Abstract
Dating the Tree of Life has now become central to relating patterns of biodiversity to key processes in Earth history such as plate tectonics and climate change. Regions with a Mediterranean climate have long been noted for their exceptional species richness and high endemism. How and when these biota assembled can only be answered with a good understanding of the sequence of divergence times for each of their components. A critical aspect of dating by using molecular sequence divergence is the incorporation of multiple suitable age constraints. Here, we show that only rigorous phylogenetic analysis of fossil taxa can lead to solid calibration and, in turn, stable age estimates, regardless of which of 3 relaxed clock-dating methods is used. We find that Proteaceae, a model plant group for the Mediterranean hotspots of the Southern Hemisphere with a very rich pollen fossil record, diversified under higher rates in the Cape Floristic Region and Southwest Australia than in any other area of their total distribution. Our results highlight key differences between Mediterranean hotspots and indicate that Southwest Australian biota are the most phylogenetically diverse but include numerous lineages with low diversification rates.
Keywords: biodiversity, diversification rates, fossil calibration, molecular dating, Proteaceae
Mediterranean-climate regions are among the most species-rich biomes on Earth (1). They are all regarded as “biodiversity hotspots,” regions that are both extraordinarily species-rich and highly threatened by human activity (2, 3). Southwest Australia (SWA) and the Cape Floristic Region (CFR) are 2 of these Mediterranean hotspots and share key features such as high levels of endemism, nutrient-poor soils, and frequent fire regimes (1) (Table 1). Both hotspots have been compared with oceanic islands for their endemism and appear to have originated after aridification and cooling from the tropical climates that they experienced in the Paleogene (5, 6). How exactly climate change has contributed to the exceptional levels of species richness and endemism observed today remains an open question. Molecular dating analyses can help us address the timing of diversification in these regions and test whether high species richness is the result of recent radiation (due to high speciation rates) or long accumulation of lineages over time. These methods have been applied to the study of these hotspots, especially the CFR (8–11). A potential problem in these studies, however, is that each analysis has relied on a single calibration point, generally from distantly related fossils, secondary calibration dates derived from previous studies, or the timing of geological events. In addition, these studies have focused on 1 or 2 species-rich Mediterranean hotspot clades and did not address the possible impact of a more complete sampling of lineages in these regions.
Table 1.
Comparison of the three Mediterranean-climate biodiversity hotspots in the Southern Hemisphere
| Biodiversity hotspot | Area, km2 | Soil fertility | Frequent fire | Total species (% endemic) | Proteaceae species (% endemic) | No. of Proteaceae lineages* |
|---|---|---|---|---|---|---|
| Cape Floristic Region (CFR) | 78,555 | very low to moderate | yes | 9,000 (69%) | 332 (99%) | 4 |
| Southwest Australia (SWA) | 356,717 | very low to low | yes | 7,380 (49%) | 697 (99%) | 7 |
| Central Chile† | 155,000 | high | no | 2,864 (30%) | 5 (0%) | 4 |
*Number of lineages defined as the number of separate lineages on the phylogeny (Fig. 1) whose immediate sister group is not found in the named area at all. Note that this may be more or less than the number of independent colonization events, depending on the biogeographic scenario and the internal history of mixed terminals.
†Hotspot defined as the winter-rainfall, Mediterranean-type climate area of the wider Chilean Winter Rainfall-Valdivian Forests biodiversity hotspot (3).
Here, we address these questions by using Proteaceae, a Southern Hemisphere flowering plant family with an exceptional fossil record and 60% of its extant diversity in SWA and the CFR. This group includes flagship taxa such as Protea in South Africa or Banksia in Western Australia. We use a rigorous approach to select adequate calibration points and date a multigene phylogenetic tree of all genera of this family. Preliminary phylogenetic analyses (12–15) have suggested that members of this group in each Mediterranean hotspot do not form a single clade but instead consist of multiple independent lineages, several of which are remarkably species-rich and others surprisingly species-poor. This pattern makes Proteaceae particularly suitable to test and compare hypotheses of diversification in the 2 regions. Specifically, we estimate absolute net diversification rates for all clades of the phylogeny and correlate the highest rates and steepest shifts in rates with occurrence in Mediterranean hotspots. We also identify extraordinarily species-rich clades, given a background diversification rate for the group under 2 models of extinction (16).
