Abstract
Homobasidiomycete fungi display many complex fruiting body morphologies, including mushrooms and puffballs, but their anatomical simplicity has confounded efforts to understand the evolution of these forms. We performed a comprehensive phylogenetic analysis of homobasidiomycetes, using sequences from nuclear and mitochondrial ribosomal DNA, with an emphasis on understanding evolutionary relationships of gilled mushrooms and puffballs. Parsimony-based optimization of character states on our phylogenetic trees suggested that strikingly similar gilled mushrooms evolved at least six times, from morphologically diverse precursors. Approximately 87% of gilled mushrooms are in a single lineage, which we call the “euagarics.” Recently discovered 90 million-year-old fossil mushrooms are probably euagarics, suggesting that (i) the origin of this clade must have occurred no later than the mid-Cretaceous and (ii) the gilled mushroom morphology has been maintained in certain lineages for tens of millions of years. Puffballs and other forms with enclosed spore-bearing structures (Gasteromycetes) evolved at least four times. Derivation of Gasteromycetes from forms with exposed spore-bearing structures (Hymenomycetes) is correlated with repeated loss of forcible spore discharge (ballistospory). Diverse fruiting body forms and spore dispersal mechanisms have evolved among Gasteromycetes. Nevertheless, it appears that Hymenomycetes have never been secondarily derived from Gasteromycetes, which suggests that the loss of ballistospory has constrained evolution in these lineages.
One of the major challenges of evolutionary biology is to understand the origin and diversification of biological form. In this endeavor, fungi have proven difficult because of their anatomical simplicity and scanty fossil record. A prime example is provided by gilled mushrooms and puffballs, which are the fruiting bodies of certain homobasidiomycetes (fungi that produce meiotic spores on nonseptate basidial cells). These are perhaps the most conspicuous and widely recognized fungal forms, yet their evolutionary origins are unresolved. Of the ≈13,500 known species of homobasidiomycetes, over half (≈8500 species) are gilled mushrooms, and ≈400 are puffballs (1). Systematic mycologists have suspected that each of these forms evolved several times, but the lack of a general phylogenetic framework for homobasidiomycetes has made it impossible to test this proposition. In this study, we used nucleotide sequences from genes encoding ribosomal RNA (rDNA) to perform the first comprehensive phylogenetic analysis of homobasidiomycetes and specifically to evaluate the evolutionary relationships of puffballs and gilled mushrooms.
Traditional 19th century classifications of fungi were based solely on macromorphology, especially that of the spore-bearing structures (the hymenophore). All fungi that produce spores on an exposed hymenophore were grouped in the class Hymenomycetes, which contained two orders: Agaricales, for gilled mushrooms, and Aphyllophorales, for polypores, toothed fungi, coral fungi, and resupinate, crust-like forms. Puffballs, and all other fungi with enclosed hymenophores, were placed in the class Gasteromycetes. Anatomical studies since the late 19th century have suggested that this traditional system is artificial (2), and recent molecular phylogenetic studies have confirmed relationships among morphologically dissimilar homobasidiomycetes. For example, it is now well established that subterranean, tuber-like “false truffles” have been derived from various lineages of epigeous mushrooms (3, 4) and that, in at least one case, gilled mushrooms have been derived from poroid ancestors (5). Although molecular studies have provided insight into the evolution of certain lineages, there has been no broad phylogenetic analysis of homobasidiomycetes.
MATERIALS AND METHODS
Taxon Sampling.
