Abstract
The extinct aurochs (Bos primigenius primigenius) was a large type of cattle that ranged over almost the whole Eurasian continent. The aurochs is the wild progenitor of modern cattle, but it is unclear whether European aurochs contributed to this process. To provide new insights into the demographic history of aurochs and domestic cattle, we have generated high-confidence mitochondrial DNA sequences from 59 archaeological skeletal finds, which were attributed to wild European cattle populations based on their chronological date and/or morphology. All pre-Neolithic aurochs belonged to the previously designated P haplogroup, indicating that this represents the Late Glacial Central European signature. We also report one new and highly divergent haplotype in a Neolithic aurochs sample from Germany, which points to greater variability during the Pleistocene. Furthermore, the Neolithic and Bronze Age samples that were classified with confidence as European aurochs using morphological criteria all carry P haplotype mitochondrial DNA, suggesting continuity of Late Glacial and Early Holocene aurochs populations in Europe. Bayesian analysis indicates that recent population growth gives a significantly better fit to our data than a constant-sized population, an observation consistent with a postglacial expansion scenario, possibly from a single European refugial population. Previous work has shown that most ancient and modern European domestic cattle carry haplotypes previously designated T. This, in combination with our new finding of a T haplotype in a very Early Neolithic site in Syria, lends persuasive support to a scenario whereby gracile Near Eastern domestic populations, carrying predominantly T haplotypes, replaced P haplotype-carrying robust autochthonous aurochs populations in Europe, from the Early Neolithic onward. During the period of coexistence, it appears that domestic cattle were kept separate from wild aurochs and introgression was extremely rare.
Keywords: ancient DNA, aurochs, starburst network, mitochondrial haplotypes, domestication
1. Introduction
The word aurochs, translated from German, is both singular and plural and literally means ‘primeval ox’ or ‘proto-ox’. In his record of the Gallic Wars, Julius Caesar wrote of them: ‘They are a little below the elephant in size, and of the appearance, colour, and shape of a bull. Their strength and speed are extraordinary; they spare neither man nor wild beast which they have espied.’ During the Pleistocene, the range of the aurochs populations in Europe expanded and contracted in response to interglacial and glacial cycles. After the last deglaciation, around 11 000 years ago, Bos primigenius primigenius was found over almost the whole continent, apart from northern Scandinavia, northern parts of Russia and Ireland. While zooarchaeological data point towards the Near East and the Indus valley as being the main domestication and diffusion centres of cattle (Helmer et al. 2005), the wide geographical distribution of the aurochs makes it tantalizing to speculate about the possibility of European centres of domestication. In addition, since osteological differentiation between European aurochs and Early Neolithic domestic cattle is mainly based on size, osteoarchaeozoological methods cannot be used to exclude the possibility of local European domestication (Vigne & Helmer 1999). This uncertainty contrasts with the case of the domesticated Caprinae, where the wild progenitors were present in the Middle East but absent in Europe during the Early Holocene (Poplin 1979; Clutton-Brock 1987).
Based upon numerous surveys of modern Bos taurus (taurine cattle) populations from the British Isles, Scandinavia, northern, central and southern Europe, the Near East, Africa and east Asia, it has been concluded that mitochondrial (mt) sequences of European and Middle Eastern taurine cattle cluster around a central sequence, the T haplotype, and that the most probable centre of domestication of European cattle was the Near East (Loftus et al. 1994; Bailey et al. 1996; Bradley et al. 1996; Cymbron et al. 1999; Mannen et al. 1998; Troy et al. 2001; Mannen et al. 2004). All modern B. taurus mtDNA sequences reported belong to the T haplogroup, which can be further subdivided into five common and phylogeographically structured haplogroups (T, T1, T2, T3 and T4), as defined by 240 bp of the mtDNA D-loop. Initial findings from the well-preserved Palaeolithic aurochs remains from the British Isles demonstrated the presence of a highly divergent mtDNA haplogroup, which is absent in modern day cattle populations, and this was designated P (Bailey et al. 1996; Troy et al. 2001). The number of differences between the P and T modal haplotype sequences is 8 bp across a 252 bp region of the mtDNA D-loop (Troy et al. 2001). The absence of the P haplogroup in any modern European B. taurus sample, together with phylogeographic analysis of the T haplogroup in Europe and the Middle East, has led to the suggestion that the domesticated cattle of Europe are descended from Near Eastern progenitors (Troy et al. 2001).