Results
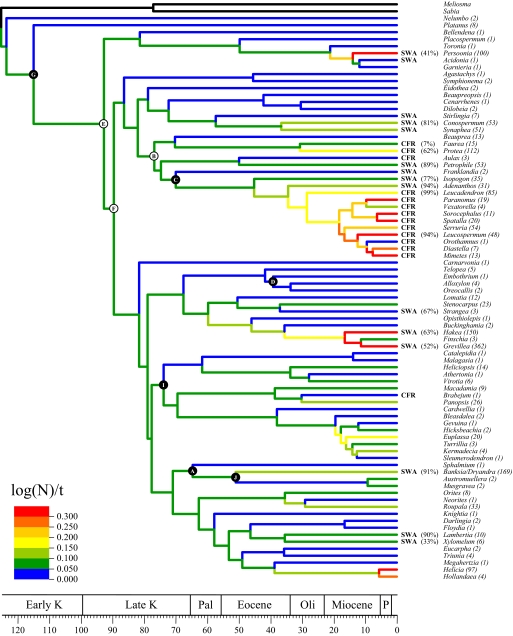
We reconstructed the phylogeny of Proteaceae by using molecular sequence data from 1 nuclear and 7 plastid regions, sampling all of the genera currently recognized. Parsimony and Bayesian analyses yielded very similar, well-supported trees [supporting information (SI) Fig. S1], which represent a great improvement on previously uncertain relationships (12–15). Taxa occurring in the CFR are scattered among 4 lineages, whereas SWA taxa have at least 7 independent origins (Fig. 1). Not all hotspot lineages are species-rich (i.e., >100 species), but they all appear well nested in the tree. To test whether species richness in these lineages is the result of rapid diversification or long accumulation over time, we dated the phylogeny.
Fig. 1.
Dated phylogeny of Proteaceae: maximum clade credibility tree with mean ages from the Bayesian uncorrelated lognormal method (implemented in BEAST) using nucleotide sequence data from 8 loci. Nodes on which fossil age constraints were applied (as uniform prior distributions) are identified with black dots (see Table S1). White dots identify additional, redundant, or uninformative age constraints obtained from the fossil analyses. Branches are colored according to absolute net diversification rate by stem age of their subtending clade. Taxa present in Mediterranean hotspots are identified with either SWA (Southwest Australia) or CFR (Cape Floristic Region). If not endemic to these hotspots, the percentage of hotspot species is mentioned in brackets. Total species numbers are indicated in brackets after the name of each taxon. Absolute ages are in million years.
Nearly three-quarters of all described fossil species of Proteaceae are dispersed pollen. We combined literature data with our own observations on pollen of >300 extant and 25 fossil palynomorph species of Proteaceae into a database of >5,000 records to compile a matrix of morphological characters (SI Materials and Methods, Dataset S1). By using the molecular phylogenetic tree as a backbone constraint, we conducted parsimony analyses to assess the phylogenetic position of each fossil. Unlike the approach followed in most of the palaeobotanical literature and molecular dating studies, where a handful of characters considered to be important is used to justify relationships to extant taxa, our analyses explore all of the possible phylogenetic positions and evaluate them by using all characters known for a fossil taxon. We find that few of the traditional literature assignments are supported (Table S1). This is a significant result because many fossil species in this group have been assigned to single extant genera and these assumptions have been used uncritically to calibrate molecular dating analyses (17) or to trace the biogeographic history of the family (18). Fossil taxa for which <10 most parsimonious positions were found were considered suitable as minimum age evidence for the most recent common ancestor of all of these positions (Fig. S2). Fossils with >10 most parsimonious positions are not suitable for calibration either because too little is known of their morphology or their combination of characters is not distinctive enough to reconstruct their position based solely on their morphology. Of the 14 fossils suitable for calibration, several gave minimum age evidence for the same node (Table S1). The oldest fossil was chosen and the remaining taxa were considered redundant calibration points. Other fossil taxa were not informative because the minimum age evidence they provided was necessarily implied by another calibration point nested higher up in the phylogeny (with equal or older minimum age). This approach identified 5 independent fossil pollen age constraints, to which a well-preserved fossil inflorescence and a maximum age constraint on the root were added (SI Materials and Methods).