To construct a comprehensive phylogenetic data set, representatives of all the major lineages of homobasidiomycetes were sampled. Exemplars were selected from 10 families of Agaricales (6), 18 families of Aphyllophorales (2), and seven families of Gasteromycetes (7); these included 20 species of gilled mushrooms, 52 nongilled Hymenomycetes, and nine Gasteromycetes, including five puffballs (a list of strains is available from D.S.H.). In modern homobasidiomycete taxonomy, there are many small families that have been segregated relatively recently on the basis of unique characters, as well as a handful of larger, older families that are united not by synapomorphies but rather by the lack of distinguishing characters that could be used to subdivide them (2, 6, 7). Single exemplars were chosen from the smaller, putatively monophyletic families (e.g., Schizophyllaceae, Fistulinaceae, and Ganodermataceae) whereas multiple species were sampled from the larger, presumably artificial families (Clavariaceae, Corticiaceae, Polyporaceae, and Tricholomataceae). Based on previous phylogenetic analyses at more inclusive levels than the present study (8), the heterobasidiomycete “jelly fungi,” Auricularia, Dacrymyces, and Tremella, were included for rooting purposes.
Molecular Techniques and Phylogenetic Analyses.
Laboratory methods for culturing, DNA isolation, PCR amplification, and DNA sequencing have been described (9, 10). Sequences of nuclear (nuc) and mitochondrial (mt) small subunit (ssu) rDNA (nuc-ssu-rDNA and mt-ssu-rDNA) were obtained using published primer sequences (10, 11). Seventy-five nuc-ssu-rDNA and 44 mt-ssu-rDNA sequences have been deposited in GenBank (accession nos. AF026567–AF026687). This study also used 40 mt-ssu-rDNA sequences from our previous work (ref. 9; GenBank accessions U27023–U27080, U59099) and one mt-ssu-rDNA and nine nuc-ssu-rDNA sequences downloaded from GenBank: Agaricus bisporus (L36658), Auricularia auricula-judae (L22254), Boletus satanas (mt-ssu-rDNA M91009 and nuc-ssu-rDNA M94337), Coprinus cinereus (M92991), Dacrymyces chrysospermus (L22257), Lepiota procera (L36659), Pleurotus ostreatus (U23544), Schizophyllum commune (X54865), and Tremella foliacea (L22262).
Parsimony analyses of manually aligned sequences were performed using paup* 4.0d53 (test version provided by D. L. Swofford, Smithsonian Institution, Washington, D.C.). All transformations were unordered and equally weighted. Three hundred replicate heuristic searches were performed, using random taxon addition sequences and TBR branch swapping. Bootstrap analyses used 100 replicates, with simple taxon addition sequence, with TBR branch swapping, and with MULPARS off. Complete nuc-ssu-rDNA coding sequences were ≈1750-bp long and were alignable over their entire length, except for three regions of 55, 95, and 83 bp in Cantharellus tubaeformis, which are highly divergent and were excluded from the analyses. Partial mt-ssu-rDNA sequences ranged from 550 to over 1000 bp, which was due to length variation in three hypervariable regions (9, 12, 13) that alternate with three conserved regions of 131, 237, and 116 bp (aligned). The conserved regions were aligned for all ingroup taxa except Sparassis spathulata, which is highly divergent and was omitted from mt-rDNA alignments. The second conserved region of mt-ssu-rDNA, termed “block 5” (9), showed greater sequence divergence than the other regions; outgroup sequences could not be aligned to the ingroup in this region. Analyses were performed that included or excluded the entire block 5 region, as well as the mt-rDNA sequence from Sparassis. Although there were some topological differences, basic conclusions regarding evolution of gilled mushrooms and puffballs were not sensitive to the inclusion or exclusion of these data (results not shown). The alignment can be obtained from TreeBASE (ref. 14) or from D.S.H.
Topologically constrained analyses were used to evaluate the hypothesis that all gilled mushrooms form a single lineage. Constraint trees were constructed using macclade (15), which forced monophyly of gilled mushrooms but which specified no other tree structure. Parsimony analyses were performed under this constraint, using the same settings as in the baseline analyses (above). The resulting trees were evaluated by the Kishino–Hasegawa maximum likelihood test, using the program dnaml of the phylip software package (16). macclade also was used to infer historical patterns of morphological transformations. Fruiting body morphology was coded as an unordered character with three-states (gilled mushroom/nongilled Hymenomycete/Gasteromycete) that were optimized onto the trees using parsimony, with all transformations equally weighted.