To date, only small numbers of geographically limited aurochs specimens have been studied (Bailey et al. 1996; Troy et al. 2001; Anderung et al. 2005; Beja-Pereira et al. 2006), and it remains questionable whether the conclusions drawn so far are based on a representative sample. Thus, the hypothesis of T haplotype-carrying B. taurus replacing P haplogroup-carrying in situ aurochs in Europe, following their domestication in the Near East and Neolithic expansion, requires further support. In this study, we have determined mtDNA D-loop sequences from a large and geographically representative sample of aurochs across northern and central Europe and also from the Near East, in order to: (i) investigate the genetic diversity and the demographic history of aurochs in Europe during the Holocene (broadly, Mesolithic to Bronze Age), (ii) present a detailed mitochondrial phylogeny of aurochs in Europe, (iii) corroborate previous evidence for the geographical origin of European domesticated taurine cattle, and (iv) examine possible interbreeding between wild and domesticated cattle during the period when the two forms coexisted in Europe.
2. Material and methods
(a) Samples
In this study, 106 Bos bones from many locations across Europe were assessed for survival of ancient DNA. Eighty-three bones had been previously differentiated as wild aurochs, rather than domestic cattle, on the basis of size or date, by the researchers who carried out the archaeozoological studies on the various sites. However, 9 bones were only tentatively labelled as aurochs, 11 either did not have determinations associated with them or were labelled as Bos (i.e. either B. p. primigenius or B. taurus), 2 were labelled as possibly bison or buffalo and 1 was differentiated as a domestic cattle bone. A further five samples, D740 (Bailey et al. 1996) and TP65, CHWF, NORF and CPC98 (Troy et al. 2001), were re-amplified as part of this study, and the aurochs sequence D812 (Bailey et al. 1996) was also included in the analyses. This made a total of 112 Bos specimens under consideration. Detailed information on sample provenance and analytical results is given in table S1 in the electronic supplementary material.
(b) Extraction and polymerase chain reaction amplification
The samples were analysed in three different laboratories: the Smurfit Institute of Genetics at Trinity College Dublin; the Institute of Anthropology at the University of Mainz; and the Henry Wellcome Ancient Biomolecules Centre at the University of Oxford. The analytical location for each sample is indicated in table S1 in the electronic supplementary material. Bone samples were prepared using previously described protocols (Yang et al. 1998; MacHugh et al. 2000; Burger et al. 2004; Shapiro et al. 2004). All primers were designed to be genus specific, if not species specific, and amplified fragments of the hyper-variable control region of the mitochondrial genome (see figure 4 in electronic supplementary material for the primer strategy employed by each of the three amplification laboratories). Polymerase chain reaction (PCR) set-up was conducted in laboratories dedicated solely to pre-amplification ancient work. PCR conditions and primer details were previously described: Bollongino et al. (2006) for Dublin and Mainz, and Shapiro et al. (2004) for Oxford. Cloning was as described in Bollongino et al. (2006). Second-round PCR was not undertaken on any samples that did not amplify in the first round. In Dublin, all non-amplifiable samples were tested for presence of inhibitors (Edwards et al. 2004).
(c) Statistical and phylogenetic analysis
The criteria for authenticating mitochondrial haplotypes were as previously described (Edwards et al. 2004; Bollongino et al. 2006). mtDNA sequences were aligned by eye. A reduced median network was constructed (figure 2) from the control region data using a median algorithm (Bandelt et al. 1995). Sequences were analysed using an HKY model of nucleotide substitution (Hasegawa et al. 1985), which was selected using the hierarchical likelihood ratio tests implemented by ModelTest v. 3.7 (Posada & Crandall 1998). The neighbour-joining method (Saitou & Nei 1987) was used to construct a dendrogram (table S4 in the electronic supplementary material) from genetic distances generated by the DNADIST program (Felsenstein 1989). Representative sequences of the five T (T, T1, T2, T3 and T4) and two Z (Z1 and Z2) cattle haplogroups, which predominate in taurine and zebu cattle, respectively, covering the 361 bp region, were located on GenBank, as well as representative outgroups of yak (Bos grunniens, AF083355), gaur (Bos gaurus, AF083371) and European bison (Bison bonasus, AF083356). Bootstraps were calculated from 1000 pseudoreplicates of the data.
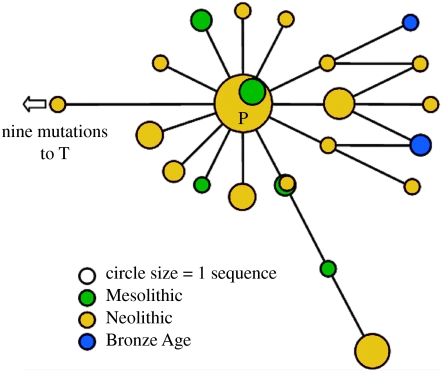
Figure 2.
Reduced control region network of the 51 P B. p. primigenius sequences obtained. The network is for positions 15 903–16 313 of the control region of the mitochondria, as denoted in the cattle mitochondrial genome sequence (Anderson et al. 1982).
