with nicked DNA (26), and it is not required for complementation of yeast cdc9 mutant strains by ligase I. Notably, this region contains sequences that are required for nuclear localization and targeting to replication factories (40–42), and it is involved in protein-protein interactions described below.
The N-terminal region of DNA ligase I is also subject to posttranslational modification. Serine residues at positions 51, 76, and 91 (Figure 4) are phosphorylated by cyclin-dependent kinase (43), and Ser66 is phosphorylated by casein kinase II (44). From low levels in the G1 phase of the cell cycle, phosphorylation increases during S phase, reaching a peak at G2/M (44). Phosphorylation of Ser91 occurs at the G1/S transition and is necessary for subsequent phosphorylation at Ser76 in the hyperphosphorylated G2/M form of DNA ligase I (43). Interestingly, the targeting of DNA ligase I to replication factories was markedly reduced when serine residues 51, 66, 76 and 91 were replaced with aspartic acid (43). Because DNA ligase I is dephosphorylated in response to both DNA damage and replication blockage (45), it appears that the participation of this enzyme in different DNA transactions is regulated by posttranslational modifications that presumably modulate interactions with different protein partners. Recent analyses of the phosphoproteome have identified additional phosphorylation sites in the N-terminal region of DNA ligase I and also in the C-terminal region beyond the catalytic core (46, 47), indicating a complex regulation by post-translational modifications.
DNA ligase I-deficient cell lines have been established from the only known case of a human with mutated LIG1 alleles (48, 49). The clinical symptoms of this individual are described in the next section. One of the mutant LIG1 alleles was inherited, whereas it is not known how the second mutant allele was acquired (48, 49). In the inherited maternal allele, the mutation results in the replacement of a conserved residue, Arg771, with a tryptophan residue within the OB domain. Although this mutant polypeptide is defective in the second step of the ligation reaction, it retains partial activity—about 10% compared with wild-type DNA ligase I (48, 50). In the other mutant allele, the mutation results in the replacement of glutamic acid 566 with a lysine residue. The nonconservative replacement of a conserved residue two amino acids away from the key active-site lysine residue almost certainly results in enzyme inactivation. Notably, although the presumed null allele has been lost from the SV40-immortalized derivative (46BR.1G1) of the primary fibroblast cell line (46BR) established from the DNA ligase I-deficient individual, the primary and immortalized fibroblast cell lines have similar phenotypes (48, 51). The 46BR.1G1 cell line has a modest reduction (about two-fold) in the amount of DNA ligase I protein compared with other SV40-immortalized fibroblasts, but a more dramatic reduction in DNA ligase I activity (10- to 20-fold) (48). In accord with its expected role in DNA replication, the DNA ligase I-deficient cells exhibit a marked defect in Okazaki fragment joining, but unexpectedly, this does not have an obvious effect on cell proliferation (52, 53). It is notable that 46BR.1G1 cells are hypersensitive to 3-amino benzamide (51), an inhibitor of poly(ADP-ribose) polymerase, which participates, together with DNA ligase IIIα, in the repair of DNA single-strand breaks (54).It appears likely that the poly(ADP-ribose) polymerase 1 (PARP1)-dependent pathway acts on single-strand breaks remaining in the lagging strand because of the DNA ligase I deficiency. The 46BR.1G1 cells are hypersensitive to simple DNA-alkylating agents because of a defect in the long-patch subpathway of base excision repair (55). In addition, 46BR cells exhibit modest sensitivity to other DNA-damaging agents, including UV-light and ionizing radiation (48, 49). An interesting, and as yet unexplained, feature of the 46BR.1G1 cells is that they are hypomutable following DNA damage (52).
Because CDC9 is an essential gene in S. cerevisiae, it was assumed that inactivation of the mammalian LIG1 gene would result in cell lethality. This notion was supported by the observation that loss of both LIG1 alleles in mouse embryonic stem cells was only observed in cells ectopically expressing full-length DNA ligase I cDNA (56). Thus, it was surprising that mouse embryos harboring homozygous deletions of the 3′ end of the LIG1 gene developed normally until midgestation (57). It is possible that the embryos were not totally devoid of DNA ligase I activity because the gene-targeting strategy did not disrupt the LIG1 gene by deleting the first exon. However, a similar result was obtained using a different targeting strategy (although again not targeting the first exon) (58). Mouse embryonic fibroblast (MEF) cell lines established from both of these animal models proliferate normally but have a defect in Okazaki fragment joining and increased genomic instability (57, 58). Thus, there is strong evidence that mammalian DNA ligase I is not absolutely required for DNA replication and for the early stages of embryo development. Recently, the Melton laboratory established a mouse knockin model that expresses a mutant lig1 allele corresponding to the 46BR.1G1 mutant allele in which Arg771 is replaced by Trp (59). The phenotype of these animals is described in the next section. As expected, MEFs established from these animals are defective in Okazaki fragment joining (59). Unlike the 46BR.1G1-immortalized human fibroblast cell line, neither these MEFs nor the MEFs established from the lig1 null mice are sensitive to DNA-damaging agents, including DNA-alkylating agents (57–59). It is possible that differences in the relative contribution of the short- and long-patch sub-pathways of base excision repair (BER) underlie the different DNA damage sensitivities of human and mouse cell lines expressing the same mutant version of DNA ligase I.
DNA Ligase III Family
As mentioned previously, the LIG3 gene appears to be restricted to vertebrates. Thus, genetically tractable lower eukaryotes such as S. cerevisiae cannot be used to delineate the cellular functions of the LIG3 gene. Moreover, conclusions regarding the cellular functions of the CDC9 and DNL4 genes, the yeast homologs of mammalian LIG1 and LIG4, cannot be extrapolated to mammalian cells because of their larger repertoire of DNA ligases. The LIG3 gene of higher eukaryotes generates three distinct DNA ligase polyeptides (Figure 5). Nuclear and mitochondrial versions of DNA ligase IIIα are ubiquitously synthesized from DNA ligase IIIα mRNA by an alternative translation initiation mechanism (60, 61). In addition, a germ cell-specific alternative splicing mechanism, in which the terminal 3′-coding exon in DNA ligase IIIα mRNA is replaced by a different exon, generates DNA ligase IIIβ mRNA (61, 62).
The polypeptides encoded by the mammalian LIG3 gene contain a catalytic region that is most closely related to that of the DNA ligases encoded by the pox viruses (5, 6). This level of homology (about 50% sequence identity) extends over the entire length of the vaccinia DNA ligase (Figure 5). As mentioned above, a unique feature of the DNA ligases encoded by the LIG3 gene is an N-terminal zinc finger, which is related to the tandemly arrayed zinc fingers at the N terminus of PARP1 (6). Although the DNA ligase III zinc finger is not required for catalytic activity, efficient DNA joining by DNA ligase III at physiological salt concentrations is dependent upon this DNA-binding region (31).
As mentioned previously, the DNA ligase IIIα and β polypeptides have different C termini (61, 62). The 77 residues that are unique to DNA ligase IIIα constitute a breast and ovarian cancer susceptibility protein 1 C-terminal (BRCT) motif, a conserved sequence found in other proteins involved in cell cycle regulation and DNA repair that was originally identified in the C-terminal region of the breast cancer susceptibility protein BRCA1 (63). In DNA ligase IIIβ, these residues are replaced by a short sequence of 17–18 residues, which acts as a nuclear localization signal (Figure 5). The BRCT motif is often involved in protein-protein interactions and, in the case of the nuclear form of DNA ligase IIIα, it interacts with the DNA repair protein XRCC1, resulting in the formation of a DNA ligase IIIα-XRCC1 complex (62, 64–66). Because XRCC1 is required to stabilize DNA ligase IIIα, XRCC1-deficient cell lines, such as the Chinese hamster ovary cell lines EM9 and EMC11, have reduced levels of DNA ligase IIIα and so are functionally DNA ligase IIIα deficient (67). The phenotype of XRCC1-deficient cell lines includes pronounced hypersensitivity to DNA-alkylating agents, modest hypersensitivity to ionizing radiation, and spontaneously elevated levels of sister chromatid exchange (68). However, it cannot be assumed that the phenotype of DNA ligase III mutant cells will recapitulate that of xrcc1 mutant cells because XRCC1 also functions independently of DNA ligase IIIα in nuclear DNA repair (69) and because DNA ligase IIIα functions independently of XRCC1 in mitochondrial DNA metabolism (70).
Deletion of either the XRCC1 or the LIG3 gene results in early embryonic lethality in the mouse, with the lig3 null embryos surviving 1–2 days longer than the xrcc1 null embryos (71, 72). Attempts to establish MEF cell lines from these embryos have failed. However, the absence of p53 extended embryonic development of xrcc1 null embryos, facilitating the establishment of mouse p53−/− xrcc1−/− embryonic fibroblasts (71) and suggesting the absence of XRCC1 and presumably DNA ligase III, resulted in the persistence of DNA damage that activated p53-dependent pathways for cell cycle checkpoints and/or apoptosis. On the basis of these results, it is evident that XRCC1 is not required for cell viability. Because mitochondrial DNA ligase IIIα functions independently of XRCC1 (70), it is possible that total inactivation of the LIG3 gene will result in cell lethality owing to defects in mitochondrial DNA metabolism. Indeed, using RNAi to reduce the cellular level of DNA ligase IIIα, Campbell and colleagues (73) showed that mitochondrial DNA ligase IIIα is critical for the function of these organelles.
