Abstract
Nα-terminal acetylation is one of the most common protein modifications in eukaryotes. The COmbined FRActional DIagonal Chromatography (COFRADIC) proteomics technology that can be specifically used to isolate N-terminal peptides was used to determine the N-terminal acetylation status of 742 human and 379 yeast protein N termini, representing the largest eukaryotic dataset of N-terminal acetylation. The major N-terminal acetyltransferase (NAT), NatA, acts on subclasses of proteins with Ser-, Ala-, Thr-, Gly-, Cys- and Val- N termini. NatA is composed of subunits encoded by yARD1 and yNAT1 in yeast and hARD1 and hNAT1 in humans. A yeast ard1-Δ nat1-Δ strain was phenotypically complemented by hARD1 hNAT1, suggesting that yNatA and hNatA are similar. However, heterologous combinations, hARD1 yNAT1 and yARD1 hNAT1, were not functional in yeast, suggesting significant structural subunit differences between the species. Proteomics of a yeast ard1-Δ nat1-Δ strain expressing hNatA demonstrated that hNatA acts on nearly the same set of yeast proteins as yNatA, further revealing that NatA from humans and yeast have identical or nearly identical specificities. Nevertheless, all NatA substrates in yeast were only partially N-acetylated, whereas the corresponding NatA substrates in HeLa cells were mainly completely N-acetylated. Overall, we observed a higher proportion of N-terminally acetylated proteins in humans (84%) as compared with yeast (57%). N-acetylation occurred on approximately one-half of the human proteins with Met-Lys- termini, but did not occur on yeast proteins with such termini. Thus, although we revealed different N-acetylation patterns in yeast and humans, the major NAT, NatA, acetylates the same substrates in both species.
Keywords: Ard1, COFRADIC, N-terminal acetylation, Nat1, NatA
Protein Nα-terminal acetylation (here referred to as N-acetylation) is one of the most common covalent modifications of eukaryotic proteins, in which an acetyl group is transferred from acetyl-CoA to the α-amino group of protein N-terminal residues. N-acetylation occurs cotranslationally on nascent polypeptide chains and almost all N-acetylations in Saccharomyces cerevisiae are catalyzed by 1 of 3 major N-terminal acetyltransferase (NAT) complexes, NatA, NatB or NatC, consisting of catalytic subunits Ard1p, Nat3p, and Mak3p, respectively, and 1 or more auxiliary subunits (1). Yeast NatA, the major and best studied NAT, is composed of the catalytic subunit Ard1p in complex with Nat1p (2). Nat1p is responsible for anchoring Ard1p to the ribosome, thus facilitating cotranslational N-acetylation (3). Both subunits are required for optimal acetyltransferase activity and yeast strains lacking either one of the subunits display the same phenotypes, indicating that both genes are also functionally linked (4). The yeast NatA, NatB and NatC complexes differ in their substrate specificities. NatA substrates represent by far the largest group and contain proteins with Ser-, Ala-, Gly-, Val-, Cys- or Thr- N termini, whereas NatB and NatC act on different protein subclasses with Met- N termini (1, 5). Higher eukaryotes and yeast have homologous NAT genes, and both have similar patterns of N-acetylated proteins, suggesting that a similar cotranslational N-acetylation system is shared by all eukaryotes (1).
Like the yeast enzyme, the human Nat1p (also denoted NATH) and hArd1p interact, associate with ribosomes and express NAT activity in vitro (6). RNAi mediated knock-down of hNAT1 or hARD1 in different human cell lines demonstrated that these proteins play an important role: the decrease in cell proliferation or increase of apoptosis observed when hARD1 or hNAT1 are knocked down indicate that these defects may be caused by insufficient levels of N-acetylation of as yet unidentified critical substrate proteins (7, 8). Studies using 2D-PAGE, HPLC separations, and mass spectrometry revealed that ≈50% of all cytosolic yeast proteins are N-acetylated (1). For mammalian proteins, early studies and database searches revealed that 80% to 90% are N-acetylated (9–11). However, more recent studies, including small-scale experiments with several mammalian proteins, indicated that this number may be closer to 30% (12). The major questions addressed in this communication are the following: Are the types and proportion of N-acetylated proteins in yeast and mammals different, and if so, what is the cause of these differences?
To gain better insight in the degree of N-acetylation by the NATs in 2 different model systems, S. cerevisiae and human HeLa cells, we initiated a global qualitative and quantitative analysis of protein N-acetylation, using the N-terminal combined fractional diagonal chromatography (COFRADIC) technology (13), which allows targeted analysis of N-terminal peptides in highly complex mixtures, whereas all internal peptides are disregarded. This COFRADIC procedure, along with stable isotope labeling by amino acids in cell culture (SILAC) (14), and in combination with stable isotope tagging N-terminal chemistries (15), allowed us to generate quantitative data on the modification status of the N termini of the proteins present in the mixture. We thus obtained a general profile of the activities and substrates of NatA and other NATs in yeast and humans. In addition, the used strategy allowed detection and estimation of partially N-acetylated proteins.