Bayesian estimates of the mutation rate and the age of the most recent common ancestor (TMRCA) of the aurochs sequences were obtained using Beast v. 1.3; available from http://evolve.zoo.ox.ac.uk/beast/ (Drummond et al. 2006), from 37 P samples, covering a comparable 360 bp region. Sequences were analysed using an HKY model of nucleotide substitution (Hasegawa et al. 1985), which was selected using the hierarchical likelihood ratio tests implemented by ModelTest v. 3.7 (Posada & Crandall 1998). As the transition: transversion ratio could not be estimated from the data due to the lack of observed transversions, three different values (5, 50 and 500) were tested and yielded almost identical results. Two demographic models were tested for the coalescent prior: exponential growth and constant population size. The two models were compared using the average marginal posterior probabilities of the data, given the model. Mutation rate estimates were calibrated using the ages of the sequences, obtained by radiocarbon dating. The incorporation of these dates serves as sufficient calibration information for the estimation of the rate and divergence times (Drummond et al. 2002), and is more appropriate than using an external calibration point (Ho & Larson 2006). Posterior estimates of the mutation rate and age of the TMRCA were obtained by Markov Chain Monte Carlo analysis, with samples drawn every 500 steps over a total of 5 000 000 steps, following a discarded burn-in of 2 000 000 steps. Two separate runs were performed and the results combined. Adequate sampling and convergence to the stationary distribution were checked using Tracer v. 1.3 (Rambaut & Drummond 2004). Posterior estimates of parameters were all found to be distinctly unimodal (although with wide 95% highest posterior densities), and all parameters appeared to be identifiable, despite the relatively low information content in the sequences and the small age range of the sequences.
3. Results
One hundred and twelve Bos bones were considered in this study. Sample locations are shown in figure 1, with details in table S1 (published as electronic supplementary material). Amplification of a 361 bp section of the control region was successful in 42 samples, with a further 17 samples yielding sequence data from at least one of the three targeted control region fragments (table 1). The 54 samples amplified for the first time here were compared with the corresponding 272 bp of the control region amplified from two previously published British Pleistocene aurochs (Bailey et al. 1996). Of the 59 control region sequences, 49 in total were classified as P haplotypes by phylogenetic criteria, and the mtDNA control region data were used to construct a phylogenetic network (figure 2).
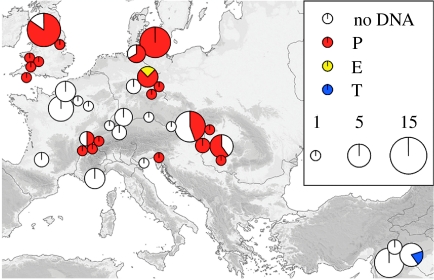
Figure 1.
Map showing locations of all sites sampled as part of this study. Map showing the relative proportions of aurochs (P and E) and taurine (T) mitotypes in the B. p. primigenius remains sampled in this study, from 34 European and 3 Near Eastern archaeological sites. Sample size is proportional to the area of the pie-circles indicated.
Table 1.
Variable positions in control region sequences of archaeological B. p. primigenius samples aligned to the aurochs consensus haplotype (CPC-05) for positions 15 903–16 313. (Differences are indicated with a period denoting identity. Missing sequence is denoted by question marks. Sequence codes are given in the first column and only variable sites are shown. The sequence positions from the BOVMT GENBANK sequence are given above each column (the reference sequence noted, accession number NC_001567 (Anderson et al. 1982)). In the final column, each sample has been assigned to have either a P haplotype or a T/T3 haplotype by means of its relative position in the full median network (not shown). The highly divergent E individual (EIL4) is not included here (see table S2 in the electronic supplementary material).)
| 15906 | 15933 | 15951 | 15953 | 15961 | 15994 | 16019 | 16022 | 16049 | 16050 | 16051 | 16055 | 16058 | 16074 | 16085 | 16109 | 16113 | 16121 | 16122 | 16133 | 16141 | 16231 | 16242 | 16247 | 16255 | 16264 | 16278 | 16301 | assignment to haplogroup | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CPC-05 | T | A | C | G | G | G | T | G | T | C | C | T | T | C | C | T | T | G | C | T | T | T | G | C | C | A | C | C | P |
| ETI1 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| ROS2 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| ROS4 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| ROS5 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| SV03 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| SZE2 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| ROS7 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| D812 | ? | ? | ? | ? | ? | ? | ? | ? | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| TP65 | ? | ? | ? | ? | ? | ? | ? | ? | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| H5 | ? | ? | ? | ? | ? | ? | ? | ? | · | · | · | · | · | · | · | · | · | · | · | · | · | ? | ? | ? | ? | ? | ? | ? | P |
| GL411 | ? | ? | ? | ? | ? | ? | ? | ? | · | · | · | · | · | · | · | · | · | · | · | · | · | ? | ? | ? | ? | ? | ? | ? | P |
| GL412 | ? | ? | ? | ? | ? | ? | ? | ? | · | · | · | · | · | · | · | · | · | · | · | · | · | ? | ? | ? | ? | ? | ? | ? | P |
| GL413 | ? | ? | ? | ? | ? | ? | ? | ? | · | · | · | · | · | · | · | · | · | · | · | · | · | ? | ? | ? | ? | ? | ? | ? | P |