In accord with the hypersensitivity of xrcc1−/− cells to DNA-alkylating agents, there is compelling evidence from biochemical and cell biology studies linking the nuclear DNA ligase IIIα-XRCC1 complex with a subpathway of BER (short patch) that is characterized by a single-nucleotide repair tract (74) and the repair of single-strand breaks (75, 76). More recently, it has been shown that the nuclear DNA ligase IIIα-XRCC1 complex is a key component of a nucleotide excision repair (NER) subpathway that operates in dividing cells and is particularly important for the repair of UV lesions in nondividing cells (77). Thus, in contrast to S. cerevisiae where Cdc9 is the only ligase active in excision repair, both DNA ligase I and DNA ligase IIIα participate in mammalian excision repair. There is also evidence that DNA ligase IIIα is a component of a backup pathway for the major DNA ligase IV-dependent nonhomologous end-joining pathway (78). Finally, it is possible that the presence of DNA ligase IIIα may compensate for the absence of DNA ligase I in lig1 null cells, allowing these cells to replicate and survive. The elucidation of the cellular and organismal roles of the nuclear and mitochondrial DNA ligase IIIα and the germ cell-specific DNA ligase IIIβ will require more sophisticated gene-targeting strategies or specific inhibitors of these enzymes.
DNA ligase IIIα is phosphorylated on Ser123, located between the N-terminal zinc finger and the catalytic region of DNA ligase III (79). This residue is progressively phosphorylated by the cyclin-dependent kinase (Cdk2) as cells progress through S phase, and it is dephosphorylated in response to DNA damage in a reaction dependent upon ATM, the ataxia-telangiectasia-mutated protein (79). The regulation of the steady-state level of phosphorylation by ATM appears to involve both inhibition of Cdk2 activity and activation of a phosphatase, most likely protein phosphatase PP1 (79). Given the marked sensitivity of cell lines deficient in DNA ligase IIIα to DNA-alkylating agents (68), it is surprising that oxidative stress, rather than DNA alkylation damage, is the most effective inducer of DNA ligase IIIα dephosphorylation (79). Analysis of the phosphoproteome has identified additional sites within the N-terminal region of DNA ligase IIIα (47). As described below, the region of DNA ligase III containing the Ser123 phosphorylation site mediates interactions with different protein partners, raising the possibility that these interactions are regulated by posttranslational modification.
DNA Ligase IV Family
In contrast to the other human DNA ligases, the DNA ligase encoded by the LIG4 gene was initially identified by a genomics approach in which a human expressed sequence tags database was searched with a sequence motif that is conserved within the catalytic domain of eukaryotic DNA ligases (6). As shown schematically in Figure 6, the polypeptides encoded by the human LIG4 and the yeast DNL4 genes have an N-terminal catalytic domain with a C-terminal extension that contains tandemly arrayed BRCT motifs (6, 80–82). Because a large fraction of DNA ligase IV molecules purified from cultured cells are adenylated, this enzyme is not robustly labeled in assays measuring enzyme-AMP formation using [α32P]ATP (83). This characteristic, combined with the similar size of the polypeptides encoded by the LIG3 and LIG4 genes, probably explains the failure to detect DNA ligase IV activity prior to its cloning. Another noteworthy feature of DNA ligase IV that distinguishes it from DNA ligases I and III is its ability to join DNA ends that are noncomplementary because of mismatches and/or short gaps (84).
DNA ligase IV interacts with human XRCC4 (85, 86), a protein involved in the repair of DNA double-strand breaks by nonhomologous end joining (NHEJ) and the completion of V(D)J recombination (87). This complex provided the initial insights into the cellular function of DNA ligase IV. Similar to the nuclear form of DNA ligase IIIα (67), DNA ligase IV is unstable in the absence of XRCC4, and so xrcc4 cell lines are functionally deficient in DNA ligase IV activity (88). The stability of DNA ligase IV is also regulated by phosphorylation at Ser650. Notably, DNA ligase IV is phosphorylated by the DNA-dependent protein kinase, DNA-PK, a key NHEJ factor that negatively regulates DNA ligase IV stability (89). As expected, targeted inactivation of LIG4 in mouse and human cells dramatically reduced V(D)J recombination and resulted in hypersensitivity to ionizing radiation because of the defect in the repair of double-strand breaks by NHEJ (90, 91). In addition, MEFs established from lig4 null embryos have a growth defect and reduced life span owing to premature senescence (90). Inactivation of the mouse LIG4 gene results in late embryonic lethality (90, 92). Prior to their demise, the developing lig4 null embryos exhibit defective lymphopoiesis, which presumably reflects their inability to carry out V(D)J recombination, but these embryos appear to die because of massive apoptosis in the central nervous system (90, 92, 93). Notably, the excessive neuronal apoptosis and embryonic lethality, caused by lig4 deficiency but not by the lymphocyte development defects, were rescued by concomitant loss of either p53 or ATM function (35, 93, 94). Thus, it appears that DNA ligase IV is involved in the repair of endogenous DNA damage that if unrepaired elicits apoptosis in neuronal cells via a pathway mediated by p53 and ATM. At the cellular level, a p53 deficiency but not ATM deficiency corrects the proliferation defects in lig4 MEFs (93, 94). The clinical symptoms of the several human individuals with mutant lig4 alleles (95, 96) are described in the section below. As expected, cell lines established from these individuals are highly radiosensitive (95, 96).
In S. cerevisiae, inactivation of the DNL4 gene does not cause significant hypersensensitivity to ionizing radiation unless homology-dependent pathways of DNA double-strand break repair are also inactivated (80–82). This reflects differences in the relative contributions of NHEJ and homology-dependent repair pathways between mammals and S. cerevisiae. In fact, yeast dnl4 strains are more resistant to agents that cause DNA double-strand breaks because Dnl4 not only catalyzes the last step of NHEJ but also suppresses homologous recombination (97).
DNA LIGASES AND HUMAN DISEASE
Since each of the three mammalian LIG genes is required for embryonic development in the mouse (57, 58, 71, 72, 90, 92), it is likely that the mutant lig alleles associated with heritable human syndromes will encode polypeptides having partial activity. As mentioned above, humans with mutant alleles of either LIG1 or LIG4 but not LIG3 have been identified. The clinical symptoms of these individuals and relevant features of DNA ligase-deficient mouse models of DNA ligase deficiency are described below. Finally, we describe emerging evidence linking neurodegeneration to DNA adducts resulting from aborted attempts to join nonligatable DNA strand breaks by DNA ligases.
DNA Ligase I Deficiency
The only known case of a human DNA ligase I deficiency caused by an inherited mutant lig1 allele was diagnosed in a patient with recurrent infections. This symptom resulted from unexplained immunodeficiencies, including hypogammaglobulinemia and poor proliferative response of lymphocytes to mitogens in vitro (48, 49). The female patient also exhibited sensitivity to sunlight, growth retardation, and delayed development. Hepatosplenomegaly was detected at age 17 with further analysis revealing lymphocyte infiltration of portal tracts, indicative of lymphoma. This individual died at age 19 from pneumonia. Although DNA ligase I-deficient cell lines established from this patient exhibit defects in DNA replication and repair (48, 50, 52, 55, 98), there is no defect in V(D)J recombination to explain the immunodeficiency (99, 100). It appears likely that the immunodeficiency was caused by a defect in the proliferation of B and T cells in response to stimuli. With only a single case, it is not possible to fully characterize the clinical symptoms caused by DNA ligase I deficiency in humans. Notably, a mouse model has been established reiterating the Arg771 to Trp change found in the DNA ligase I-deficient human (59). As in the human, this resulted in delayed growth. In young mutant mice, hematopoietic defects were observed, including enlargement of the spleen (59). The mutant mice did not exhibit signs of a compromised immune system, although this may result from housing the animals in a clean environment. Increased levels of spontaneous genomic instability were evident in spleen cells from the mutant mice, and there was also a marked increase in the incidence of spontaneous tumors in the mutant mice (59). Thus, it appears that the DNA replication defect caused by DNA ligase I deficiency in mammals results in delayed growth and increased cancer incidence because of genomic instability. This proliferation defect also appears to underlie abnormalities in hematopoiesis and lymphopoiesis (49, 59).