We estimated divergence times by using 3 relaxed clock methods and this set of 7 fossil age constraints. Penalized likelihood (PL) (19), the Bayesian autocorrelation method (20–22) (implemented in multidivtime), and the uncorrelated lognormal (UCLN) method (23) (implemented in BEAST) produced similar age estimates for most nodes (Table S2). This was verified by correlation analysis of each pair of methods (PL vs. UCLN, R2 = 0.98, slope = 1.03; multidivtime vs. UCLN, R2 = 0.98, slope = 1.05; PL vs. multidivtime, R2 = 0.99, slope = 0.98; Fig. S3 a–c). Because ages obtained in BEAST take into account phylogenetic uncertainty, we refer to these in the following discussion.
We calculated absolute net diversification rates (speciation minus extinction) for all clades in the phylogeny by using stem (rst) and crown (rcr) group ages. The diversification rate for Proteaceae (rst = 0.066; rcr = 0.074) is comparable to that of angiosperms as a whole (rcr = 0.089) (16). The highest diversification rates (rst >0.200) are all found in Mediterranean hotspot lineages, with the exception of the predominantly Southeast Asian genus Helicia (Fig. 1). This is also true of the most important increases in diversification rates (Table S3). These high rates are comparable to the highest rates found in angiosperm clades such as Lamiales (rcr = 0.212) or Asterales (rcr = 0.330) (16). However, not all hotspot lineages appear to have experienced unusually high diversification rates. In addition, the link between hotspots and diversification rates is difficult to appreciate because many terminal taxa have species both in and outside these hotspots.
To compensate for this problem, we plotted diversification rates against the proportion of Mediterranean hotspot species for all clades in the phylogeny (Fig. S3e). We obtained a highly significant positive correlation (P < 0.0001) between these 2 variables, indicating that Proteaceae have indeed diversified faster in Mediterranean hotspots than anywhere else. To test the effect of other climatic conditions on diversification rates in this group, we also looked at the proportion of tropical species in each clade and found a significant negative correlation (P = 0.0133). These relationships remain significant when only terminal taxa are taken into account or when sampling all lineages present 20 or 40 mya. We also tested this correlation by using the method of independent contrasts of both relative and absolute rate differences (24) and obtained the same results, although the effect of tropical climates is no longer significant (Fig. S3 f and g).
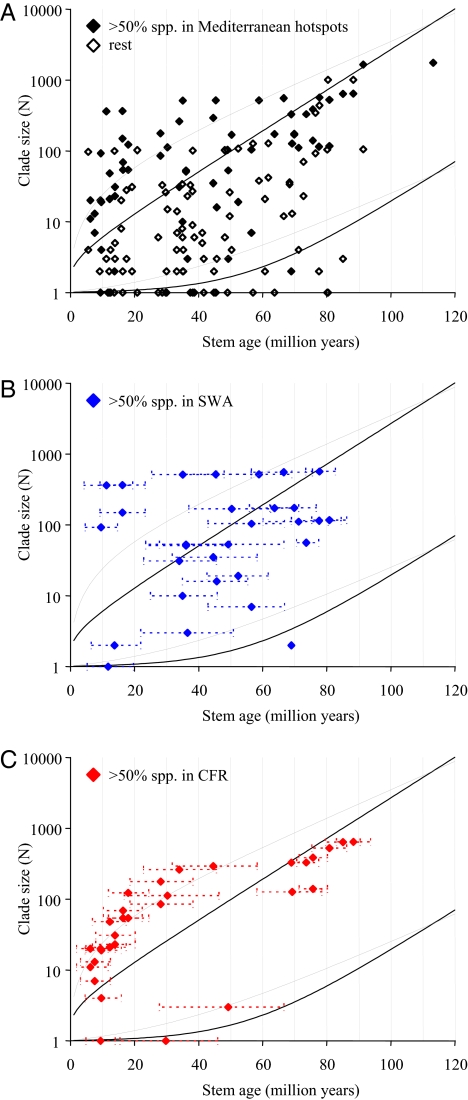
We followed the approach of Magallón and Sanderson (16) to identify extraordinarily species-rich clades in Proteaceae. We calculated 95% confidence intervals for expected clade size given a stem age and the background rate of Proteaceae, assuming either a pure birth model (no extinction, κ = 0) or a high rate of extinction relative to speciation (κ = 0.9). We find that all of the clades that are larger than expected under the most conservative model (κ = 0.9) have species occurring in Mediterranean hotspots (>50% for most of them), with the exception of Helicia (Fig. 2A). Whereas clades with >50% of their species in SWA are scattered above, within, and below these intervals (Fig. 2B), most of the clades with >50% of their species in the CFR concentrate above the upper limit of expected clade size under a pure birth model (κ = 0; Fig. 2C). Tropical clades are dispersed, but few appear to be larger than expected under either model.