| ROS1 | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| ROS10 | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| ROS11 | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| CPC-03 | · | G | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| GL418 | · | G | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| LJU3 | · | · | · | A | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| H3 | ? | ? | ? | ? | ? | ? | ? | ? | C | · | · | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | P |
| CHWF | · | · | · | · | · | · | · | · | · | T | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| CPC-02 | ? | ? | ? | ? | ? | ? | ? | ? | · | · | T | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| HAL1 | · | · | · | · | · | · | · | · | · | · | T | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| CPC-12 | · | · | · | · | A | · | · | · | · | · | T | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| CPC-13 | · | · | · | · | A | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| EIL14 | · | · | · | · | A | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| NES2 | · | · | · | · | A | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| CPC-11 | · | · | · | · | A | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| ALB2 | · | · | · | · | A | · | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | P |
| ALB4 | · | · | · | · | A | · | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | P |
| ROS3 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | T | · | · | · | · | P | |
| CAT1 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| H1 | · | · | · | · | A | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | ? | ? | ? | ? | ? | ? | ? | P |
| ALL 1 | · | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| D740 | ? | ? | ? | ? | ? | ? | ? | ? | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | P |
| NORF | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | P |
| CPC-14 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | A | · | · | · | · | · | · | · | · | · | · | P |
| CPC-06 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | P |
| CPC-07 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | P |
| CPC-08 | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | · | G | · | · | P |
| CPC98 | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | C | T | P | ||||||
| EIL05 | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | T | P |
| EIL06 | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | T | P |
| NES1 | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | T | P |
| PAR1 | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | T | P |
| ROS12 | · | · | · | · | · | · | C | · | · | · | · | · | · | · | · | · | · | · | · | · | C | · | · | · | · | · | · | T | P |
| GL411 | ? | ? | ? | ? | ? | ? | ? | ? | · | · | · | · | · | · | · | · | · | · | · | · | C | ? | ? | ? | ? | ? | ? | ? | P |
| RUF4 | ? | ? | ? | ? | ? | ? | ? | ? | · | · | · | · | · | · | · | · | · | · | · | · | C | ? | ? | ? | ? | ? | ? | ? | P |
| Ref. seq. | · | · | T | C | · | A | · | · | C | · | T | · | C | T | T | · | · | · | T | · | · | C | · | · | T | G | · | · | T3 |
| CPC-10 | · | · | T | C | · | A | · | · | C | · | T | · | C | T | T | · | · | · | T | · | · | C | · | · | T | G | · | · | T3 |
| WH10 | · | · | T | C | · | A | · | · | C | · | T | · | C | T | T | · | · | · | T | · | · | C | · | · | T | G | · | · | T3 |
| NOY02 | ? | ? | ? | ? | ? | ? | ? | ? | C | · | T | · | C | T | T | · | · | · | T | · | · | C | · | · | T | G | · | · | T3 |
| WH06 | ? | ? | ? | ? | ? | ? | ? | ? | C | · | T | · | C | T | T | · | · | · | T | · | · | C | · | · | T | G | · | · | T3 |
| Syria17 | ? | ? | ? | ? | ? | ? | ? | ? | C | · | T | · | C | T | T | · | · | · | T | · | · | C | · | · | T | G | ? | ? | T3 |
| EF02 | ? | ? | ? | ? | ? | ? | ? | ? | C | · | T | · | C | T | T | · | · | · | T | · | · | ? | ? | ? | ? | ? | ? | ? | T3 |
| ALB3 | · | · | T | C | · | A | · | A | C | · | T | · | C | T | T | · | · | · | T | · | · | C | · | · | T | G | · | · | T3 |
| CPC-04 | · | · | T | C | · | A | · | · | C | · | T | · | C | T | T | · | · | · | T | · | · | C | · | · | T | G | · | · | T3 |
| MAR10 | · | · | T | · | · | A | · | · | C | · | T | · | C | T | T | · | · | · | T | · | · | C | · | · | · | G | · | · | T |
| IRN02 | ? | ? | ? | ? | ? | ? | ? | ? | C | · | T | · | C | T | T | · | · | · | T | · | · | ? | A | · | · | G | · | · | T |
As can be seen in figure 2, similar to the five haplogroups found in extant domestic taurine cattle, the P sequences form a network that is reminiscent of a starburst, or uncorrelated genealogy (figure 2), suggestive of a past population expansion (Bradley et al. 1996). To investigate this further, a Bayesian demographic inference analysis was performed on the 37 P sequences that were at least 360 bp in length. A comparison of marginal posterior probabilities of demographic parameters indicated that an exponential growth model (ln L=−1058.9) gave a significantly better fit to the data than a constant population size model (ln L=−1065.7). From the same analysis, the time to TMRCA of all the P sequences was estimated to be 17 230 years, with a 95% highest posterior density (HPD) interval of between 10 050 and 30 230 years. The estimate of the mutation rate for this 360 bp of control region was 77.2% per Myr, with a 95% HPD interval of 18.2–139.1% per Myr. Although wide, this range encompasses previous mutation rate estimates of 38 and 32%, determined using a fossil calibration date (Troy et al. 2001) and direct Bayesian phylogenetic analysis of an extensive radiocarbon-dated bison dataset (Shapiro et al. 2004), respectively. The wide 95% HPDs are an inevitable consequence of the low information content of the sequences.