RESULTS AND DISCUSSION
Evolution of Gilled Mushrooms.
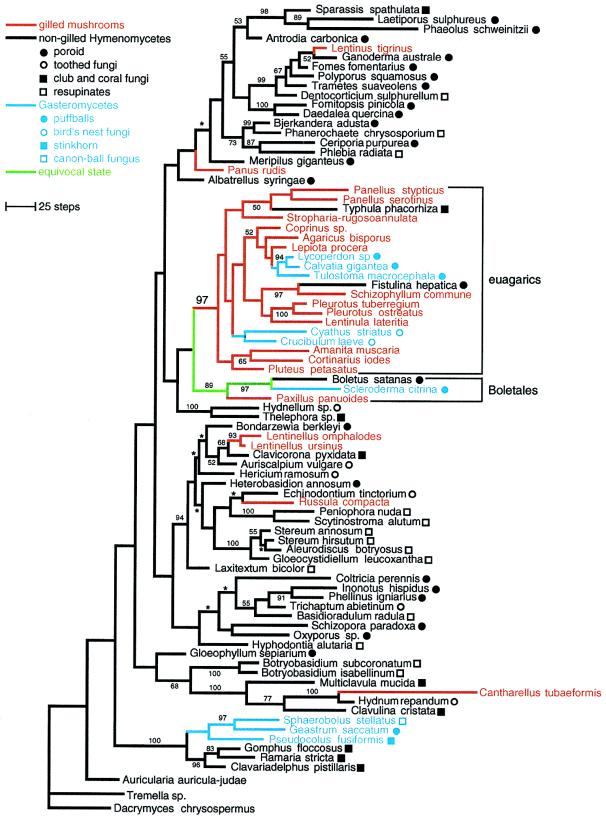
Phylogenetic analysis resulted in 52 equally parsimonious trees of 4650 steps (consistency index = 0.273, retention index = 0.496), which differ only by minor rearrangements in three clades (Fig. 1). Thirteen of the 20 species of gilled mushrooms in this study are contained in a single lineage that is present in all equally parsimonious trees and was strongly supported by bootstrap analysis (97%; Fig. 1). For purposes of discussion, we refer to this clade as the “euagarics.” Extrapolating from current taxonomy (1, 6) and other molecular phylogenetic studies (9, 17–19), we estimated that the euagarics clade contains ≈7,400 (87%) of the recognized species of gilled mushrooms, which is over half of all known species of homobasidiomycetes. This main radiation of gilled mushrooms has produced such familiar forms as the cultivated button mushroom, Agaricus bisporus, and the poisonous “fly agaric,” Amanita muscaria, as well as two of the best studied fungal model systems for developmental and mating genetics, Coprinus cinereus and Schizophyllum commune.
Figure 1.
Phylogeny of homobasidiomycetes inferred from nuc-ssu-rDNA and mt-ssu-rDNA sequences. One of 52 equally parsimonious trees. Branches with asterisks collapse in the strict consensus tree. Numbers by nodes are bootstrap frequencies (values <50% not shown). Branch colors represent morphological character state optimizations. Symbols by taxon names indicate specific fruiting body types of Gasteromycetes and nongilled Hymenomycetes.