Our results reveal that hNatA is functionally active in yeast and displays similar specificities and kinetics as compared with yNatA when acting on identical substrates. However, yeast proteins are generally not acetylated to the same extent as human proteins. These findings are discussed in terms of a global functional analysis.
Results
Human hARD1-hNAT1 Genes Complement the Yeast ard1-Δ nat1-Δ Phenotypes.
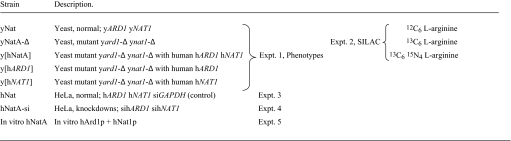
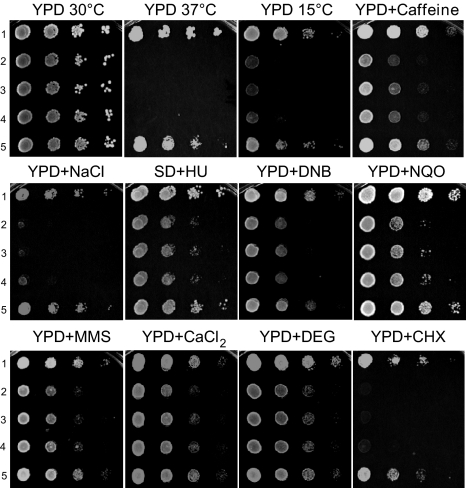
Previously, the ard1-Δ NAT1, ARD1 nat1-Δ and ard1-Δ nat1-Δ yeast mutants were shown to have a variety of phenotypes, including defects in general growth, growth on nonfermentable carbon sources, transition to Go phase, sporulation, cell growth on salts, caffeine, hydroxyurea and SDS containing media, and in derepression of silent loci (16, 17). These defects are due to the lack of N-acetylation of numerous proteins, including Orc1p and Sir3p (18, 19). The following yeast strains (Exp. 1 in Table 1) were used to demonstrate that the human ARD1-NAT1 genes can complement the yeast defects: the wild type strain yNat; yNatA-Δ containing the deletion yard1-Δ ynat1-Δ; and the y[hNatA] strain containing yard1-Δ ynat1-Δ hARD1 hNAT1. Thus, yNatA-Δ lacks NatA, whereas y[hNatA] lacks yeast NatA, but has human NatA. Expression of both hARD1 and hNAT1 fully complements all observed yNatA-Δ defects, whereas expression of either of the single genes, hARD1 or hNAT1 does not (Fig. 1). Furthermore, heterologous combinations, hARD1 yNAT1 and yARD1 hNAT1, were not functional in yeast, suggesting significant structural subunit differences between the species.
Table 1.
Strains and datasets
Fig. 1.
Complementation of the S. cerevisiae ard1-Δ nat1-Δ phenotypes by hARD1 and hNAT1. The following yeast strains were grown to early log phase and serial 1/10 dilutions containing the same number of cells were spotted on various media: 1, the normal strain (yNat); 2, the ard1-Δ nat1-Δ strain (yNatA-Δ); 3, the ard1-Δ nat1-Δ strain expressing hARD1 (y[hARD1]); 4, the ard1-Δ nat1-Δ strain expressing hNAT1 (y[hNAT1]); 5, the ard1-Δ nat1-Δ strain expressing hARD1 and hNAT1 (y[hNatA]). The plates containing the following media were incubated at 30 °C for 3 days unless indicated otherwise: YPD 15 °C, incubated for 6 days; YPD + caffeine, 0.1% caffeine; YPD + NaCl, 0.75 M NaCl; SD + HU, 75 mM hydroxyurea; YPD + DNB, 250 μM o-dinitrobenzene; YPD + NQO, 0.1 μg/mL 4-nitroquinoline oxide; YPD + MMS, 0.1% methyl methanesulfonate; YPD + CaCl2, 0.3 M CaCl2; YPD + DEG, 6.7% diethylenglycol; and YPD + CHX, 0.1 μg/mL cycloheximide.
Human hARD1-hNAT1 Genes and the Yeast yARD1-yNAT1 Genes Encode NATs That N-Acetylate the Same Set of Yeast Proteins.