There were nine European specimens dating to the Neolithic and Bronze Age that, while being considered originally as aurochs specimens, possessed T haplotypes (ALB3, EF02, MAR10, NOY02, CPC-04, CPC-10, SV03, WH06 and WH10). However, subsequent re-measurements indicated that the previous classifications should be treated with caution, as these samples are just as likely to be from domestic cattle as from aurochs (table S3 in the electronic supplementary material). A sample from the German Neolithic site of Eilsleben (EIL4) displayed a previously undescribed divergent haplotype (termed here as E; table S2 in the electronic supplementary material). EIL4 was radiocarbon dated to 5830±29 years BP (6733–6557 cal. years BP, KIA24758), a date consistent with the archaeological finds typologically attributed to the Neolithic Bandkeramik complex. The bone analysed was an unusually robust distal humerus (for measurements, see table S3 in the electronic supplementary material). For the purposes of authentication, the sample was independently extracted, amplified and sequenced at both Mainz and Dublin. Cloning, and subsequent sequencing of various clones, was undertaken on the most divergent third fragment, which includes an insertion at base 16 200 (table S2, electronic supplementary material) and, using the Consensus Confidence program v. 1.12 (Bower et al. 2005), the consensus sequence from 15 clones was found to be correct with a confidence value of greater than 95%. To check that this novel haplotype, E, was not a nuclear insertion sequence from the mitochondrion, both BLAST and BLAT searches of each of the PCR fragments were performed against the draft cattle nuclear genome (Assembly March 2005, approximate coverage=6.2×) and did not return any significant hits of non-trivial length. Owing to this, the low copy number of nuclear sequences and the absence of the E haplotype in potentially contaminating modern cattle, we are satisfied that this sequence represents a novel mitochondrial haplotype.
A neighbour-joining tree (figure 3) was constructed using the novel sequences described here, as well as reference sequences from each of the five taurine and two zebu cattle haplogroups, and representative outgroups. As shown previously (Troy et al. 2001), taurine (T) and zebu (Z) sequences form monophyletic clades, with P sequences more closely related to T sequences rather than to Z sequences. The E haplotype is distinct from both the P and T sequences, but branches closer to these than to Z sequences (figure 3; table S2 in the electronic supplementary material). Bayesian analysis of the 360 bp E sequence length, when compared with 37 samples with P haplotype sequences, supports this: the P and T sequences group together, to the exclusion of E, with a posterior probability of 100%. The time since the divergence between E and P lineages was estimated to be 52 700 years (95% HPD: 16 020–108 000 years).
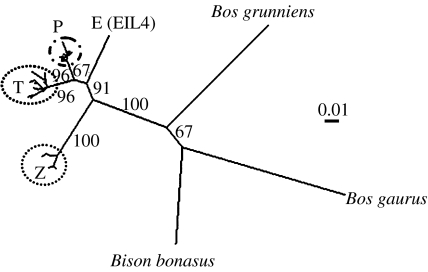
Figure 3.
Neighbour-joining tree of ancient aurochs and extant domestic cattle mitochondrial haplogroups. Unrooted tree based on 361 bp of the control region. Bootstrap values (threshold of 60% after 1000 replicates) are reported on the nodes. T denotes taurine sequences (including T, T1, T2, T3 and T4 individuals determined from concordance with reference sequences (Mannen et al. 1998; Troy et al. 2001)); Z denotes zebu sequences (including Z1 and Z2 individuals determined from concordance with reference sequences (Baig et al. 2005)); P denotes representative aurochs-like sequences from this study; EIL4 (E) is the novel aurochs sequence from an Early Neolithic site in Germany. The branch leading to the three outgroups, yak (B. grunniens), gaur (B. gaurus) and European bison (B. bonasus), indicates the ancestral node of the cattle groups. Scale bar denotes units of genetic distance.
4. Discussion
(a) Population history of the aurochs
This study is the most comprehensive, to date, of aurochs mtDNA sequence variation and has increased the number of available aurochs sequences to around fourfold. Previous studies have found the P haplogroup in aurochs from Britain and Iberia (Bailey et al. 1996; Troy et al. 2001; Anderung et al. 2005), but here we document its prevalence across the European continent (Hungary, Slovenia, Slovakia, Austria, Germany and France), dating from the Mesolithic to the Bronze Age. All P haplotypes fall into a star-like phylogeny, consistent with a past population expansion. Through Bayesian phylogenetic dating analysis, the coalescence of all P haplotypes was estimated to occur between 10 050 and 30 230 cal. years BP; a finding that is reconcilable with Late Glacial expansion from a refugial population.