Although most euagarics are gilled mushrooms, our results imply that this clade also has given rise to certain coral fungi (Typhula phacorhiza) and polypores (Fistulina hepatica), as well as two lineages of Gasteromycetes, one containing puffballs in the Lycoperdales (Lycoperdon, Calvatia) and Tulostomatales (Tulostoma) and the other containing “bird’s nest fungi” in the Nidulariales (Cyathus, Crucibulum; Fig. 2). [Placement of Nidulariales in the euagarics is also supported by independent analyses of nuc-ssu-rDNA sequences (A. Gargas, University of Copenhagen, personal communication) and nuc large subunit rDNA sequences (R. G. Thorn, University of Wyoming, personal communication).] Despite these parallelisms, parsimony-based character state optimizations suggest that the stem species of the euagarics was a gilled mushroom (Fig. 1). The oldest unambiguous homobasidiomycete fossils are 90–94 million-year-old gilled mushrooms that are strikingly similar to certain extant euagarics (20, 21). This suggests that (i) the origin of the euagarics must have been no later than the mid-Cretaceous and (ii) the gilled mushroom morphology has been maintained in some lineages for tens of millions of years.
Figure 2.
Fruiting body forms in homobasidiomycetes. (A–C) Independently derived gilled mushrooms. (A) Pleurotus ostreatus. (B) Lentinellus ursinus. (C) Panus rudis. (D–F) Independently derived puffballs. (D) Lycoperdon perlatum. (E) Scleroderma citrina (Boletales). (F) Geastrum saccatum, an earthstar. (G–I) Uniquely evolved fruiting body forms of Gasteromycetes. (G) Crucibulum laeve, a bird’s nest fungus. (H) Pseudocolus fusiformis, a stinkhorn. (I) Sphaerobolus stellatus, the cannon ball fungus (fruiting bodies are ≈1.5 mm in diameter). Pleurotus (A), Lycoperdon (D), and Crucibulum (G) are in euagarics; Geastrum (F), Pseudocolus (H), and Sphaerobolus (I) form a monophyletic group (see Fig. 1).
Seven species of gilled mushrooms in this analysis were placed outside of the euagarics. Alternate tree topologies in which all gilled mushrooms form a single clade are 217 steps (4.7%) longer than the unconstrained trees and are rejected by maximum likelihood (16). Parsimony-based optimizations of morphological character states suggest that gilled mushrooms evolved at least six times although the precise location on the tree of some changes is equivocal (Fig. 1). Developmental studies have demonstrated that there are ontogenetic differences among gills of two of the independently derived gilled mushroom lineages in this analysis, the genera Lentinus and Panus, which supports the view that they are not homologous (Fig. 1; ref. 22). The results presented here provide a phylogenetic framework for additional comparative studies of hymenophore development.
The sister group of the euagarics in all equally parsimonious trees is the Boletales clade (Boletus, Paxillus, and Scleroderma; Fig. 1), but this is not resolved with confidence, and the evolutionary precursor of the gills of euagarics therefore remains unclear. Even if monophyly of the euagarics plus the Boletales was strongly supported, however, the plesiomorphic morphology of the Boletales is unresolved (Fig. 1). In our study, the Boletales were represented by poroid, gilled, and puffball forms. This is consistent with the findings of Bruns and colleagues (3, 12, 18), who also have shown that the Boletales includes false truffles and resupinate forms. The closest relatives of the other lineages of gilled mushrooms in our analysis are various nongilled Hymenomycetes. For example, the gilled mushroom Lentinus tigrinus is nested in a clade of polypores whereas the closest relatives of the gilled mushrooms Lentinellus omphalodes and L. ursinus are a coral fungus and a toothed fungus (Fig. 1). These results indicate that gilled mushrooms have been derived from morphologically diverse precursors.
Evolution of Gasteromycetes.
The nine species of Gasteromycetes that we examined occur in four separate lineages and appear to have been derived from both gilled and nongilled Hymenomycetes (Fig. 1). In addition, anatomical studies have suggested that as many as 14 lineages of Hymenomycetes have given rise to gasteromycetous false truffles and “secotioid” fungi, which are epigeous Gasteromycetes that resemble unexpanded mushrooms (23, 24). In several cases, such hypotheses have been supported by molecular studies. For example, previous studies have suggested that (i) the false truffles Rhizopogon and Chamonixia and the secotioid fungus Gastrosuillus are derived from within the Boletales (3, 12, 18, 25), (ii) the false truffle Hydnangium is closely related to the gilled mushroom Laccaria (4), and (iii) the secotioid fungi Podaxis and Montagnea are nested in the gilled mushroom family Coprinaceae (19). Taken together with our results, this suggests that Gasteromycetes have been repeatedly derived from Hymenomycetes, but there is no evidence that this transformation has ever been reversed.