DNA Ligase IV Deficiency
Five individuals with DNA ligase IV deficiency have been identified (95, 96, 101). These individuals exhibited a range of clinical symptoms (95, 96, 101) that appear to reflect differences in the residual DNA ligase IV activity encoded by the hypomorphic lig4 alleles (102). Notably, reduced DNA ligase IV activity has a more dramatic effect on the repair of double-strand breaks by NHEJ than V(D)J recombination (95, 96). The first case of DNA ligase IV deficiency was detected because of an extreme reaction to radiation treatment for leukemia (96). In contrast to the first DNA ligase IV-deficient individual, who apart from the leukemia was clinically normal, the other four individuals had more severe and diverse symptoms that included distinctive facial features, microcephaly, skin abnormalities, growth and developmental deficiencies, and immunodeficiency (95, 101). In the DNA ligase IV-deficient syndrome, the immunodeficiency is caused by defective V(D)J recombination. There is considerable overlap between the symptoms associated with DNA ligase IV deficiency and those of the DNA damage response disorder, Nijmegen breakage syndrome (95, 103, 104). Although these syndromes are characterized by radiation sensitivity at the cellular level, the radiosensitivity in nbs1 mutant cells is primarily a consequence of a checkpoint defect, whereas it is caused by a defect in the repair of DNA double-strand breaks by NHEJ in lig4 mutant cells (95, 96). At the present time, there are too few cases to determine whether DNA ligase IV deficiency causes cancer predisposition in humans. In the mouse, however, there is evidence that DNA ligase IV deficiency increases cancer formation in nonlymphoid tissues. Specifically, lig4 heterozygosity in the tumor-prone ink4a/arf mouse strain promotes the development of soft tissue sarcomas (105).
DNA Ligation Intermediates in Human Disease
It is evident from the studies described above that DNA ligase deficiency causes genomic instability, which in turn increases susceptibility to cancer. During DNA replication and DNA excision repair, it is possible that DNA ligases will encounter DNA nicks with mismatches because of misincorporation by a DNA polymerase. DNA joining in vitro by DNA ligases I and III is inhibited by 3′ mismatches at DNA nicks (106), providing an opportunity to remove and then replace the mismatched nucleotide. Thus, DNA ligases can actively contribute to the fidelity of DNA replication and repair. DNA ligases may also encounter damaged DNA termini, such as those generated when oxygen free radicals cause a DNA strand break. Recent work by West and colleagues (9) has implicated an intermediate of the DNA ligation reaction, the DNA adenylate, in the pathology of certain human neurodegenerative diseases. Specifically, they have shown that the DNA adenylate intermediate accumulates when DNA ligase IIIα attempts to join DNA strand breaks with modified termini, such as those generated by oxygen free radicals, and following abasic site cleavage by the major human AP endonuclease (9, 11). Under normal circumstances, the adenylate group is removed by aprataxin (9), which is linked to DNA ligase IIIα by a physical interaction with its partner XRCC1 (107). In addition, aprataxin also interacts with XRCC4, the partner protein of DNA ligase IV (108). The gene encoding aprataxin is mutated in the inherited human neurological disorder ataxia oculomotor apraxia-1, characterized by early-onset cerebellar ataxia, oculomotor apraxia, and late peripheral neuropathy (12, 13). Because aprataxin appears to be the only enzyme that can remove an adenylate group from a 5′ phosphate terminus, the loss of aprataxin is particularly deleterious in postmitotic cells, such as those of the adult nervous system, owing to their more limited repertoire of DNA repair pathways compared with proliferating cells. Attempts to ligate DNA breaks with damaged termini in the absence of aprataxin results in the accumulation of the unrepairable and cytotoxic DNA adenylate in postmitotic neuronal cells (9).
PROTEIN PARTNERS OF DNA LIGASES
The unique regions flanking the conserved catalytic domain of eukaryotic DNA ligases (Figures 4–6) mediate protein-protein interactions that target these isozymes to different DNA transactions. In the following sections, we describe the various protein partners of eukaryotic DNA ligases and their role in directing the cellular functions of these enzymes.
DNA Ligase I
Several DNA ligase I-interacting proteins have been identified by DNA ligase I affinity chromatography (45, 109, 110). Among these interacting proteins are two DNA-sliding clamps, the homotrimeric proliferating cell nuclear antigen (PCNA) and the heterotrimeric hRad9-hRad1-hHus1 (9-1-1) complex, which are involved in DNA replication and cell cycle checkpoints, respectively. The interaction with PCNA but not 9-1-1 is primarily mediated by a conserved PCNA-binding motif, or PIP box, at the N terminus of DNA ligase I (45, 55). This same region is required for the targeting of DNA ligase I to replication factories (Figure 4). The interaction with PCNA is required for localization of DNA ligase I at the sites of DNA replication because inactivation of PCNA binding also abolishes targeting to replication factories (41). As with other PCNA interacting proteins, the PIP box of human DNA ligase I and yeast Cdc9 DNA ligase binds to the interdomain connector loop (IDCL) of PCNA and, at least for Cdc9, involves formation of an additional β−zipper structure between residues adjacent to the PIP box and residues within the C-terminal domain of PCNA (111). This interaction is necessary for complex formation both in the absence of DNA and when PCNA is topologically linked to DNA. The DNA ligase I–PCNA complex formed on DNA contains one molecule of DNA ligase I per PCNA trimer, suggesting that the binding of DNA ligase I to one PCNA monomer occludes the IDCL-binding sites on the other two PCNA subunits of the trimer (109). Inactivation of the DNA ligase I PIP box not only abolishes the targeting of DNA ligase I to replication foci but also the complementation of the Okazaki fragment joining and long-patch base excision repair defects in the DNA ligase I-deficient cell line 46BR.1G1 (41, 55).
The similarity in the size and shape of the rings formed by PCNA and the catalytic fragment of DNA ligase I suggested that the PCNA ring encircling DNA may facilitate the transition of DNA ligase I from an extended conformation into the ring shape via interactions between one of the surfaces of the PCNA ring and regions within the catalytic fragment of DNA ligase I (26). In accord with this idea, a direct interaction between the DBD of DNA ligase I and PCNA was detected (112). Thus, it appears that DNA ligase I initially docks with a PCNA trimer via the interaction between the DNA ligase I PIP box and the IDCL of one of the PCNA monomers (Figure 7a). We envision that the region adjacent to the PIP box is flexible, allowing the catalytic fragment to engage both the PCNA ring and DNA. Despite these protein-protein interaction interfaces, a large molar excess of PCNA is required for significant stimulation of DNA ligase I activity (113). In contrast, the heterotrimeric 9-1-1 clamp is much more effective at stimulating DNA ligase I catalytic activity (114, 115). Because the heterotrimeric PCNA clamp of Sulfolbus solfataricus also stimulates its cognate DNA ligase (116), the ability to stimulate DNA ligation appears to be a common feature of heterotrimeric clamps. Although the mechanism by which the heterotrimeric clamps stimulate DNA joining has not been elucidated, it does not require the topological linkage of the clamp to DNA (115). Interactions between heterotrimeric DNA sliding clamps and the DBD domain of DNA ligases have varying degrees of specificity. The Sulfolobus DNA ligase preferentially interacts with one subunit of the heterotrimeric clamp, whereas human DNA ligase I interacts similarly with each of the 9-1-1 subunits (45, 116). Small-angle X-ray scattering studies with the Sulfolobus ligase and PCNA revealed that the DNA ligase remains in an extended conformation when it is complexed with the heterotrimeric PCNA in the absence of DNA (112). Thus, when the DBD domain of DNA ligase I engages the PCNA ring after the initial PIP box-mediated docking, the catalytic region may still be in an extended conformation (Figure 7b). In this scenario, the conversion of the catalytic fragment presumably involves additional contacts with nicked DNA (Figure 7c).
Figure 7.
Model for the interaction of human DNA ligase I with DNA-linked homotrimeric proliferating cell nuclear antigen (PCNA). (a) DNA ligase I in an elongated conformation docks onto the PCNA ring via an interaction between the N-terminal PIP box of DNA ligase I (light blue) and the interdomain connector loop of a PCNA monomer. (b) The initial docking by the PIP box that is flexibly linked to the catalytic core of ligase I facilitates an interaction between the DNA-binding domain (DBD) (red) domain and the PCNA ring. At this stage, the catalytic region remains in an extended conformation. (c) Subsequent interactions with nicked DNA orchestrate the transition of the catalytic region of DNA ligase I from the extended to a closed ring conformation with each of the domains, DBD (red), adenylation (green), and OB-fold (yellow) contacting the DNA. This model is based on structural studies of human ligase I bound to DNA (26) and the Sulfolobus DNA ligase complexed with PCNA (112).