Fig. 2.
Confidence intervals on expected clade diversity (number of extant species) according to stem group age, given a fixed background diversification rate (r) equal to that of Proteaceae as a whole. The black lines are the 95% confidence intervals in the absence of extinction (r0 = 0.066) and the gray lines under a model of high relative extinction rate (r0.9 = 0.046). (A) All clades on the phylogeny; clades with >50% of their species in Mediterranean hotspots are marked with filled diamonds. (B) Clades with >50% of their species in Southwest Australia; horizontal dashed lines are the 95% highest probability densities of ages estimated in BEAST. (C) Clades with >50% of their species in the Cape Floristic Region; horizontal lines as above.
Finally, we tested whether the speciation rate (λ) was significantly higher in clades with >50% of their species in Mediterranean hotspots than in other clades, following the method of Ricklefs (25). By using nonlinear least-squares regression, we estimated the relative extinction rate (κ) for the sample of Proteaceae genera with >1 species (Fig. S3h). We then calculated the speciation rate (λ) and net diversification rate (r) for each clade (genus), given a fixed value of κ, and we ran Kruskal–Wallis tests on these rates comparing clades from Mediterranean hotspots against other clades. κ was estimated to whichever upper limit value <1 this parameter was allowed to reach in the nonlinear regression (e.g., if κ ≤ 0.99, κ = 0.99 ± 0.085, P < 0.0001). However, regardless of the value of κ that was used for the tests, the speciation rate was always significantly higher in clades with >50% of their species in Mediterranean hotspots than in other clades, by a factor of 1.9 to 2.6 (P = 0.0004–0.0038; Table S4). This result was also true when including monospecific clades, using a different sample of all independent (non nested) clades present 20 my ago, or comparing net diversification rates instead of speciation rates. However, although the speciation rate is on average 0.8 to 2.0 times higher in clades with >50% of their species in the CFR than those in SWA, this result was only significant for lower values of κ (P < 0.05).
The fact that our attempt to model the observed diversity by fixing the speciation and extinction rates leads to estimates of the relative extinction rate tending to 1 may be interpreted as an indication that these parameters have probably not remained constant through time and across lineages (26). The difference we found between the standing diversity of Mediterranean hotspots and that of other areas may either be the result of higher speciation rates in the CFR and SWA or higher extinction rates elsewhere (25).
Discussion
Our results show that net diversification rates are significantly higher in some (but not all) Mediterranean hotspot lineages, as might be expected from the high species richness and occurrence of large genera in both SWA and the CFR. This study also highlights an important difference between the 2 regions. There are more separate lineages in SWA, and only a few of them are responsible for the remarkable diversity of the group in the region (Fig. 2 B and C; Table 1). Biogeographic reconstruction by using Fitch parsimony infers Australia as the center of origin and diversity of Proteaceae (Fig. S1). It is therefore not surprising that, with intracontinental immigration, SWA has more lineages and exhibits a more complex pattern of diversity than the CFR. Nearly all of the SWA lineages have sister groups elsewhere in Australia, suggesting that they arose either from the fragmentation of ancient pan-Australian distributions before the aridification of Central Australia or by more recent dispersal from Southern or Eastern Australian ancestors. In contrast, all of the CFR lineages have sister groups outside Africa. Two of them (Leucadendreae and Aulax) actually appear to have originated as the results of long-distance dispersal of Australian ancestors, whereas the other 2 (Proteeae and Brabejum) might have come about as the combination of Gondwana breakup and long-distance dispersal. In this respect, the CFR appears more “insular” than SWA. Studies in other plant groups have demonstrated similar phylogenetic isolation of the CFR from the rest of Africa (27, 28). In a recent study, Linder (26) compared speciation rates among plant clades of 6 large regions and found that Australia as a whole had the signature of an old radiation, whereas the Cape flora could be seen as the result of a combination of both old and recent and rapid radiations. Our results suggest that recent and rapid radiations may also have taken place in Southwest Australia, in combination with older radiations.