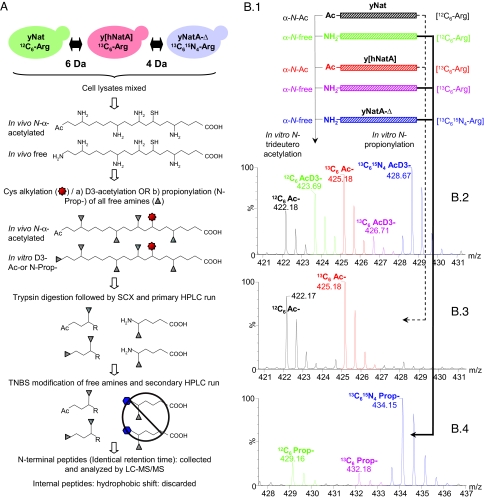
We used a COFRADIC technique to isolate N-terminal peptides (13) and to further determine the N-acetylated and unacetylated proteins from HeLa cells (hNat) (see below), a normal yeast strain (yNat), a mutant of yeast deficient in NatA (yNatA-Δ), and the deficient yeast mutant expressing human NatA (y[hNatA]). Proteome comparison of the yNat and y[hNatA] strains revealed that the yeast NatA and the human NatA act on the same set of proteins. The COFRADIC analysis performed on the 3 yeast strains, yNat, yNatA-Δ, and y[hNatA], is schematically outlined in Fig. 2. As briefly described in the figure, this COFRADIC technique can be used to enrich for N-terminal peptides ending in an arginine residue of α-amino blocked proteins before LC-MS/MS analysis. By mass tagging protein amino groups, including free protein α-amino groups, before the actual COFRADIC step, mass spectrometry can be used to determine the in vivo protein acetylation status and the degree of acetylation. In this study, the following 2 different amino-directed mass tagging strategies were used: trideutero-acetylation [AcD3] and propionylation [N-Prop] (see Fig. 2B). In the former case, acetylated peptides from in vivo blocked proteins and trideutero-acetylated peptides from in vivo free proteins coeluted during RP-HPLC and were used to calculate the degree of acetylation. In the latter case however, propionylated peptides, indicative for in vivo free protein forms, segregated by RP-HPLC from their acetylated counterparts and were used to point to Nat substrates, but not to quantify the degree of acetylation. To further distinguish N-terminal peptides isolated from the 3 yeast strains, Arg-SILAC labeling (20) was performed after introducing the arg4-Δ marker (Exp. 2 in Table 1 and Fig. 2). Such yeast strains are dependent on an external arginine source for growth (Fig. S1), enabling the incorporation of isotopic variants of arginine. Yeast strains were cultured in the presence of 1 of 3 different stable l-arginine isotopic variants (12C6 l-Arg, 13C6 l-Arg, or 13C615N4 l-Arg) (Table 1). Such an approach permits triplexed proteomics; all isolated N-terminal peptides end on arginine and the mass labels provide ample separations, thus allowing distinction of the 3 N-terminal peptide forms by MS (Fig. S2).
Fig. 2.
COFRADIC and mass spectrometry scheme for separating and identifying N-terminal peptides. (A) The experimental strategy used to determine N-terminal acetylation of proteins from normal yeast (yNat) or yeast expressing human NatA (y[hNatA]) by N-terminal COFRADIC is illustrated. After 2 protein modification steps (Cys alkylation and α- and ε-NH2 acetylation) and trypsin digestion, SCX enrichment (pH 3.0) for amino-blocked peptides is carried out. Two consecutive RP-HPLC runs are applied to isolate N termini after modification of trypsin-generated new α-NH2 groups with 2,4,6-trinitrobenzenesulfonic acid (indicated by a hexagon) between the 2 peptide separation steps. When combined with differential isotope labeling strategies, the exact origin of N termini can be determined. (B) Comparison of protein N-trideuteroacetylation and N-propionylation, and representative MS-spectra of isolated N-terminal peptides originating from yNatA and y[hNatA]. (1) A protein that contains in vivo free and N-acetylated termini (partially in vivo α-N-acetylated) can be modified in vitro by N-trideuteroacetylation or N-propionylation. (2) When trideuteroacetylation is used, the RP-HPLC elution profiles of the α-N-acetylated and α-N-trideuteroacetylated variants are indistinguishable and the peptide variants only segregate upon MS analysis by their 3-Da mass difference. (3 and 4) In contrast, when propionylation is used, the α-N-acetylated (3) and α-N-propionylated (4) variants segregate upon RP-HPLC, with the propionylated variant generally eluting at a later time because of increased hydrophobicity. MS spectra of doubly charged peptide ions originating from the in vivo acetylated (Ac) and/or free form of the α-N terminus from the vacuolar ATP synthase catalytic subunit A (P17255) are shown. The peptide was identified as 2AGAIENAR12. When trideutero-acetylation was applied at the protein level (2), this N terminus was found to be partially acetylated (upper MS-spectral) in the control yeast strain (43% α-N-acetylated) (12C6) and the y[hNatA] strain (82% α-N-acetylated) (13C6), whereas it was found completely free in the yNatA-Δ deficient strain (0% α-N-acetylated) (13C615N4 l-arginine). The theoretical % of α-N-acetylation was calculated by use of the MS-isotope pattern calculator (http://prospector.ucsf.edu). (3 and 4) Upon applying N-propionylation at the protein level, in vivo α-N-acetylated variants and free (in vitro α-N-propionylated) variants segregate during RP-HPLC separation. Here, in vivo acetylated (Ac) NatA substrate peptides appear as singletons in their 12C6 and 13C6 forms (3) and when partially in vivo α-N-acetylated their propionylated forms appear in all 3 setups analyzed (as shown in 4).