The PCNA and 9-1-1 clamps are loaded onto DNA in an ATP-dependent reaction catalyzed by replication factorC (RFC) and by an alternative RFC complex containing the human homolog of the Schizosaccharomyces pombe Rad17 protein (hRad17), respectively (117). DNA ligase I interacts with the unique large subunits, RFC p140 and hRad17, of these alternative RFC heteropentamers, which share in common the four small subunits (45, 110). RFC inhibits DNA ligase I, although preincubation of RFC with PCNA alleviates this inhibition, provided that DNA ligase I has a functional PIP box (110). Similar physical and functional interactions occur among the S. cerevisiae homologs, suggesting that this interplay is biologically relevant (111). On the basis of these results, DNA ligase I may be held in an inactive complex with RFC before it is delivered to a PCNA trimer located at a DNA nick that has already been processed by DNA replication and/or excision repair activities. Alternatively, DNA ligase I may interact with a complex of RFC and PCNA left behind at the DNA nick after gap-filling synthesis and end processing. Finally, it is possible that ligation by DNA ligase I is the signal for unloading of the PCNA trimer, thereby recycling PCNA for subsequent Okazaki fragments or repair events. In contrast to the inhibitory effect of RFC, the checkpoint clamp loaders (hRad17-RFC and S. cerevisiae Rad24-RFC) stimulate DNA joining by the replicative DNA ligase of humans and yeast (45). Further work is needed to elucidate the functional and biological relevance of the interactions between DNA ligase I and the clamp loaders.
In addition to interacting with DNA sliding clamps and clamp loaders, DNA ligase I also interacts with DNA polymerase β within a base excision repair complex that was purified from bovine testis (118). This appears to be an alternative version of the major DNA ligase IIIα-dependent short-patch BER pathway that may operate in specific tissues and/or cell types.
DNA Ligase III
As mentioned above, the LIG3 gene encodes four different polypeptides (60, 62). To date, no unique protein partners have been identified for the germ cell-specific isoform, DNA ligase IIIβ, which lacks the C-terminal BRCT motif of polypeptides encoded by the ubiquitously expressed DNA ligase IIIα mRNA (62). The mitochondrial and nuclear versions of DNA ligase IIIα are distinguished by an N-terminal mitochondrial targeting sequence that is proteolytically removed during transport into mitochondria (60). Although XRCC1 forms a complex with nuclear DNA ligase IIIα (64), it is absent from mitochondria (70). Within these organelles, DNA ligase IIIα interacts with DNA polymerase γ, the mitochondrial DNA polymerase (119). This interaction, which presumably integrates DNA ligase IIIα into mitochondrial DNA replication and repair, is not mediated by the BRCT domain of DNA ligase IIIα but instead involves the catalytic region (119).
The discovery of the interaction between XRCC1 and nuclear DNA ligase IIIα (64), which occurs between the C-terminal BRCT domains of these proteins (Figure 8), provided the first clues as to the cellular functions of the polypeptides encoded by the LIG3 gene. Moreover, the structure of the C-terminal BRCT domain of XRCC1 was the first of its type to be determined and has provided the framework for studying the function of this domain in other proteins, including DNA repair and cell cycle regulatory proteins as well as the amino acid changes in the BRCT domain of the BRCA1 protein that are linked to breast cancer (120). In addition to DNA ligase IIIα, XRCC1 interacts with several other proteins involved in base excision and single-strand break repair (Figure 8), suggesting that XRCC1 may act as a scaffold for the assembly of multiprotein repair complexes. By interacting with the hOGG1 DNA glycosylase (121),AP endonuclease (122), and DNA polymerase β (123) in addition to DNA ligase IIIα, XRCC1 potentially contributes at every step of the short-patch base excision repair pathway in which a single nucleotide is inserted in place of the damaged base (123). In vivo XRCC1 is recruited to single-strand breaks by an interaction with automodified PARP1 (75). Polynucleotide kinase (PNK) (124), aprataxin (107), and aprataxin- and PNK-like factor (APLF) (125, 126) share a similar fork head-associated (FHA) domain that interacts with phosphorylated XRCC1. All of these proteins appear to contribute to the processing of DNA termini during single-strand break repair.
Figure 8.
Protein partners of the DNA ligase IIIα/XRCC1 complex. Diagram showing the regions of the DNA ligase IIIα (pink) and XRCC1 (blue) that are involved in interactions with other DNA repair proteins (54, 62, 107, 121–126). Abbreviations: APE1, apurinic/apyrimidinic site endonuclease 1; APLF, aprataxin- and PNK-like factor; BRCT I and II, breast and ovarian cancer susceptibility protein 1 C-terminal domains I and II; DBD, DNA-binding domain; DNA Polβ, DNA polymerase β; NEIL1/2, nei endonuclease VIII-like 1/2; NLS, nuclear localization signal; NTD, nucleotidyl transferase domain; OGG1, 8-oxoguanosine DNA glycosylase; PARP1, poly(ADP-ribose) polymerase 1; PCNA, proliferating cell nuclear antigen; PNK, polynucleotide kinase; TDP1, tyrosyl-DNA phosphodiesterase 1; XRCC1, X-ray cross-complementing 1; ZnF, zinc finger.
Similar to XRCC1, DNA ligase IIIα preferentially interacts with automodified PARP1 and is recruited to DNA single-strand breaks (76). As mentioned above, DNA ligase IIIα is associated with aprataxin, a DNA repair protein that protects against neurodegeneration (9, 107). DNA ligase IIIα also directly interacts with TDP1, a protein involved in the repair of covalent toposiomerase I-DNA adducts and DNA single-strand breaks generated by reactive oxygen species (127). Notably, TDP1 is defective in the neurodegenerative disease spinocerebellar ataxia with axonal neuropathy-1 (127), a second example emphasizing the importance of the DNA ligase IIIα-dependent repair of DNA single-strand breaks generated by reactive oxygen species in neuronal cells. In addition, DNA ligase IIIα interacts directly with the NEIL1 and NEIL2DNA glycosylases that repair oxidized bases in a subpathway of BER, which is dependent upon PNK but not APE1 (128, 129). This complex network of protein-protein interactions involving nuclear DNA ligase IIIα and XRCC1 raises many questions because XRCC1 has repair functions that are independent of DNA ligase IIIα (69). Although XRCC1 and DNA ligase IIIα participate together in different nuclear DNA repair pathways, including single-strand break repair and BER subpathways, it appears likely that different overlapping subsets of the interacting proteins will define the various repair pathways. Indeed, there is evidence that the DNA ligase IIIα-XRCC1 complex is present in different larger multiprotein complexes (107), indicating that XRCC1 and DNA ligase IIIα can simultaneously interact with several repair proteins and suggesting that some of the repair transactions may be carried out by preexisting complexes. Alternatively, repair reactions may be coordinated by sequential handovers of intermediates between repair proteins that were recruited individually to the sites of damage. In particular, this is likely to be the case for PNK, aprataxin, and APLF, which are all involved in cleaning termini at DNA breaks but have overlapping binding sites onXRCC1 (107, 124–126) (Figure 8).
DNA Ligase IV
As mentioned above, DNA ligase IV is unstable in the absence of its partner protein XRCC4 (88). Although the C-terminal extension from the catalytic region of DNA ligase IV contains two adjacent BRCT domains (Figure 6), it is the linker sequence between the BRCT domains that interacts with XRCC4 (130). Interestingly, XRCC4 resembles structural maintenance of chromosome proteins in that it has an N-terminal globular head and a C-terminal coiled-coil domain (131). Although XRCC4 alone forms dimers and tetramers, structural studies indicate that a single DNA ligase IV polypeptide interacts with the dimeric form in the DNA ligase IV-XRCC4 complex (28, 131). Within the XRCC4 dimer, there are interactions between the globular head domain and the regions of the coiled-coil tail adjacent to the head domains (131). The linker region between the BRCT domains of DNA ligase IV binds asymmetrically to the same region of the two coiled coils and induces a large change in the arrangement of the coiled coils (28, 132). To date, there are no available structures of fragments of DNA ligase IV containing the catalytic region, so the position of the catalytic domains relative to the C-terminal BRCT domains and XRCC4 is not known.
Unlike DNA ligase I and nuclear DNA ligase IIIα, which participate in several DNA repair pathways, DNA ligase IV appears to function only in NHEJ, except in specialized cells of the immune system where it also completes V(D)J recombination (91). As shown in Figure 9a, there are physical and functional interactions between DNA ligase IV-XRCC4 and both the Ku proteins and the catalytic subunit of DNA-dependent protein kinase (DNA PKcs) in NHEJ complexes (133–136). In humans, DNA PK mediates end bridging or synapsis of two DNAs (137), an activity that promotes intermolecular joining by DNA ligase IV-XRCC4 (134). The ends at most DNA double-strand breaks generated in vivo will require processing prior to ligation. In Figure 9b, interactions between DNA ligase IV-XRCC4 and various end-processing factors are shown. Notably, DNA ligase IV-XRCC4 associates with the Pol X family polymerases, Pol μ and Pol λ, as well as with terminal transferase (138, 139). These versatile enzymes fill in the short gaps generated during DNA end alignment and processing. In some circumstances, they add nontemplated nucleotides, suggesting that changes in nucleotide sequence at the break site are tolerated as long as the ends are joined. The joining of incompatible DNA ends has been reconstituted with these purified proteins plus Artemis, a nuclease that interacts with DNA PKcs (138, 140). DNA ligase IV-XRCC4 contributes to this activity by joining two aligned DNA ends having a short gap in one strand or mismatched termini (84). Interestingly, the recently identified NHEJ factor Cernunnos/XLF, which interacts with XRCC4 (141, 142), stimulates the joining of incompatible DNA ends by DNA ligase IV-XRCC4 (143). The DNA ligase IV-XRCC4 complex also interacts with the end-processing activities, PNK (144), and aprataxin (108).