Although the highest net diversification rates in Proteaceae are clearly associated with Mediterranean hotspots, our analyses rule out a contemporaneous origin for these species-rich lineages. This indicates that diversification in these hotspots increased gradually over an extended period, starting as far back as 40 mya (11, 27). Recent work has shown that the transition from tropical or subtropical to Mediterranean climates in these regions during the Cenozoic has also been gradual, even though the stabilization of modern climates is fairly recent (5, 6).
Adaptation to frequent fire regimes (e.g., serotiny) and nutrient-poor soils (e.g., cluster roots), pollinator shifts, and ant dispersal have long been considered to promote speciation in Mediterranean hotspots (1, 29–31). These are well known in Proteaceae but also occur in many other plant groups with high diversity in SWA and the CFR. This replicated pattern, both within Proteaceae and across angiosperms, suggests a common, convergent response to climate change in numerous distinct lineages. The ability to adapt to this change, more than the change itself, has probably been an important determinant in limiting the number of lineages that survived in these new environments. The lack of competition from other lineages may have resulted in lower extinction rates for those surviving and therefore higher net diversification rates. However, it is very difficult to disentangle extinction and speciation rates and further data are needed to determine whether the observed pattern is a result of higher speciation rates, lower extinction rates, or a combination of both.
In comparison with Southwest Australia and the Cape Floristic Region, the Mediterranean-climate region of Central Chile has fewer plant species and a lower rate of endemism (Table 1). One of the most notable differences between this region and the other 2 southern Mediterranean hotspots is its higher soil fertility as a result of mountain building and increased volcanic activity in the Cenozoic (1, 32). We speculate that this difference may have resulted in higher competition among local taxa and therefore higher rates of extinction. This might explain why Proteaceae, although present in other environments across South America, have only 5 species today in Central Chile, none of which is endemic to this particular region (Table S5).
Our results from a robust dated phylogeny in a model group show that the outstanding species richness of both SWA and the CFR has been produced by greater diversification rates. They also suggest that SWA biota are less isolated (i.e., have more biogeographical connections with their continent) and more phylogenetically diverse (i.e., have more lineages with sister groups outside the region) than those of the CFR. The quantification of diversity production over time is only as good as the dating analyses. We suggest that fine tuning of age estimates in other components of the Mediterranean hotspot biomes, resulting from morphological research to place fossils on phylogenies, will ultimately result in a much better global understanding of the evolutionary forces that have shaped such spectacular biodiversity.
Materials and Methods
Phylogenetic Analyses.
DNA sequences from 1 nuclear (ITS) and 7 plastid markers (atpB, atpB-rbcL, matK, rbcL, rpl16 intron, trnL intron, trnL-trnF) were aligned and combined into a dataset of 97 taxa and 9,914 characters, which was analyzed by using parsimony and Bayesian methods (SI Materials and Methods, Table S6).
Fossil Calibration.
Twenty-five fossil palynomorph species were analyzed phylogenetically by using a morphological matrix of 22 pollen characters and 113 taxa (SI Materials and Methods, Dataset S1) and the Bayesian majority-rule consensus tree from the molecular analyses (Fig. S1) as a backbone constraint. Following protocols detailed in SI Materials and Methods, this approach produced a set of 5 fossil pollen minimum age constraints (Table S1; Fig. S2).
Dating Analyses.
Penalized likelihood (PL) and Bayesian autocorrelation dating were performed as outlined in SI Materials and Methods. The uncorrelated lognormal (UCLN) dating method (23) was implemented in BEAST v. 1.4.7 (33) after partitioning the data to use the optimal models selected by MrModeltest for each partition. Age constraints were applied as uniform priors with hard bounds. To conform to these constraints, the PL chronogram was used as a starting tree. Ten independent runs of 4 × 106 generations each were performed, sampling every 1,000th generation. Careful examination of all parameters in Tracer v. 1.4 indicated that each run converged immediately (because the starting tree was already near-optimal) and that parameter estimates were consistent among multiple runs. All runs were therefore combined to produce the maximum clade credibility tree (Fig. 1) and calculate the 95% highest probability density for the age of each node on the phylogeny.