In total, 3 different and independent N-terminal COFRADIC analyses were performed: in vitro AcD3-acetylation, N-propionylation (N-Prop-), or no modification (N-noMod) of free protein amino groups (Fig. 2B). The ratio of in vivo acetylated (Ac termini) versus nonacetylated N termini was only inferred from the AcD3-experiment (see above). However, because of the high frequency of partially acetylated N termini, the resulting complex MS-spectra sometimes precluded straightforward analysis. Therefore, we used N-propionylation (N-Prop-) to extend our dataset (Fig. 2B). Finally, in vivo acetylated peptides were also detected in the N-noMod setup. The overall results of these analyses are discussed below.
The AcD3 incorporation data, comprising 262 unique N termini, indicated that 12% of the identified proteins were fully acetylated at their N terminus, whereas 45% were partially acetylated and 43% were nonacetylated (Table 2). The N-propionylation and N-noMod setups confirmed several AcD3 results, which, when combined, comprised 379 unique N termini (Table S1A). Combining these results with previously catalogued yeast N termini (1), a compilation of 614 different endogenous yeast proteins are included, of which at least 58% are partially or fully N-terminally acetylated (Table S1B). Our data further revealed that 163 unique yeast proteins were N-acetylated in the 2 strains yNat or y[hNatA], whereas these proteins were not acetylated in the strain not expressing any NatA (yNatA-Δ) (Table S1C). Furthermore, yNatA and y[hNatA] substrates had a distinct MS-profile as compared with NatB or NatC substrates and to the non-Nat substrates, thus allowing straightforward detection upon examination of MS spectra (Fig. S2 A–C).
Table 2.
Number and proportions of completely and partially acetylated human and yeast proteins with various N termini
| Yeast (yNat) |
Human (hNat) |
|||||
|---|---|---|---|---|---|---|
| No. | Completely, % | Completely and partially, % | No. | Completely, % | Completely and partially, % | |
| NatA substrates | ||||||
| Ala- | 40 | 0.0 | 42.5 | 289 | 87.2 | 96.2 |
| Cys- | 1 | 0.0 | 0.0 | 1 | 100.0 | 100.0 |
| Gly- | 13 | 0.0 | 0.0 | 27 | 22.2 | 40.7 |
| Ser- | 93 | 0.0 | 96.8 | 111 | 98.2 | 99.1 |
| Thr- | 22 | 0.0 | 36.4 | 26 | 80.8 | 92.3 |
| Val- | 22 | 0.0 | 0.0 | 31 | 3.2 | 19.4 |
| NatB substrates | ||||||
| Met-Asn- | 7 | 100.0 | 100.0 | 14 | 85.7 | 100.0 |
| Met-Asp- | 14 | 100.0 | 100.0 | 48 | 97.9 | 100.0 |
| Met-Glu- | 4 | 100.0 | 100.0 | 79 | 97.5 | 100.0 |
| NatC substrates | ||||||
| Met-Ile- | 2 | 0.0 | 0.0 | 4 | 50.0 | 75.0 |
| Met-Leu- | 8 | 12.5 | 25.0 | 10 | 40.0 | 70.0 |
| Met-Phe | 2 | 50.0 | 50.0 | 10 | 70.0 | 80.0 |
| Other | ||||||
| Asp- and Glu- (actin) | 0 | — | — | 2 | 100.0 | 100.0 |
| Met-Gln- | 4 | 50.0 | 75.0 | 10 | 70.0 | 90.0 |
| Met-Lys- | 10 | 0.0 | 0.0 | 19 | 10.5 | 42.1 |
| Met-Met- | 0 | — | — | 7 | 100.0 | 100.0 |
| Met-Ala- | 2 | 0.0 | 0.0 | 2 | 50 | 50 |
| Met-Gly- | 2 | 0.0 | 0.0 | 1 | 0 | 100 |
| Met-Ser- | 1 | 0.0 | 0.0 | 1 | 100 | 100 |
| Met-Thr- | 1 | 0.0 | 0.0 | 2 | 100 | 100 |
| Met-Val- | 1 | 0.0 | 100.0 | 6 | 50 | 66.7 |
| Met-Pro- | 1 | 0.0 | 0.0 | 0 | — | — |
| Met-Tyr- | 0 | — | — | 1 | 100 | 100 |
| Pro- | 11 | 0.0 | 0.0 | 40 | 0.0 | 0.0 |
| Ile- | 1 | 0.0 | 0.0 | 1 | 0.0 | 0.0 |
| Total | 262 | 11.8 | 56.9 | 742 | 76.1 | 84.4 |
No yeast protein was N-acetylated in yNat that was not N-acetylated in y[hNatA], and vice versa, although there was variation in the degree of N-acetylation by yNat versus y[hNatA], which was determined for 81 substrates identified in the AcD3 setup (Table S1C). Up to 85% of the partially acetylated NatA-type substrates were acetylated to a higher degree in the y[hNatA] strain as compared with the yNat strain, as exemplified by the La protein homolog (Table S1C and Fig. S2D). However, some substrates were more prone to acetylation in the yNat strain compared with the y[hNatA] strain, as for example, the 60S ribosomal protein L1 (Table S1C and Fig. S2E).