Derivation of Gasteromycetes from Hymenomycetes involves the evolution of an enclosed hymenophore. In the gilled mushroom Lentinus tigrinus, there is a naturally occurring developmental mutant in which a recessive allele at a single locus confers a Gasteromycete-like enclosed hymenophore (26). Although the genetic basis of gasteromycetization in other lineages is unknown, the situation in L. tigrinus suggests that such transformations could be mediated by one or a small number of mutations in genes that have large phenotypic effects. The resemblance of secotioid Gasteromycetes to unopened mushrooms has led to suggestions that the initial steps in transformations from Hymenomycetes to Gasteromycetes are mutations that confer loss of function in developmental pathways, resulting in pedomorphosis (3, 23, 25). This view is consistent with observations of low levels of rDNA sequence divergence between certain secotioid fungi and closely related Hymenomycetes (3, 25).
In addition to the evolution of an enclosed hymenophore, derivation of Gasteromycetes from Hymenomycetes entails changes in the mechanisms of spore dispersal. Hymenomycetes discharge spores by a forcible mechanism, termed “ballistospory,” that is absent in Gasteromycetes. Structural features associated with ballistospory include short, curved sterigmata (the stalks that bear the spores), asymmetrical spores, and formation of a droplet of liquid at the base of the spore at the time of discharge (27). It appears that the suite of characters involved in ballistospory, once lost, has never been regained, which may explain why forms with exposed hymenophores have never been secondarily derived from Gasteromycetes.
In the absence of ballistospory, diverse spore dispersal mechanisms have evolved among Gasteromycetes (28). In puffballs, spores are produced internally and sift into the air through cracks or pores in the outer wall of the fruiting body (Fig. 2 D–F). Our results suggest that the puffball type fruiting body has evolved at least three times (Figs. 1 and 2). This is a conservative estimate because the taxonomically controversial puffballs Astraeus and Calostoma were not included in the analysis. In false truffles, spores are produced internally and are disseminated into soil as fruiting bodies break down and may also be dispersed by rodents that eat the fruiting bodies (29). As discussed above, molecular and morphological evidence suggests that false truffles also have multiple origins.
Other “solutions” to nonballistosporic dispersal appear to have arisen only once. In Nidulariales, spores are contained in packets (peridioles) that are dislodged from an upturned, concave fruiting body by a splash-cup mechanism (Figs. 1 and 2G; ref. 30). The dislodged peridioles adhere to vegetation by means of a specialized hyphal cord and are thought to be dispersed by herbivores (30). In Phallales (represented in this study by Pseudocolus fusiformis), spores are dispersed by insects, especially Diptera. Spores develop within an initially enclosed fruiting body primordium but become exposed as the fruiting body expands. At maturity, a showy, flower-like structure is produced, which is lined by a dark, fetid slime in which the spores are suspended (Fig. 2H). Finally, in Sphaerobolus, spores are produced in a glebal mass inside minute (≈1.5 mm in diameter) fruiting bodies. At maturity, the outer wall of the fruiting body splits open, and the inner wall suddenly evaginates, ejecting the spore mass up to 6 m (Fig. 2I; ref. 31).