Diversification Rates.
Total species numbers for each genus of Proteaceae were compiled from the literature (Table S5; references available in ref. 15). Stem group diversification rates were calculated as rst = log(N)/Tst, where N is the clade size (number of extant species) and Tst the stem age. Crown group diversification rates were calculated as rcr = (log(N) − log (2))/Tcr where Tcr is the crown age. Stem and crown group diversification rates under a high relative rate of extinction (κ = λ/μ = 0.9) and 95% confidence intervals on clade size based on stem age were calculated by using equations 6, 7, and 10 of Magallón and Sanderson (16). We did not use equation 11 of Magallón and Sanderson (confidence intervals based on crown ages), which was corrected in an erratum published in 2006 (16).
In addition, we followed the methods of Ricklefs (25) and Ricklefs et al. (34) to estimate global rates of speciation and extinction for Proteaceae and test whether speciation rates are significantly higher in Mediterranean hotspot clades than in other clades (SI Materials and Methods; Table S4; Fig. S3 h–j).
Supplementary Material
Acknowledgments.
We thank Timothy Barraclough, Mark Chase, Félix Forest, Susana Magallón, the Editor, and 3 anonymous reviewers for their comments; Katherine Downs, Simon Gilmore, Nahid Heidari, and John Thomson for assistance in DNA sequencing; and numerous colleagues for providing plant material for DNA extraction. This work was supported by the Royal Botanic Gardens and Domain Trust, Sydney, the Swedish Research Council, National Science Foundation Grant DEB-0516340, and European Commission EST HOTSPOTS and Marie Curie Outgoing International Fellowship 021943-HOTMED.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU169608–EU169673 and EU676050–EU676122). For a list of accession numbers, see Table S6.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805607106/DCSupplemental.
References
- 1.Cowling RM, et al. Plant diversity in Mediterranean-climate regions. Trends Ecol Evol. 1996;11:362–366. doi: 10.1016/0169-5347(96)10044-6. [DOI] [PubMed] [Google Scholar]
- 2.Myers N, et al. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 3.Mittermeier RA, et al. Hotspots Revisited: Earth's Biologically Richest and Most Endangered Terrestrial Ecoregions. Mexico City: CEMEX; 2004. [Google Scholar]
- 4.Cowling RM, Lamont BB. On the nature of Gondwanan species flocks: Diversity of Proteaceae in Mediterranean South-western Australia and South Africa. Aust J Bot. 1998;46:335–355. [Google Scholar]
- 5.Linder HP. The radiation of the Cape flora, southern Africa. Biol Rev. 2003;78:597–638. doi: 10.1017/s1464793103006171. [DOI] [PubMed] [Google Scholar]
- 6.Hopper SD, Gioia P. The Southwest Australian Floristic Region: Evolution and conservation of a global hot spot of biodiversity. Annu Rev Ecol Evol Syst. 2004;35:623–650. [Google Scholar]
- 7.Arroyo MTK, et al. La flora de Chile central y su protección: Antecedentes y prioridades para el establecimiento del Jardín Botánico Chagual. Chagual. 2003;1:31–40. [Google Scholar]
- 8.Richardson JE, et al. Rapid and recent origin of species richness in the Cape flora of South Africa. Nature. 2001;412:181–183. doi: 10.1038/35084067. [DOI] [PubMed] [Google Scholar]
- 9.Goldblatt P, et al. Radiation in the Cape flora and the phylogeny of peacock irises Moraea (Iridaceae) based on four plastid DNA regions. Mol Phylogenet Evol. 2002;25:341–360. doi: 10.1016/s1055-7903(02)00235-x. [DOI] [PubMed] [Google Scholar]
- 10.Linder HP, Eldenäs P, Briggs BG. Contrasting patterns of radiation in African and Australian Restionaceae. Evolution (Lawrence, Kans) 2003;57:2688–2702. doi: 10.1111/j.0014-3820.2003.tb01513.x. [DOI] [PubMed] [Google Scholar]
- 11.Edwards D, Hawkins JA. Are Cape floral clades the same age? Contemporaneous origins of two lineages in the genistoids s. l. (Fabaceae) Mol Phylogenet Evol. 2007;45:952–970. doi: 10.1016/j.ympev.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Hoot SB, Douglas AW. Phylogeny of the Proteaceae based on atpB and atpB-rbcL intergenic spacer region sequences. Aust Syst Bot. 1998;11:301–320. [Google Scholar]