Determination of Protein N-Acetylation in HeLa Cells and Analysis of hARD1-hNAT1 Knockdown Effects.
To investigate the level of N-acetylation in humans and elaborate on the substrate specificity of hNatA in its physiological context, we determined the N-terminal sequences of acetylated and nonacetylated soluble proteins from the HeLa cell lines listed in Table 1 (Expts. 3 and 4). hNat denotes the normal HeLa cell line and hNatA-si denotes a cell line in which the expression of hARD1 and hNAT1 was reduced by sihARD1 and sihNAT1 knockdown (7). N-terminal COFRADIC analyses were carried out using trideutero-acetylation to block free protein N termini (similar to Fig. 2). Here, again, AcD3- peptides and their in vivo acetylated counterparts could be distinguished after MS analysis by their 3-Da mass difference. As illustrated in Fig. S3, the N terminus of the ADP-ribosylation factor GTPase-activating protein 3 was in vivo acetylated (A), the pirin N terminus was partially acetylated (B), whereas the developmentally-regulated GTP-binding protein 2 carries an in vivo free N terminus (C). In this experiment, we identified the N termini and the proportion of acetylation of 742 different proteins from the normal hNat HeLa strain (Tables 2 and S2A-B). These results constitute the largest dataset of N-acetylation of human proteins and reveal that ≈76% of HeLa proteins are fully acetylated at their N terminus, whereas 8% are partially acetylated and 16% are unacetylated. A comparison of N-acetylated and unacetylated proteins from yeast and humans reveal that the distribution N termini of naturally occurring substrates differs significantly (Table 2). Most N-acetylated proteins from yeast begin with serine, whereas most N-acetylated proteins from humans begin with alanine. A more striking difference is the N-acetylation of 8 of 19 Met-Lys- proteins from humans and none from yeast (Table 2). The enzyme acetylating Met-Lys- proteins may represent a NAT found in humans but not in yeast.
The overall protein N-acetylation patterns from the hNat and hNatA-si cell lines (Exps. 3 and 4 in Table 1) were surprisingly similar. Reduced expression of hARD1 and hNAT1 in the hNatA-si strain was verified by Western blot analysis of hArd1p and hNat1p, using aliquots from the samples analyzed by COFRADIC (Fig. S4A). The hNatA-si strain contained ≈30% of the normal amount of Nat1p, and ≈5% of the normal amount of Ard1p. Unexpectedly this low amount of hNatA is still sufficient to acetylate most of the NatA substrates, and the acetylation of only a few of the NatA substrates appeared to be reduced. We here considered a hNatA substrate to be affected by knockdown of the hNatA complex when the difference in percentage of N-acetylation was >5% between Exp. 3 and Exp. 4 (Table 1). Only 16 hNatA substrates were susceptible to the diminished level of hNatA out of a total pool of 242 acetylated NatA type substrates (i.e., 6.6%) identified in both setups (Table S2C and Fig. S4 B–R). Interestingly, 13 of the 16 affected N termini having diminished levels of acetylation in Exp. 4 (hNatA knock-down) were also found to be only partially acetylated in Exp. 3 (control setup). In contrast, nearly all NatA type substrates with completely acetylated termini from Exp. 3 were also found fully acetylated in Exp. 4. It therefore appears that N termini can be assigned to different classes with various degrees of efficacy for hNatA acetylation: (i) completely N-acetylated and nearly unaffected by hNatA-si; (ii) partially acetylated and N termini of which the degree of acetylation is not affected by hNatA-si; (iii) partially acetylated and N termini of which acetylation is diminished by NatA-si, and (iv) those that are not acetylated.
In Vitro Acetylation by hArd1p + hNat1p.