Our results suggest that Phallales, Sphaerobolus, and the puffball Geastrum form a monophyletic group (Fig. 1). With three radically different spore dispersal mechanisms, this clade provides a remarkable example of functional and morphological diversification. Although this group is not strongly supported by bootstrapping, it is nevertheless nested in a strongly supported, slightly more inclusive clade, which also includes the Hymenomycetes Gomphus, Ramaria, and Clavariadelphus. [This is consistent with results of unpublished analyses of nuc-lsu-rDNA and mt-lsu-rDNA sequences that suggest that Ramaria, Phallales, and Sphaerobolus are monophyletic (R. G. Thorn, personal communication, and J. Spatafora, Oregon State University, personal communication).]
CONCLUSIONS
Our results support the view that homobasidiomycete evolution has been marked by extensive convergence and parallelism in fruiting body morphology (Fig. 1). A prime example is provided by gilled mushrooms, which evidently evolved at least six times. In addition to demonstrating such patterns, our results provide a phylogenetic framework for studying mechanisms of morphological evolution, as well as for discovering correlations between the evolution of fruiting body forms and other morphological features. For example, in Gasteromycetes, our observation that each separate origin of an enclosed hymenophore is associated with the loss of ballistospory strengthens the view that they are causally related, as has been suggested (23). Similarly, the inference that Gasteromycetes have never given rise to Hymenomycetes suggests that loss of ballistospory has constrained evolution in certain lineages. Under this constraint, diverse nonballistosporic dispersal mechanisms have evolved. Puffballs, false truffles, and secotioid fungi apparently have evolved repeatedly, and there may be simple genetic and developmental bases for their derivation from Hymenomycetes (3, 23, 26). In contrast, there is no model, nor is there any empirical evidence, for developmental modifications that could result in the direct transformation of a Hymenomycete into a bird’s nest fungus, stinkhorn, or cannon ball fungus. These uniquely derived Gasteromycete forms (Fig. 2 G–I) are among the most complex structures in fungi, involving a high degree of developmental integration and a large number of differentiated tissues. It is most likely that these forms evolved by elaboration from simpler Gasteromycetes with puffball, false truffle, or secotioid morphologies. Although this study did not include any false truffles or secotioid fungi, with further sampling of these groups it will be possible to test this and other hypotheses concerning pathways of morphological evolution in homobasidiomycetes.
Acknowledgments
We thank Andrea Gargas, Jean-Marc Moncalvo, Joey Spatafora, Greg Thorn, and Rytas Vilgalys for sharing unpublished results, David Swofford for providing a test version of paup* 4.0d53, Tom Bruns and Meredith Blackwell for helpful comments, Jean-Marc Moncalvo, Ronald Petersen, Joost Stalpers, and Rytas Vilgalys for certain fungal materials and DNAs, Kathie Hodge for the image in Fig. 2G, Beth Brantley for the image in Fig. 2I, and Josef Breitenbach (Verlag Mykologia Luzern) for permission to reprint the images in Fig. 2 B, D, and E. Support was provided by National Science Foundation Grants DEB-930268 to D.S.H. and DEB-9629427 to M.J.D. and D.S.H. and by Ford Foundation and Sigma-Xi grants to E.M.P.
ABBREVIATIONS
- rDNA
ribosomal RNA
- mt
mitochondrial
- nuc
nuclear
- ssu
small subunit
Footnotes
References
- 1.Hawksworth D L, Kirk P M, Sutton B C, Pegler D N. Dictionary of the Fungi. 8th Ed. Oxon: CAB International; 1995. [Google Scholar]
- 2.Donk M A. Persoonia. 1964;3:199–324. [Google Scholar]
- 3.Bruns T D, Fogel R, White T J, Palmer J. Nature (London) 1989;339:140–142. doi: 10.1038/339140a0. [DOI] [PubMed] [Google Scholar]
- 4.Mueller G M, Pine E M. McIlvainea. 1994;11:61–74. [Google Scholar]
- 5.Hibbett D S, Vilgalys R. Syst Bot. 1993;18:409–433. [Google Scholar]
- 6.Singer R. The Agaricales in Modern Taxonomy. 4th Ed. Koenigstein, Germany: Koeltz; 1986. [Google Scholar]
- 7.Dring D M. In: The Fungi. Ainsworth G C, Sparrow F K, Sussman A S, editors. IVB. New York: Academic; 1973. pp. 451–478. [Google Scholar]
- 8.Swann E C, Taylor J W. Mycologia. 1993;85:923–936. [Google Scholar]
- 9.Hibbett, D. S. & Donoghue, M. J. (1995) Can. J. Bot. 73, Suppl., s853–s861.