- 13.Barker NP, Weston PH, Rourke JP, Reeves G. The relationships of the southern African Proteaceae as elucidated by internal transcribed spacer (ITS) DNA sequence data. Kew Bull. 2002;57:867–883. [Google Scholar]
- 14.Barker NP, Weston PH, Rutschmann F, Sauquet H. Molecular dating of the ‘Gondwanan’ plant family Proteaceae is only partially congruent with the timing of the break-up of Gondwana. J Biogeogr. 2007;34:2012–2027. [Google Scholar]
- 15.Weston PH, Barker NP. A new suprageneric classification of the Proteaceae, with an annotated checklist of genera. Telopea. 2006;11:314–344. [Google Scholar]
- 16.Magallón S, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution (Lawrence, Kans) 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. and erratum (2006) 60:2411. [DOI] [PubMed] [Google Scholar]
- 17.Warren BH, Hawkins JA. The distribution of species diversity across a flora's component lineages: Dating the Cape's ‘relicts.’. Proc R Soc Lond B. 2006;273:2149–2158. doi: 10.1098/rspb.2006.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dettmann ME, Jarzen DM. The early history of the Proteaceae in Australia: The pollen record. Aust Syst Bot. 1998;11:401–438. [Google Scholar]
- 19.Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- 20.Thorne JL, Kishino H, Painter IS. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- 21.Kishino H, Thorne JL, Bruno WJ. Performance of a divergence time estimation method under a probabilistic model of rate evolution. Mol Biol Evol. 2001;18:352–361. doi: 10.1093/oxfordjournals.molbev.a003811. [DOI] [PubMed] [Google Scholar]
- 22.Thorne JL, Kishino H. Divergence time and evolutionary rate estimation with multilocus data. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- 23.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isaac NJB, Agapow P-M, Harvey PH, Purvis A. Phylogenetically nested comparisons for testing correlates of species richness: A simulation study of continuous variables. Evolution (Lawrence, Kans) 2003;57:18–26. doi: 10.1111/j.0014-3820.2003.tb00212.x. [DOI] [PubMed] [Google Scholar]
- 25.Ricklefs RE. Global variation in the diversification rate of passerine birds. Ecology. 2006;87:2468–2478. doi: 10.1890/0012-9658(2006)87[2468:gvitdr]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 26.Linder HP. Plant species radiations: Where, when, why? Phil Trans R Soc Lond B. 2008;363:3097–3105. doi: 10.1098/rstb.2008.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linder HP. Evolution of diversity: the Cape flora. Trends Plants Sci. 2005;10:536–541. doi: 10.1016/j.tplants.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Galley C, Linder HP. Geographical affinities of the Cape flora, South Africa. J Biogeogr. 2006;33:236–250. [Google Scholar]
- 29.Verdú M, Pausas JG, Segarra-Moragues JG, Ojeda F. Burning phylogenies: Fire, molecular evolutionary rates, and diversification. Evolution (Lawrence, Kans) 2007;61:2195–2204. doi: 10.1111/j.1558-5646.2007.00187.x. [DOI] [PubMed] [Google Scholar]
- 30.Barraclough TG. What can phylogenetics tell us about speciation in the Cape flora? Divers Distrib. 2006;12:21–26. [Google Scholar]
- 31.Lambers H, et al. Root structure and functioning for efficient acquisition of phosphorus: Matching morphological and physiological traits. Ann Bot. 2006;98:693–713. doi: 10.1093/aob/mcl114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ege H, Sobel ER, Scheuber E, Jacobshagen V. Exhumation history of the southern Altiplano plateau (southern Bolivia) constrained by apatite fission track thermochronology. Tectonics. 2007;26:TC1004. [Google Scholar]
- 33.Drummond AJ, Rambaut A BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricklefs RE, Losos JB, Townsend TM. Evolutionary diversification of clades of squamate reptiles. J Evol Biol. 2007;20:1751–1762. doi: 10.1111/j.1420-9101.2007.01388.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.