(b) A novel aurochs haplotype
Bayesian phylogenetic analysis of the novel E haplotype, from the Early Neolithic German site of Eilsleben, estimates the age of the split between E and P lineages to be 52 700 years ago (95% HPD: 16 020–108 000 years). The bone analysed was an unusually robust distal humerus and was determined to be definitely a Bos sp. individual from morphological analysis. As this novel E sequence is an outlier to the P lineage, but groups within the B. taurus part of the tree (figure 3), it is plausible that its presence in Europe points to a greater variability of cattle prior to the Last Glacial Maximum. The location of E, between the P/T haplogroups and the Z haplogroup (figure 3), could also indicate a glacial refugial origin further to the East.
(c) Prior demonstration of pre-Neolithic T3 aurochs in Italy
The recent reporting of five T haplotypes in southern Italian aurochs (Beja-Pereira et al. 2006) is surprising and, on first viewing, contradictory to our data from northern and central Europe and the Near East. At the last glacial maximum, Italy was one of the three Southern European refugia for the Holocene fauna that subsequently repopulated Europe. Postglacial expansion of the majority of Italian refugial species was restricted by the barrier of the Alps to the North (Hewitt 1999). Therefore, with the presence of T haplotypes in the Italian aurochs population, we would expect them to remain there until the local aurochs population died out, rather than contributing to the northwards postglacial expansion of aurochs (characterized by P haplotypes in central and northern Europe), which instead is more likely to have been derived from the Iberian and Balkan refuges (Taberlet et al. 1998). A similar case is found in the native Italian wild boar, which are genetically distinct from the rest of Europe (Larson et al. 2005), supporting the notion that many Italian wild faunas are phylogenetically distinct. It is also possible that, at some point in the past, the Near Eastern and Italian aurochs belonged to the same ancestral population, perhaps connected via the Northern Adriatic basin, which remained submerged late during the Late Glacial transgression (van Andel 1989). Although we examined a Mesolithic aurochs sample from northern Italy, this bone yielded no reproducibly amplifiable DNA. Therefore, our one sequence from the region that could have connected the Near Eastern and Italian populations is the Mesolithic LJU3 from Slovenia, which exhibited a P haplotype. However, as it is situated at the northern extremity of the Balkan Peninsula, additional sampling from Italy and the Balkans is required to fully address this possibility. Given the above, and the results presented here, the unexpected haplotypes found in the Italian aurochs reported by Beja-Pereira et al. (2006) raise many questions and warrant additional verification.
(d) Post-domestication introgression between wild and domestic Bos populations
Eight European samples, classified on morphological grounds as aurochs by the corresponding archaeologists, yielded a T haplotype. These may have arisen due to: (i) morphological misclassification of domestic samples, as already postulated (Anderung et al. 2005; Bollongino et al. 2006), (ii) sporadic introgression from domestic cattle, or (iii) low-frequency haplotypes that survived in the predominantly P haplotype northern and eastern European aurochs population. Introgression (scenario (ii)) is certainly possible and plausible. Zooarchaeological diagnoses are inherently error prone, as they are based on size differences, and bone size depends upon many different genetic and environmental factors. It is conceivable that introgression might have been more frequent in the Early Neolithic, which would explain the predominantly Early Neolithic dates for the anomalous specimens. Scenario (iii) is also possible. However, as there are no pre-domestication T haplotypes apart from Italy and we did not find T haplotypes in our morphologically robust specimens, we favour hypotheses (i) and (ii).
Critical archaeozoological and archaeological examination of each of the eight discordant European samples leads to the conclusion that a domestic origin cannot be entirely excluded for any of them (see electronic supplementary material), again favouring the morphological misclassification explanation (scenario (i)). As the P haplogroup is not observed in extant cattle, births arising from hybridization are likely to have been rare, suggesting that domestic cattle were kept isolated from wild aurochs by humans. Even if P haplotypes were present in domestic cattle, but were subsequently lost by genetic drift, it is unlikely that such haplotypes were ever at high frequency in the B. taurus population. In sum, in all the samples that were morphologically identifiable, there is a clear-cut division between P haplotypes, found in morphologically robust specimens classified as aurochs, which were already present in Europe during the Pleistocene, and T haplotypes, found in less robust specimens that arrived in Europe during the Early Holocene. This shows that introgression was unlikely to have been a common event. However, as this study is based on mitochondrial data alone, we cannot rule out any introgression of male aurochs. Nevertheless, stable isotope data support the notion that these two subpopulations did not interact extensively. 13C/12C ratios yielded by bovine remains, from several Neolithic contexts in the Paris Basin (Balasse et al. 1997; Bocherens et al. 2005) and Denmark (Noe-Nygaard et al. 2005), clearly show that aurochs and domestic cattle, at least in these places, were feeding on distinct plant sources, the former in the forest and the latter in more open environments.