Figure 9.
DNA end-bridging and end-processing complexes formed by human NHEJ proteins. (a) A model of the end-bridging complex formed by heterodimeric Ku, DNA PKcs/Artemis, and DNA ligase IV/XRCC4. (b) Diagram showing the protein-protein interactions among the NHEJ factors involved in the end-processing and ligation steps of NHEJ (130–142). Pol X refers to Pol λ, Pol μ, and terminal transferase (138, 139). Abbreviations: DNA PKcs, catalytic subunit of the DNA-dependent protein kinase; PNK, polynucleotide kinase; X4, X-ray cross-complementing 4 or XRCC4; XLF, XRCC4-like factor. This protein is also known as Cernunnos.
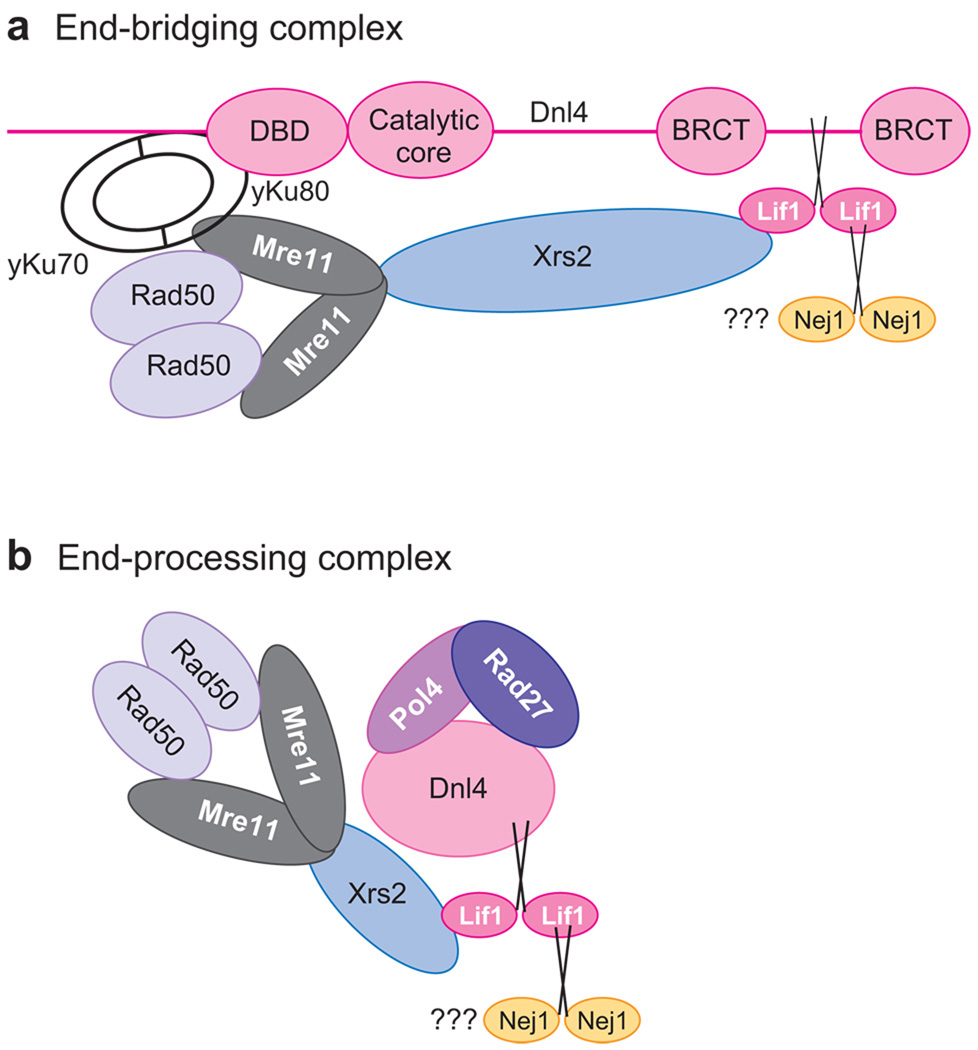
In S. cerevisiae, Dnl4-Lif1, which is the homolog of human DNA ligase IV-XRCC4, interacts with yeast Ku, and as with the homologous human proteins, yeast Ku recruits Dnl4-Lif1 to DNA ends in vitro (97, 145). The major difference between the primary NHEJ pathways of humans and S. cerevisae is the absence of a yeast homolog of DNA PKcs. In the yeast pathway, the Rad50/Mre11/Xrs2 complex is the end-bridging factor (146). Rad50/Mre11/Xrs2 interacts with and specifically stimulates intermolecular joining by Dnl4/Lif1 (146). At physiological salt concentrations, the joining of linear DNA molecules with DNA ends by Rad50/Mre11/Xrs2, Dnl4/Lif1, and Ku is dependent upon pair-wise, species-specific protein-protein interactions among these factors (Figure 10a). In addition, as shown in Figure 10b, Dnl4/Lif1 interacts with the processing activities, Pol4, and FEN-1 (Rad27) (147), as well as with Nej1, an ortholog of human Cernunnos/XLF (148–150).
Figure 10.
End-bridging and end-processing complexes formed by yeast nonhomologous end-joining (NHEJ) proteins. (a) Diagram showing the protein-protein interaction among the core NHEJ factors, heterodimeric Ku, Dnl4/Lif1, and Mre11/Rad50/Xrs2. (b) Diagram showing the protein-protein interactions among the NHEJ factors involved in the end-processing and ligation steps of NHEJ. The role of Nej1, an ortholog of human XLF/Cernunnos, has yet to be determined (148–150). Abbreviations: BRCT, breast and ovarian cancer susceptibility protein 1 C-terminal; DBD, DNA-binding domain; Dnl4, homolog of human DNA ligase IV; Lif1, ligase interacting factor 1, an ortholog of human XRCC4; Mre11, Rad50, Xrs2, subunits of the Mre11/Rad50/Xrs2 protein complex that processes double-strand DNA breaks to enable repair; yKu70 and yKu80, homologs of human Ku70 and Ku80.
Although DNA ligase IV/XRCC4 catalyzes the last step in the major NHEJ pathway in humans, there is emerging evidence from in vitro and in vivo studies that this complex acts at earlier stages in this repair pathway and plays a key structural role in the assembly of functional NHEJ complexes. Studies by the Chu lab (151) with fractionated cell extracts have shown that DNA ligase IV/XRCC4 is required for processing of DNA molecules with incompatible ends. This implies that DNA ligase IV/XRCC4 acts prior to and is necessary for the subsequent recruitment of end-processing activities, providing an explanation as to why DNA molecules with cohesive ligatable ends are predominantly joined without processing (152). Although Ku is required for the recruitment of DNA ligase IV/XRCC4 to double-strand breaks, there are contradictory reports as to the role DNA PKcs in this recruitment (153, 154). In yeast, the recruitment of Dnl4/Lif1 is dependent upon Ku but not Rad50/Mre11/Xrs2 (97). Interestingly, the binding of Ku to in vivo double-strand breaks is dynamic (97, 154) and is stabilized, at least in yeast, by Dnl4/Lif1 (97). Regardless of the DNA PKcs contribution, it is evident that the DNA ligase that completes NHEJ has an earlier role in engaging the ends of the DNAs to be joined. The interaction of either DNA PKcs or DNA ligase IV/XRCC4 with Ku at a DNA end results in the translocation of Ku along the DNA away from the end (133, 155). This presents a problem because Ku is a ring that slides onto the DNA end (156), so ligation is likely to result in the trapping of Ku molecules on the DNA. Given the intertwining of the subunits of the Ku ring (156), a proteolytic mechanism, perhaps triggered by the ligation step of NHEJ, may be required to remove Ku from the ligated DNA.
CONCLUDING REMARKS
DNA ligases are attractive enzymes to study because of the almost ubiquitous requirement for DNA joining in replication, repair, and recombination. Although the three steps of the ligation reaction were described 40 years ago, it is only recently that we have gained significant insights into the molecular mechanism of this fundamentally important reaction through a rapidly growing number of published DNA ligase structures. The cloning of three human LIG3 genes in the 1990s prompted studies to determine the cellular functions of the different species of DNA ligase. This was particularly challenging for the DNA ligases encoded by LIG3 because this gene is not present in the lower eukaryotes that are commonly used as model organisms because of their ease of genetic manipulation. Indeed, there is emerging evidence that DNA ligase III isozymes may participate in a broader range of DNA transactions than DNA ligases I and IV.