It is essential to determine whether the different classes of hNatA-type N termini identified in our HeLa experiments, from fully acetylated to nonacetylated, are actually differentially preferred substrates of the hNatA complex per se. If a positive correlation is observed, this would establish that the observed differences in N-acetylation of NatA type N termini in human cells are directly linked to the substrate preferences of the hNatA complex. We addressed this question by carrying out in vitro N-acetylation (Exp. 5 in Table 1) of representatives of the following 4 groups of synthetic 24-mer peptides with an immunoprecipitated hArd1p and hNat1p complex (6): group A, a previously described in vitro hNatA substrate (SYSM-); group B (class i above), hNatA substrates found to be fully acetylated and not affected in Exp. 4 (SESS-, STPD-, ANSA-); group C (class iii above), hNatA substrates found to be partially acetylated and affected in Exp. 4 (TSAL-, AVFA-, ADGK-); and group D (class iv above), a peptide (SPTP-) found to be unacetylated in vivo. The results demonstrate that the hArd1p-hNat1p complex acetylates in vitro all 3 class i N termini (Fig. S5). Two of the in vivo partially acetylated class iii substrates, TSAL and ADGK, were also found to be acetylated in vitro, but generally at a lower level compared with their fully in vivo acetylated counterparts, whereas AVFA was not significantly acetylated. The class iv peptide starting with serine (SPTP-), found to be unacetylated in vivo (Table S2A), was also not acetylated in vitro by hNatA.
Discussion
The COFRADIC procedure was used to determine the N-acetylation status of 742 proteins from HeLa (hNat), and of 379 proteins from yeast (yNat). In general, the N-acetylation of specific N-terminal sequences in yeast and humans described herein are in good accordance with those published (1). However, the current study also allows estimates of partially N-acetylated proteins. A compilation of all yeast and human proteins with N termini known to be fully acetylated, partially acetylated or free, including those determined in this study, is presented in Table S3 and Table S4. Although both partially and completely N-acetylated proteins are found in both yeast and humans, it is noteworthy that all NatA substrates in yeast are only partially N-acetylated, whereas most NatA substrates in humans are completely or nearly completely acetylated (Table 2). In contrast, NatB substrates are completely acetylated in yeast, and are almost completely acetylated in humans. Furthermore, by considering the values presented in Table 2, the proportion of the NatA and NatC substrates are comparable in yeast and humans; 73% and 65% NatA substrates in yeast and humans, respectively; and 4.5% and 3.2% NatC substrates in yeast and humans, respectively. However, there are 9.5% and 19% NatB substrates in yeast and humans, respectively, thus indicating a higher proportion of NatB substrates in humans. Overall, there are 57% and 84% N-acetylated proteins in yeast and humans, respectively. Meinnel et al. (12) suggested that more abundant proteins are more likely to be N-terminally acetylated. We therefore performed an unbiased estimation of N-terminal acetylation for all yeast and human SwissProt entries (version 56.0) based on the nature of the N-terminal amino acids and the N-terminal acetylation status uncovered in this study (Table 2, Table S1A, and Table S2A). In this way, we predict that ≈50% of all yeast proteins and 77% of all human proteins may be completely or partially N-terminal acetylated, values that closely resemble our experimental findings and indicate that the proteins identified here, are unbiased representatives. The slight experimental overestimation is likely because of an overrepresentation of NatA substrates and underrepresentation of NatC substrates in the datasets and to the fact that Met-His- and Met-Arg- N termini are under-sampled in our analyses. The higher value in humans could be partially accounted for by higher proportion of NatB substrates and the presence of a larger variety of N-terminal sequences that are acetylated in human; in particular, a significantly higher percentage of N-alanine residues are acetylated in human; the same is true for glycine, valine and threonine residues. The biological significance of such evolved differences remains to be determined. In addition, in humans there is a presence of N-acetylated proteins that have not been assigned to a specific NAT. Most notably, 8 of 19 of the human proteins with Met-Lys- were N-acetylated, whereas none of 10 yeast proteins with this terminus were N-acetylated (Table 2). It is possible that Met-Lys- proteins in human are modified by a new class of NATs; or possibly one of the major NATs, NatB or NatC, both acetylating different subsets of Met- proteins in yeast, diverged to acetylate Met-Lys- N termini in humans. Although the acetylation patterns in yeast and human are very similar, it appears that the human protein N termini evolved to be more prone to acetylation. Perhaps, evolution of both the N-terminal protein sequences and NAT complexes caused the overall increased proportion of N-terminally acetylated proteins in human.
Recently, an N-acetylation prediction program, TermiNator2 (www.isv.cnrs-gif.fr/terminator2/index.html), was described. When applied to our dataset, correct prediction was obtained in ≈80% of the cases (Table S3). It should also be noted that partial acetylation is not predicted, emphasizing the need for experimental analyses.