(e) Near Eastern origin of taurine haplotype
Modern genetic and archaeological evidence suggests that the domestication centre of European cattle was in the Near East. Here, we present, for the first time, ancient biomolecular data that establish the presence of the T family of haplogroups in an archaic Near Eastern population. The sample Syria17 (table 1; table S1 in the electronic supplementary material) is a Bos specimen from Dja'de el Mughara in the Middle Euphrates Valley. This site was occupied mainly during the Early Pre-Pottery Neolithic (PPNB), ca 10 650–10 250 cal. BP (Coqueugniot 2000). Archaeozoological Bos material from this site has long been considered as wild, but recent morphometric studies have revealed a very small decrease of size and overall a slight and significant decrease of the sexual dimorphism that convincingly suggest an incipient domestication (Helmer et al. 2005). This result is in concordance with the observation that domestic cattle were transferred to Cyprus from the mainland as early as ca 10 250–10 150 cal. BP (Vigne et al. 2000, 2003). The Dja'de sample, which lay undisturbed 3 m beneath the surface of a Tell, yielded a T3 haplotype, a result that was replicated in both Dublin and Mainz. Although previous work has shown the occurrence of the T haplotype in early Near Eastern cattle (Edwards et al. 2004), and we report here a Bronze Age aurochs specimen from Maral Tappeh, Iran, with a T haplotype, this Syrian sample is the earliest dated DNA evidence for the occurrence of this haplogroup in the Near East. A taxonomic discrimination between B. taurus and B. p. primigenius forms that are chronologically and geographically close to the centre of domestication is arbitrary, and thus the bone sample Syria17 may be an example of a very early taurine domesticate, or alternatively an Early Holocene aurochs. In either case, this is the first confirmation of the presence of a common B. taurus haplotype close to the suggested centre of domestication, at the very beginning of the Neolithic.
5. Conclusions
In summary, we observe three different mitochondrial haplogroups in Eurasian aurochs populations: a P haplogroup predominant across Europe; an E haplotype (evidenced from a single sample, EIL4) in Neolithic Germany; and a T3 haplotype from a sample in the Near East. From these and other mtDNA sequence data, we conclude that the domestic gene pool was established from animals with T haplotypes around 10 000 years ago, and that this haplogroup was predominant in the aurochs populations of the Near East at this time, concordant with an Early Neolithic T3 haplotype in Syria. The T haplogroups were then introduced to Europe by early farming populations ca 8800 cal. BP, following which domestic (T haplogroup-carrying) cattle coexisted in the same habitat with local (P haplogroup-carrying) aurochs that previously had experienced a population expansion, possibly at the end of the Last Glacial Maximum. Although a small amount of introgression of T haplotypes into P herds is indicated (and perhaps even probable), the evidence from Neolithic and modern cattle populations, and ancient dietary surveys, supports the notion that domestic herds were largely separate from aurochs. Aurochs probably remained relatively genetically distinct until they became extinct in the seventeenth century.
Acknowledgments
We thank David Magee and Brian McEvoy for critically reviewing this paper, Umberto Albarella, Matthew Collins and Helmut Hemmer for discussion, and François Giligny, Bassam Jamous, Yves Lanchon, Claude and Daniel Mordant, and Danielle Stordeur for additional sampling and access to material. C.J.E. is supported by the Irish Research Council for Science Engineering and Technology Basic Research Grant Scheme (project numbers SC/1999/409 and SC/2002/510). The project in Mainz was funded by the Bundesministerium für Bildung und Forschung (03BUX1MZ). Both laboratories were part funded from a Eurocores OMLL Programme Grant via CNRS, Natural History Museum, Paris, France. G.L. and S.Y.W.H. were both funded by the Leverhulme Trust.
Accession Numbers: the sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ915519–DQ915576).