There is a growing number of DNA replication and repair proteins that have been shown to interact with one or more of the eukaryotic DNA ligases. Further work is needed to delineate the functional and biological significance of these interactions. The importance of understanding the mechanism and regulation of DNA ligation is highlighted by the link between defects in DNA ligation and human diseases, including cancer and neurodegeneration. Finally, with availability of structural information and our current understanding of the cellular functions of human DNA ligases, this research area is poised for the development of DNA ligase inhibitors, which will not only be useful for delineating catalytic mechanism and cellular functions, but may also have utility as cancer therapeutics.
SUMMARY POINTS.
Eukaryotic and bacterial DNA ligases encircle their DNA substrates, stabilizing a distorted conformation of the DNA that exposes the ligatable ends to the active site.
DNA ligases change conformation during the DNA joining reaction in order to accommodate multiple reactions with the nucleotide and DNA substrates (Figure 1). The multidomain architecture of these enzymes provides the necessary flexibility and probably enables loading on and off of the DNA.
There are three families of eukaryotic DNA ligases. Members of the DNA ligase I and IV families are found in all eukaryotes, whereas DNA ligase III family members are restricted to vertebrates.
Members of the three families of eukaryotic DNA ligases are directed to participate in replication, repair, and recombination pathways by specific protein-protein interactions.
DNA ligase deficiences are associated with genomic instability.
In mammals, deficiencies in either DNA ligase I or DNA ligase IV result in an increased incidence of cancer.
Attempts by vertebrate-specific DNA ligase IIIα to repair single-strand breaks with damaged termini generate DNA lesions that, if unrepaired, are cytotoxic to neuronal cells and cause neurodegeneration.
FUTURE ISSUES.
The influence of protein-protein interactions on the conformation and catalytic activities of DNA ligases within multiprotein complexes is an active area of investigation. These protein interactions are generally thought to be important for cellular localization and for coordinating the activities of multiple enzymes acting in a repair or replication pathway. The interactions of DNA ligases with partner proteins may also modify intrinsic DNA-joining activity.
Although significant progress has been made in defining the interactions between DNA ligase I and DNA sliding clamps, important questions remain. What are the relative orientations of the DNA ligase and clamp rings in the complex formed on nicked DNA, and does the clamp contribute to the transition of the DNA ligase from an extended to a closed conformation? What is the mechanism by which heterotrimeric clamps stimulate DNA joining?
DNA ligase I interacts with the clamp loaders involved in DNA replication and cell cycle checkpoints. Further work is needed to determine the functional and biological relevance of these interactions and the regulation of these interactions by posttranslational modification of DNA ligase I.
The vertebrate-specific LIG3 gene encodes multiple DNA ligase polypeptides. Additional studies are needed to elucidate the roles of these enzymes.
There is emerging evidence that DNA ligase IV family members not only catalyze the last step of DNA double-strand break repair but also play an important structural role at an early stage when the repair complex is being assembled on the DNA end. Further studies are needed to define the assembly and architecture of these repair complexes formed on DNA ends.
ACKNOWLEDGMENTS
We are grateful to members of the Ellenberger and the Tomkinson labs and in particular to Melissa Hefferin and Elizabeth Cotner-Gohara for their help with the preparation of this manuscript and the figures. We apologize to our colleagues that much of the primary literature on DNA ligases could not be cited because of space limitations. Studies on DNA ligases in the Tomkinson and Ellenberger laboratories are supported by the Structural Cell Biology of DNA Repair Program Grant (P01 CA92584) and other research grants from the National Institutes of Health (GM47251, GM57479, and ES12512 to A.E.T. and GM52504 to T.E.). T.E. is the Raymond H. Wittcoff Professor at Washington University School of Medicine, St. Louis.
Glossary
- Nucleotidyltransferases (NTases)
a superfamily of phosphotransferase enzymes with conserved sequence motifs that includes DNA and RNA ligases and mRNA capping enzymes (15)
- Nick sensing
the interaction of a ligase with a nicked DNA substrate or other interruption in the DNA backbone
- Zinc finger
a zinc-binding motif present in ligase III that binds specifically to DNA nicks
- BER
base excision repair
- BRCT
breast and ovarian cancer susceptibility protein 1 C-terminal
- NHEJ
nonhomologous end joining
- DNA-sliding clamp
ring-shaped protein complexes consisting of two or three subunits that encircle duplex DNA, allowing the protein to slide freely
- PCNA
proliferating cell nuclear antigen
- IDCL
interdomain connector loop
- DNA end bridging or synapsis
apposition and alignment of DNA ends through the interactions of bound proteins
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any biases that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Engler MJ, Richardson CC. In: The Enzymes. Boyer PD, editor. New York: Academic; 1982. pp. 3–29. [Google Scholar]
- 2.Lehman IR. Science. 1974;186:790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T, Barnes DE. Annu. Rev. Biochem. 1992;61:251–281. doi: 10.1146/annurev.bi.61.070192.001343. [DOI] [PubMed] [Google Scholar]
- 4.Barnes DE, Johnston LH, Kodama K, Tomkinson AE, Lasko DD, Lindahl T. Proc. Natl. Acad. Sci. USA. 1990;87:6679–6683. doi: 10.1073/pnas.87.17.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Tomkinson AE, Ramos W, Mackey ZB, Danehower S, et al. Mol. Cell. Biol. 1995;15:5412–5422. doi: 10.1128/mcb.15.10.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei YF, Robins P, Carter K, Caldecott KW, Papin DJC, et al. Mol. Cell. Biol. 1995;15:3206–3216. doi: 10.1128/mcb.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson A, Day J, Bowater R. Mol. Microbiol. 2001;40:1241–1248. doi: 10.1046/j.1365-2958.2001.02479.x. [DOI] [PubMed] [Google Scholar]
- 8.Sriskanda V, Shuman S. Nucleic Acids Res. 1998;26:525–531. doi: 10.1093/nar/26.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, et al. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 10.el-Khamisy SF, Caldecott KW. Neuroscience. 2007;145:1260–1266. doi: 10.1016/j.neuroscience.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 11.Rass U, Ahel I, West SC. J. Biol. Chem. 2007;282:9469–9474. doi: 10.1074/jbc.M611489200. [DOI] [PubMed] [Google Scholar]
- 12.Date H, Onodera O, Tanaka H, Iwabuchi K, Uekawa K, et al. Nat. Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 13.Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, et al. Nat. Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 14.Nandakumar J, Shuman S, Lima CD. Cell. 2006;127:71–84. doi: 10.1016/j.cell.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 15.Shuman S, Schwer B. Mol. Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 16.Nair PA, Nandakumar J, Smith P, Odell M, Lima CD, Shuman S. Nat. Struct. Mol. Biol. 2007;14:770–778. doi: 10.1038/nsmb1266. [DOI] [PubMed] [Google Scholar]
- 17.Odell M, Malinina L, Sriskanda V, Teplova M, Shuman S. Nucleic Acids Res. 2003;31:5090–5100. doi: 10.1093/nar/gkg665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Odell M, Sriskanda V, Shuman S, Nikolov DB. Mol. Cell. 2000;6:1183–1193. doi: 10.1016/s1097-2765(00)00115-5. [DOI] [PubMed] [Google Scholar]
- 19.Sriskanda V, Shuman S. Nucleic Acids Res. 1998;26:3536–3541. doi: 10.1093/nar/26.15.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sriskanda V, Shuman S. Nucleic Acids Res. 1998;26:4618–4625. doi: 10.1093/nar/26.20.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho CK, Van Etten JL, Shuman S. J. Virol. 1997;71:1931–1937. doi: 10.1128/jvi.71.3.1931-1937.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hakansson K, Doherty AJ, Shuman S, Wigley DB. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 23.Hakansson K, Wigley DB. Proc. Natl. Acad. Sci. USA. 1998;95:1505–1510. doi: 10.1073/pnas.95.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murzin AG. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nandakumar J, Nair PA, Shuman S. Mol. Cell. 2007;26:257–271. doi: 10.1016/j.molcel.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 26.Pascal JM, O’Brien PJ, Tomkinson AE, Ellenberger T. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 27.Akey D, Martins A, Aniukwu J, Glickman MS, Shuman S, Berger JM. J. Biol. Chem. 2006;281:13412–13423. doi: 10.1074/jbc.M513550200. [DOI] [PubMed] [Google Scholar]