In this regard, human cells may have additional NATs with overlapping specificities as compared with the described yeast NAT complexes and the described major human homologues. The hArd1p-hNat1p complex was proposed to be the human NatA complex based on homology with the yeast Nat1p and Ard1p proteins and on biochemical evidence (6). However, in contrast to yeast, human has 2 orthologues both for Nat1p, hNat1p and hNat2p, and for Ard1p, hArd1p and hArd2p. hArd2p was recently shown to potentially interact with hNat1p and to express acetyltransferase activity (21). However, only hArd1p, and not hArd2p was detected by mass spectrometry when endogenous hNat1p was immunoprecipitated from human cells. Furthermore, the expression levels of hARD2 and hNAT2 appear to be quite low in most tissues; therefore the exact contribution from these proteins to N-acetylation remains unclear. Our data clearly establishes that hArd1p-hNat1p is the major NatA type complex in human cells. When considering all human SwissProt entries (v. 56.0) and our experimentally observed level of acetylation for the different hNatA subgroups, we estimate that ≈8,400 of the total 19,903 unique proteins are partially or completely N-acetylated by the hArd1p-hNat1p complex.
Our experimental data further revealed that different patterns of N-acetylation in humans can occur on proteins having identical sequences of up to the first 7 residues (position 1–7) (Table S2D), indicating that N-acetylation can be influenced by residues distant from the N terminus. Cotranslational N-terminal acetylation occurs when 25–50 aa protrude from the ribosome (22), which clearly makes it possible that amino acid residues within this range influence N-terminal acetylation. However, our data suggest that for an absolute majority of proteins both in yeast and human, only the first and second amino acid residues determine the efficiency of N-acetylation (Tables 2 and S3). The in vitro acetylation assays, using peptides differing in the first 7 residues support this observation (Fig. S5).
Although a significant hARD1-hNAT1 knockdown was achieved in our HeLa experiments (95% reduction of hArd1p level, Fig. S4A), the hNatA identified substrates represent only a small proportion of the estimated hNatA proteins, and these were predominantly partially acetylated N termini. The discrepancy between potential substrates and identified substrates is probably due to the high efficiency of the N-acetylation reaction, resulting only in partial loss of N-terminal acetylation on proteins having suboptimal sequences. The observed apoptotic phenotype of the hARD1-hNAT1 knockdown (7) may be mechanistically linked to the rather small decrease of N-acetylation on these suboptimal hNatA substrates.
In conclusion, we here present the most comprehensive experimental study of protein N-terminal acetylation to date, demonstrating that ≈84% of human proteins carry this modification, whereas the corresponding number for yeast proteins is ≈57%, probably reflecting both slight differences in intrinsic enzymatic properties and differences in sequence distributions between the 2 species.
Materials and Methods
Strains and datasets are presented in Table 1. The experimental setup of SILAC labeling, COFRADIC and Mass Spectrometry are briefly outlined in Fig. 2 and in Results. Further information on cell lines, yeast strains, plasmids, antibodies, chemicals, protein preparation, COFRADIC isolation of N-terminal peptides, mass spectrometry, isotopic peak distribution, Western blot analysis, and in vitro acetyltransferase assays are provided in SI Materials and Methods, Table S5, and Fig. S3 D and E. Excel files are available online at http://genesis.ugent.be/public_data/arnesen09. All original mass spectrometer data are available online at PRIDE (23) (www.ebi.ac.uk/pride) with accession numbers 8636, 8637 and 8638.
Supplementary Material
Acknowledgments.
We thank D. Gromyko, C. Hoff, E. Timmerman, A. Staes, and J. Van Damme for technical assistance. This work was supported by the Norwegian Health region West (to T.A. and J.E.V.), the Norwegian Research Council (to T.A), the Meltzer Foundation (to T.A), the Norwegian Cancer Society (to J.E.V), National Institutes of Health Grant R01 GM12702 (to F.S.), a Ph.D grant of the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen) (K.H.). The laboratory in Ghent us supported by Fund for Scientific Research–Flanders (Belgium) Projects G.0156.05, G.0077.06, and G.0042.07; Concerted Research Actions Project BOF07/GOA/012 from Ghent University; Interuniversity Attraction Poles Phase VI Research Project P6/28; and the European Union Interaction Proteome 6th Framework Program.
Note added in Proof.
Recently B. Polevoda, T. Arnesen, and F. Sherman (24) introduced the following revised genetic nomenclature for the NAT genes: NAA10 (ARD1); NAA15 (NAT1); NAA20 (NAT3); NAA25 (MDM20); NAA30 (MAK3); NAA35 (MAK10); NAA38 (MAK31); NAA40 (NAT4); and NAA50 (NAT5).
Footnotes
The authors declare no conflict of interest.
Data deposition: The mass spectrometry data reported in this paper have been deposited in the PRIDE database, www.ebi.ac.uk/pride (accession nos. 8636, 8637, and 8638).
This article contains supporting information online at www.pnas.org/cgi/content/full/0901931106/DCSupplemental.