- 10.Hibbett D S. Mol Biol Evol. 1996;13:903–917. doi: 10.1093/oxfordjournals.molbev.a025658. [DOI] [PubMed] [Google Scholar]
- 11.White T J, Bruns T D, Lee S B, Taylor J W. In: PCR Protocols. Innis M A, Gelfand D H, Sninsky J J, White T J, editors. San Diego: Academic; 1990. pp. 315–322. [Google Scholar]
- 12.Bruns T D, Szaro T M. Mol Biol Evol. 1992;9:836–855. doi: 10.1093/oxfordjournals.molbev.a040760. [DOI] [PubMed] [Google Scholar]
- 13.Cummings D J, Domenico J M, Nelson J, Sogin M L. J Mol Evol. 1989;28:232–241. doi: 10.1007/BF02102481. [DOI] [PubMed] [Google Scholar]
- 14.Sanderson M J, Donoghue M J, Piel W, Eriksson T. Am J Bot. 1994;81:183. (abstr.). [Google Scholar]
- 15.Maddison W P, Maddison D R. macclade. Sunderland, MA: Sinauer; 1992. , Ver. 3. [Google Scholar]
- 16.Felsenstein J. phylip. Seattle, WA: University of Washington; 1993. [Google Scholar]
- 17.Chapela I H, Rehner S A, Schultz T R, Mueller U G. Science. 1994;226:1691–1694. doi: 10.1126/science.266.5191.1691. [DOI] [PubMed] [Google Scholar]
- 18.Bruns T D, Fogel R, Taylor J W. Mycologia. 1990;82:175–184. [Google Scholar]
- 19.Hopple J S, Jr, Vilgalys R. Mycologia. 1994;86:96–107. [Google Scholar]
- 20.Hibbett D S, Donoghue M J, Grimaldi D A. Nature (London) 1995;377:487. [Google Scholar]
- 21.Hibbett D S, Donoghue M J, Grimaldi D A. Am J Bot. 1997;84:981–991. [PubMed] [Google Scholar]
- 22.Hibbett D S, Murakami S, Tsuneda A. Am J Bot. 1993;80:1336–1348. [Google Scholar]
- 23.Thiers H D. Mycologia. 1984;76:1–8. [Google Scholar]
- 24.Smith A H. In: The Fungi. Ainsworth G C, Sparrow F K, Sussman A S, editors. IVB. New York: Academic; 1973. pp. 421–450. [Google Scholar]
- 25.Baura G, Szaro T M, Bruns T D. Mycologia. 1992;84:592–597. [Google Scholar]
- 26.Hibbett D S, Tsuneda A, Murakami M. Am J Bot. 1994;81:466–478. [Google Scholar]
- 27.Webster J, Davey R A, Duller G A, Ingold C T. Trans Br Mycol Soc. 1984;82:13–29. [Google Scholar]
- 28.Ingold C T. Fungal Spores: Their Liberation and Dispersal. Oxford: Clarendon; 1971. [Google Scholar]
- 29.Miller S L, Torres P, McClean T. Mycologia. 1994;86:89–95. [Google Scholar]
- 30.Brodie H J. The Bird’s Nest Fungi. Toronto: Univ. of Toronto Press; 1975. [Google Scholar]
- 31.Ingold G T. Trans Br Mycol Soc. 1972;58:179–195. [Google Scholar]