- 28.Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, et al. Nat. Struct. Biol. 2001;8:1015–1019. doi: 10.1038/nsb725. [DOI] [PubMed] [Google Scholar]
- 29.Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. Chem. Rev. 2006;106:687–699. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- 30.Taylor RM, Whitehouse CJ, Caldecott KW. Nucleic Acids Res. 2000;28:3558–3563. doi: 10.1093/nar/28.18.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackey ZB, Niedergang C, Murcia JM, Leppard J, Au K, et al. J. Biol. Chem. 1999;274:21679–21687. doi: 10.1074/jbc.274.31.21679. [DOI] [PubMed] [Google Scholar]
- 32.Kulczyk AW, Yang JC, Neuhaus D. J. Mol. Biol. 2004;341:723–738. doi: 10.1016/j.jmb.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 33.Taylor RM, Whitehouse J, Cappelli E, Frosina G, Caldecott KW. Nucleic Acids Res. 1998;26:4804–4810. doi: 10.1093/nar/26.21.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deleted in proof
- 35.Lee JY, Chang C, Song HK, Moon J, Yang JK, et al. EMBO J. 2000;19:1119–1129. doi: 10.1093/emboj/19.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartwell LH. Bacteriol. Rev. 1974;38:164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnston LH. Mol. Gen. Genet. 1979;170:89–92. doi: 10.1007/BF00268583. [DOI] [PubMed] [Google Scholar]
- 38.Johnston LH, Nasmyth KA. Nature. 1978;274:891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- 39.Willer M, Rainey M, Pullen T, Stirling CJ. Curr. Biol. 1999;9:1085–1094. doi: 10.1016/s0960-9822(99)80477-1. [DOI] [PubMed] [Google Scholar]
- 40.Cardoso MC, Joseph C, Rahn HP, Reusch R, Nadal-Ginard B, Leonhardt H. J. Cell Biol. 1997;139:579–587. doi: 10.1083/jcb.139.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montecucco A, Rossi R, Levin DS, Gary R, Park MS, et al. EMBO J. 1998;17:3786–3795. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montecucco A, Savini E, Weighardt F, Rossi R, Ciarrocchi G, et al. EMBO J. 1995;14:5379–5386. doi: 10.1002/j.1460-2075.1995.tb00222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrari G, Rossi R, Arosio D, Vindigni A, Biamonti G, Montecucco A. J. Biol. Chem. 2003;278:37761–37767. doi: 10.1074/jbc.M304462200. [DOI] [PubMed] [Google Scholar]
- 44.Rossi R, Villa A, Negri C, Scovassi I, Ciarrocchi G, et al. EMBO J. 1999;18:5745–5754. doi: 10.1093/emboj/18.20.5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song W, Levin DS, Varkey J, Post S, Bermudez VP, et al. J. Biol. Chem. 2007;282:22721–22730. doi: 10.1074/jbc.M703774200. [DOI] [PubMed] [Google Scholar]
- 46.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, et al. Proc. Natl. Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, et al. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 48.Barnes DE, Tomkinson AE, Lehmann AR, Webster AD, Lindahl T. Cell. 1992;69:495–503. doi: 10.1016/0092-8674(92)90450-q. [DOI] [PubMed] [Google Scholar]
- 49.Webster AD, Barnes DE, Arlett CF, Lehmann AR, Lindahl T. Lancet. 1992;339:1508–1509. doi: 10.1016/0140-6736(92)91266-b. [DOI] [PubMed] [Google Scholar]
- 50.Prigent C, Satoh MS, Daly G, Barnes DE, Lindahl T. Mol. Cell. Biol. 1994;14:310–317. doi: 10.1128/mcb.14.1.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lehmann AR, Willis AE, Broughton BC, James MR, Steingrimsdottir H, et al. Cancer Res. 1988;48:6343–6347. [PubMed] [Google Scholar]
- 52.Henderson LM, Arlett CF, Harcourt SA, Lehmann AR, Broughton BC. Proc. Natl. Acad. Sci. USA. 1985;82:2044–2048. doi: 10.1073/pnas.82.7.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lonn U, Lonn S, Nylen U, Winblad G. Carcinogenesis. 1989;10:981–985. doi: 10.1093/carcin/10.6.981. [DOI] [PubMed] [Google Scholar]
- 54.Leppard JB, Dong Z, Mackey ZB, Tomkinson AE. Mol. Cell. Biol. 2003;23:5919–5927. doi: 10.1128/MCB.23.16.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Curr. Biol. 2000;10:919–922. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- 56.Petrini JH, Xiao Y, Weaver DT. Mol. Cell. Biol. 1995;15:4303–4308. doi: 10.1128/mcb.15.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bentley D, Selfridge J, Millar JK, Samuel K, Hole N, et al. Nat. Genet. 1996;13:489–491. doi: 10.1038/ng0896-489. [DOI] [PubMed] [Google Scholar]
- 58.Bentley DJ, Harrison C, Ketchen AM, Redhead NJ, Samuel K, et al. J. Cell Sci. 2002;115:1551–1561. doi: 10.1242/jcs.115.7.1551. [DOI] [PubMed] [Google Scholar]
- 59.Harrison C, Ketchen AM, Redhead NJ, O’Sullivan MJ, Melton DW. Cancer Res. 2002;62:4065–4674. [PubMed] [Google Scholar]
- 60.Lakshmipathy U, Campbell C. Mol. Cell. Biol. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perez-Jannotti RM, Klein SM, Bogenhagen DF. J. Biol. Chem. 2001;276:48978–48987. doi: 10.1074/jbc.M107177200. [DOI] [PubMed] [Google Scholar]
- 62.Mackey ZB, Ramos W, Levin DS, Walter CA, McCarrey JR, Tomkinson AE. Mol. Cell. Biol. 1997;17:989–998. doi: 10.1128/mcb.17.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 64.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. Mol. Cell. Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dulic A, Bates PA, Zhang X, Martin SR, Freemont PS, et al. Biochemistry. 2001;40:5906–5913. doi: 10.1021/bi002701e. [DOI] [PubMed] [Google Scholar]
- 66.Nash RA, Caldecott KW, Barnes DE, Lindahl T. Biochemistry. 1997;36:5207–5211. doi: 10.1021/bi962281m. [DOI] [PubMed] [Google Scholar]
- 67.Caldecott KW, Tucker JD, Stanker LH, Thompson LH. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson LH, Brookman KW, Jones NJ, Allen SA, Carrano AV. Mol. Cell. Biol. 1990;10:6160–6171. doi: 10.1128/mcb.10.12.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taylor RM, Moore DJ, Whitehouse J, Johnson P, Caldecott KW. Mol. Cell. Biol. 2000;20:735–740. doi: 10.1128/mcb.20.2.735-740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lakshmipathy U, Campbell C. Nucleic Acids Res. 2000;28:3880–3886. doi: 10.1093/nar/28.20.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, et al. Dev. Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 72.Puebla-Osorio N, Lacey DB, Alt FW, Zhu C. Mol. Cell. Biol. 2006;26:3935–3941. doi: 10.1128/MCB.26.10.3935-3941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lakshmipathy U, Campbell C. Nucleic Acids Res. 2001;29:668–676. doi: 10.1093/nar/29.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frosina G, Fortini P, Rossi O, Carrozzino F, Raspaglio G, et al. J. Biol. Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 75.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Mol. Cell. Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Okano S, Lan L, Tomkinson AE, Yasui A. Nucleic Acids Res. 2005;33:422–429. doi: 10.1093/nar/gki190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moser J, Kool H, Giakzidis I, Caldecott K, Mullenders LH, Fousteri MI. Mol. Cell. 2007;27:311–323. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 78.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, et al. Cancer Res. 2005;65:4020–4230. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 79.Dong Z, Tomkinson AE. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkl705. 5721-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wilson TE, Grawunder U, Lieber MR. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 81.Schar P, Herrmann G, Daly G, Lindahl T. Genes Dev. 1997;11:1912–1924. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Teo SH, Jackson SP. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robins P, Lindahl T. J. Biol. Chem. 1996;271:24257–24261. doi: 10.1074/jbc.271.39.24257. [DOI] [PubMed] [Google Scholar]
- 84.Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Critchlow SE, Bowater RP, Jackson SP. Curr. Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 86.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, et al. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 87.Li Z, Otevrel T, Gao Y, Cheng HL, Seed B, et al. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 88.Bryans M, Valenzano MC, Stamato TD. Mutat. Res. 1999;433:53–58. doi: 10.1016/s0921-8777(98)00063-9. [DOI] [PubMed] [Google Scholar]
- 89.Wang YG, Nnakwe C, Lane WS, Modesti M, Frank KM. J. Biol. Chem. 2004;279:37282–37290. doi: 10.1074/jbc.M401217200. [DOI] [PubMed] [Google Scholar]