References
- 1.Polevoda B, Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J Mol Biol. 2003;325:595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- 2.Park EC, Szostak JW. ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. EMBO J. 1992;11:2087–2093. doi: 10.1002/j.1460-2075.1992.tb05267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gautschi M, et al. The yeast Nα-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol Cell Biol. 2003;23:7403–7414. doi: 10.1128/MCB.23.20.7403-7414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullen JR, et al. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. EMBO J. 1989;8:2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polevoda B, Norbeck J, Takakura H, Blomberg A, Sherman F. Identification and specificities of N-terminal acetyltransferases from Saccharomyces cerevisiae. EMBO J. 1999;18:6155–6168. doi: 10.1093/emboj/18.21.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnesen T, et al. Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. Biochem J. 2005;386:433–443. doi: 10.1042/BJ20041071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnesen T, et al. Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein N-alpha-acetyltransferase complex. Oncogene. 2006;25:4350–4360. doi: 10.1038/sj.onc.1209469. [DOI] [PubMed] [Google Scholar]
- 8.Fisher TS, Etages SD, Hayes L, Crimin K, Li B. Analysis of ARD1 function in hypoxia response using retroviral RNA interference. J Biol Chem. 2005;280:17749–17757. doi: 10.1074/jbc.M412055200. [DOI] [PubMed] [Google Scholar]
- 9.Driessen HP, de Jong WW, Tesser GI, Bloemendal H. The mechanism of N-terminal acetylation of proteins. CRC Crit Rev Biochem. 1985;18:281–325. doi: 10.3109/10409238509086784. [DOI] [PubMed] [Google Scholar]
- 10.Jornvall H. Acetylation of protein N-terminal amino-groups structural observations on alpha-amino acetylated proteins. J Theor Biol. 1975;55:1–12. doi: 10.1016/s0022-5193(75)80105-6. [DOI] [PubMed] [Google Scholar]
- 11.Persson B, Flinta C, von Heijne G, Jornvall H. Structures of N-terminally acetylated proteins. Eur J Biochem. 1985;152:523–527. doi: 10.1111/j.1432-1033.1985.tb09227.x. [DOI] [PubMed] [Google Scholar]
- 12.Meinnel T, Peynot P, Giglione C. Processed N-termini of mature proteins in higher eukaryotes and their major contribution to dynamic proteomics. Biochimie. 2005;87:701–712. doi: 10.1016/j.biochi.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Gevaert K, et al. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- 14.Impens F, et al. Mechanistic insight into taxol-induced cell death. Oncogene. 2008;27:4580–4591. doi: 10.1038/onc.2008.96. [DOI] [PubMed] [Google Scholar]
- 15.Staes A, et al. Improved recovery of proteome-informative, protein N-terminal peptides by combined fractional diagonal chromatography (COFRADIC) Proteomics. 2008;8:1362–1370. doi: 10.1002/pmic.200700950. [DOI] [PubMed] [Google Scholar]
- 16.Whiteway M, Szostak JW. The ARD1 gene of yeast functions in the switch between the mitotic cell cycle and alternative developmental pathways. Cell. 1985;43:483–492. doi: 10.1016/0092-8674(85)90178-3. [DOI] [PubMed] [Google Scholar]
- 17.Whiteway M, Freedman R, Van Arsdell S, Szostak JW, Thorner J. The yeast ARD1 gene product is required for repression of cryptic mating-type information at the HML locus. Mol Cell Biol. 1987;7:3713–3722. doi: 10.1128/mcb.7.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geissenhoner A, Weise C, Ehrenhofer-Murray AE. Dependence of ORC silencing function on NatA-mediated Nα acetylation in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:10300–10312. doi: 10.1128/MCB.24.23.10300-10312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Connelly JJ, Wang CL, Sternglanz R. Importance of the Sir3 N terminus and its acetylation for yeast transcriptional silencing. Genetics. 2004;168:547–551. doi: 10.1534/genetics.104.028803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 21.Arnesen T, et al. Characterization of hARD2, a processed hARD1 gene duplicate, encoding a human protein N-alpha-acetyltransferase. BMC Biochem. 2006;7:13. doi: 10.1186/1471-2091-7-13. –. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pestana A, Pitot HC. Acetylation of nascent polypeptide-chains on rat-liver polyribosomes in vivo and in vitro. Biochemistry. 1975;14:1404–1412. doi: 10.1021/bi00678a010. [DOI] [PubMed] [Google Scholar]
- 23.Martens L, et al. PRIDE: The proteomics identifications database. Proteomics. 2005;5:3537–3545. doi: 10.1002/pmic.200401303. [DOI] [PubMed] [Google Scholar]
- 24.Polevoda B, Arnesen T, Sherman F. A synopsis of eukaryotic Na-terminal acetyltransferases: Nomenclature, subunits and substrates. BMC Proc. 2009 doi: 10.1186/1753-6561-3-S6-S2. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.