- 90.Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, et al. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 91.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber MR. Mol. Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 92.Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Curr. Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- 93.Frank KM, Sharpless NE, Gao Y, Sekiguchi JM, Ferguson DO, et al. Mol. Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 94.Sekiguchi J, Ferguson DO, Chen HT, Yang EM, Earle J, et al. Proc. Natl. Acad. Sci. USA. 2001;98:3243–3248. doi: 10.1073/pnas.051632098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O’Driscoll M, Cerosaletti KM, Girard PM, Dai Y, Stumm M, et al. Mol. Cell. 2001;8:1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 96.Riballo E, Critchlow SE, Teo SH, Doherty AJ, Priestley A, et al. Curr. Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 97.Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, et al. Nat. Struct. Mol. Biol. 2007;14:639–646. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
- 98.Teo IA, Arlett CF, Harcourt SA, Priestley A, Broughton BC. Mutat. Res. 1983;107:371–386. doi: 10.1016/0027-5107(83)90177-x. [DOI] [PubMed] [Google Scholar]
- 99.Petrini JH, Donovan JW, Dimare C, Weaver DT. J. Immunol. 1994;152:176–183. [PubMed] [Google Scholar]
- 100.Hsieh CL, Arlett CF, Lieber MR. J. Biol. Chem. 1993;268:20105–20109. [PubMed] [Google Scholar]
- 101.O’Driscoll M, Gennery AR, Seidel J, Concannon P, Jeggo PA. DNA Repair. 2004;3:1227–1235. doi: 10.1016/j.dnarep.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 102.Girard PM, Kysela B, Harer CJ, Doherty AJ, Jeggo PA. Hum. Mol. Genet. 2004;13:2369–2376. doi: 10.1093/hmg/ddh274. [DOI] [PubMed] [Google Scholar]
- 103.Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, et al. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 104.Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, et al. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 105.Sharpless NE, Ferguson DO, O’Hagan RC, Castrillon DH, Lee C, et al. Mol. Cell. 2001;8:1187–1196. doi: 10.1016/s1097-2765(01)00425-7. [DOI] [PubMed] [Google Scholar]
- 106.Bhagwat AS, Sanderson RJ, Lindahl T. Nucleic Acids Res. 1999;27:4028–4033. doi: 10.1093/nar/27.20.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Luo H, Chan DW, Yang T, Rodriguez M, Chen BP, et al. Mol. Cell. Biol. 2004;24:8356–8365. doi: 10.1128/MCB.24.19.8356-8365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Clements PM, Breslin C, Deeks ED, Byrd PJ, Ju L, et al. DNA Repair. 2004;3:1493–1502. doi: 10.1016/j.dnarep.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 109.Levin DS, Bai W, Yao N, O’Donnell M, Tomkinson AE. Proc. Natl. Acad. Sci. USA. 1997;94:12863–12868. doi: 10.1073/pnas.94.24.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Levin DS, Vijayakumar S, Liu X, Bermudez VP, Hurwitz J, Tomkinson AE. J. Biol. Chem. 2004;279:55196–55201. doi: 10.1074/jbc.M409250200. [DOI] [PubMed] [Google Scholar]
- 111.Vijayakumar S, Chapados BR, Schmidt KH, Kolodner RD, Tainer JA, Tomkinson AE. Nucleic Acids Res. 2007;35:1624–1637. doi: 10.1093/nar/gkm006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, et al. Mol. Cell. 2006;24:279–291. doi: 10.1016/j.molcel.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 113.Tom S, Henricksen LA, Park MS, Bambara RA. J. Biol. Chem. 2001;276:24817–24825. doi: 10.1074/jbc.M101673200. [DOI] [PubMed] [Google Scholar]
- 114.Smirnova E, Toueille M, Markkanen E, Hubscher U. Biochem. J. 2005;389:13–17. doi: 10.1042/BJ20050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang W, Lindsey-Boltz LA, Sancar A, Bambara RA. J. Biol. Chem. 2006;281:20865–20872. doi: 10.1074/jbc.M602289200. [DOI] [PubMed] [Google Scholar]
- 116.Dionne I, Nookala RK, Jackson SP, Doherty AJ, Bell SD. Mol. Cell. 2003;11:275–282. doi: 10.1016/s1097-2765(02)00824-9. [DOI] [PubMed] [Google Scholar]
- 117.Majka J, Burgers PM. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:227–260. doi: 10.1016/S0079-6603(04)78006-X. [DOI] [PubMed] [Google Scholar]
- 118.Prasad R, Singhal RK, Srivastava DK, Molina JT, Tomkinson AE, Wilson SH. J. Biol. Chem. 1996;271:16000–16007. doi: 10.1074/jbc.271.27.16000. [DOI] [PubMed] [Google Scholar]
- 119.De A, Campbell C. Biochem. J. 2007;402:175–186. doi: 10.1042/BJ20061004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang X, Morera S, Bates PA, Whitehead PC, Coffer AI, et al. EMBO J. 1998;17:6404–6411. doi: 10.1093/emboj/17.21.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marsin S, Vidal AE, Sossou M, Menissier-de Murcia J, Le Page F, et al. J. Biol. Chem. 2003;278:44068–44074. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 122.Vidal AE, Boiteux S, Hickson ID, Radicella JP. EMBO J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 124.Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, et al. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 125.Iles N, Rulten S, El-Khamisy SF, Caldecott KW. Mol. Cell. Biol. 2007;27:3793–3803. doi: 10.1128/MCB.02269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kanno S, Kuzuoka H, Sasao S, Hong Z, Lan L, et al. EMBO J. 2007;26:2094–2103. doi: 10.1038/sj.emboj.7601663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, et al. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 128.Das A, Wiederhold L, Leppard JB, Kedar P, Prasad R, et al. DNA Repair. 2006;5:1439–1448. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, et al. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 130.Grawunder U, Zimmer D, Leiber MR. Curr. Biol. 1998;8:873–876. doi: 10.1016/s0960-9822(07)00349-1. [DOI] [PubMed] [Google Scholar]
- 131.Junop MS, Modesti M, Guarne A, Ghirlando R, Gellert M, Yang W. EMBO J. 2000;19:5962–5970. doi: 10.1093/emboj/19.22.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dore AS, Furnham N, Davies OR, Sibanda BL, Chirgadze DY, et al. DNA Repair. 2006;5:362–368. doi: 10.1016/j.dnarep.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 133.Kysela B, Doherty AJ, Chovanec M, Stiff T, Ameer-Beg SM, et al. J. Biol. Chem. 2003;278:22466–22474. doi: 10.1074/jbc.M303273200. [DOI] [PubMed] [Google Scholar]
- 134.Chen L, Trujillo K, Sung P, Tomkinson AE. J. Biol. Chem. 2000;275:26196–26205. doi: 10.1074/jbc.M000491200. [DOI] [PubMed] [Google Scholar]
- 135.Hsu HL, Yannone SM, Chen DJ. DNA Repair. 2002;1:225–235. doi: 10.1016/s1568-7864(01)00018-0. [DOI] [PubMed] [Google Scholar]
- 136.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Mol. Cell. Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.DeFazio LG, Stansel RM, Griffith JD, Chu G. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, et al. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 139.Mahajan KN, Nick McElhinny SA, Mitchell BS, Ramsden DA. Mol. Cell. Biol. 2002;22:5194–5202. doi: 10.1128/MCB.22.14.5194-5202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ma Y, Pannicke U, Schwarz K, Lieber MR. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 141.Ahnesorg P, Smith P, Jackson SP. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 142.Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, de Villartay JP. J. Biol. Chem. 2006;281:13857–13860. doi: 10.1074/jbc.C500473200. [DOI] [PubMed] [Google Scholar]
- 143.Tsai CJ, Kim SA, Chu G. Proc. Natl. Acad. Sci. USA. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Koch CA, Agyei R, Galicia S, Metalnikov P, O’Donnell P, et al. EMBO J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Palmbos PL, Daley JM, Wilson TE. Mol. Cell. Biol. 2005;25:10782–10790. doi: 10.1128/MCB.25.24.10782-10790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Mol. Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 147.Tseng HM, Tomkinson AE. J. Biol. Chem. 2004;279:47580–47588. doi: 10.1074/jbc.M404492200. [DOI] [PubMed] [Google Scholar]
- 148.Frank-Vaillant M, Marcand S. Genes. Dev. 2001;15:3005–3012. doi: 10.1101/gad.206801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ooi SL, Shoemaker DD, Boeke JD. Science. 2001;294:2552–2556. doi: 10.1126/science.1065672. [DOI] [PubMed] [Google Scholar]
- 150.Valencia M, Bentele M, Vaze MB, Herrmann G, Kraus E, et al. Nature. 2001;414:666–669. doi: 10.1038/414666a. [DOI] [PubMed] [Google Scholar]
- 151.Budman J, Kim SA, Chu G. J. Biol. Chem. 2007;282:11950–11959. doi: 10.1074/jbc.M610058200. [DOI] [PubMed] [Google Scholar]
- 152.Budman J, Chu G. EMBO J. 2005;24:849–860. doi: 10.1038/sj.emboj.7600563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Calsou P, Delteil C, Frit P, Drouet J, Salles B. J. Mol. Biol. 2003;326:93–103. doi: 10.1016/s0022-2836(02)01328-1. [DOI] [PubMed] [Google Scholar]
- 154.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, et al. Proc. Natl. Acad. Sci. USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dynan WS, Yoo S. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Walker JR, Corpina RA, Goldberg J. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 157.Cotner-Gohara E, Kim IK, Tomkinson AE, Ellenberger T. J. Biol. Chem. 2008 doi: 10.1074/jbc.M708175200. PMID: 18238776. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]