Supplementary Material
Discussion of the T haplotypes in morphological aurochs specimens. Figure 4. Primer strategy view for the amplification of the control region. Table S1. Table of archaeological Bos samples studied, with associated codes, skeletal element used and extra information supplied. Table S2. The variable positions in control region sequences of the aurochsen consensus haplotype (P), novel aurochs haplotype (E). Table S3. Assessment of individuals whose haplotype did not match with the archaeozoological assessment of the remains. Table S4. Genetic distance matrix generated by DNADIST to determine the position of the novel E haplotype. Table S5. Radiocarbon dates conducted as part of this study, with associated collagen values. Table S6. Osteological measurement of the distal breadth of the radius CPC-04
References
- Anderson S, de Bruijn M.H, Coulson A.R, Eperon I.C, Sanger F, Young I.G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J. Mol. Biol. 1982;156:683–717. doi: 10.1016/0022-2836(82)90137-1. doi:10.1016/0022-2836(82)90137-1 [DOI] [PubMed] [Google Scholar]
- Anderung C, et al. Prehistoric contacts over the Straits of Gibraltar indicated by genetic analysis of Iberian Bronze Age cattle. Proc. Natl Acad. Sci. USA. 2005;102:8431–8435. doi: 10.1073/pnas.0503396102. doi:10.1073/pnas.0503396102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig M, Beja-Pereira A, Mohammad R, Kulkarni K, Farah S, Luikart G. Phylogeography and origin of Indian domestic cattle. Curr. Sci. 2005;89:38–40. [Google Scholar]
- Bailey J.F, Richards M.B, Macaulay V.A, Colson I.B, James I.T, Bradley D.G, Hedges R.E, Sykes B.C. Ancient DNA suggests a recent expansion of European cattle from a diverse wild progenitor species. Proc. R. Soc. B. 1996;263:1467–1473. doi: 10.1098/rspb.1996.0214. doi:10.1098/rspb.1996.0214 [DOI] [PubMed] [Google Scholar]
- Balasse M, Bocherens H, Tresset A, Vigne J.-D, Mariotti A. Emergence de la production laitière au Néolithique? Contribution de l'analyse isotopique d'ossements de bovins archéologiques. Comptes rendus de l'Académie des Sciences de Paris. Sciences de la Terre et des Planètes. 1997;325:105–110. [Google Scholar]
- Bandelt H.J, Forster P, Sykes B.C, Richards M.B. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beja-Pereira A, et al. The origin of European cattle: evidence from modern and ancient DNA. Proc. Natl Acad. Sci. USA. 2006;103:8113–8118. doi: 10.1073/pnas.0509210103. doi:10.1073/pnas.0509210103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocherens H, Billiou D, Tresset A. Approche biogéochimique (13C, 15N) de l'exploitation de l'environnement par les humains. In: Giligny F, editor. Louviers La Villette (Eure). Un site Néolithique moyen en zone humide. Documents Archéologiques de l'Ouest; Rennes, France: 2005. pp. 265–269. [Google Scholar]
- Bollongino R, Edwards C.J, Alt K.W, Burger J, Bradley D.G. Early history of European domestic cattle as revealed by ancient DNA. Proc. R. Soc. B. 2006;2:155–159. doi: 10.1098/rsbl.2005.0404. doi:10.1098/rsbl.2005.0404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower M.A, Spencer M, Matsumura S, Nisbet R.E, Howe C.J. How many clones need to be sequenced from a single forensic or ancient DNA sample in order to determine a reliable consensus sequence? Nucleic Acids Res. 2005;33:2549–2556. doi: 10.1093/nar/gki550. doi:10.1093/nar/gki550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D.G, MacHugh D.E, Cunningham P, Loftus R.T. Mitochondrial diversity and the origins of African and European cattle. Proc. Natl Acad. Sci. USA. 1996;93:5131–5135. doi: 10.1073/pnas.93.10.5131. doi:10.1073/pnas.93.10.5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger J, et al. Molecular phylogeny of the extinct cave lion Panthera leo spelaea. Mol. Phylogenet. Evol. 2004;30:841–849. doi: 10.1016/j.ympev.2003.07.020. doi:10.1016/j.ympev.2003.07.020 [DOI] [PubMed] [Google Scholar]
- Clutton-Brock J. Cambridge University Press; Cambridge, UK: 1987. A natural history of domesticated mammals. [Google Scholar]
- Coqueugniot E. Dja'de (Syrie), un village à la veille de la domestication. In: Guilaine J, editor. Premiers paysans du monde. Naissance de agricultures. Errance; Paris, France: 2000. pp. 55–71. [Google Scholar]
- Cymbron T, Loftus R.T, Malheiro M.I, Bradley D.G. Mitochondrial sequence variation suggests an African influence in Portuguese cattle. Proc. R. Soc. B. 1999;266:597–603. doi: 10.1098/rspb.1999.0678. doi:10.1098/rspb.1999.0678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J, Nicholls G.K, Rodrigo A.G, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J, Ho S.Y, Phillips M.J, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. doi:10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards C.J, et al. Ancient DNA analysis of 101 cattle remains: limits and prospects. J. Archaeol. Sci. 2004;31:695–710. doi:10.1016/j.jas.2003.11.001 [Google Scholar]
- Felsenstein J. Phylip—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of the human–ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. doi:10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- Helmer D, Gourichon L, Monchot H, Peters J, Saña Segui M. Identifying early domestic cattle from Pre-Pottery Neolithic sites on the Midddle Euphratesusing sexual dimorphism. In: Vigne J.-D, Helmer D, Peters J, editors. The first steps of animal domestication: new archaeozoological approaches. Oxbow Books; Oxford, UK: 2005. pp. 86–95. [Google Scholar]
- Hewitt G.M. Post-glacial re-colonization of European biota. Biol. J. Linn. Soc. 1999;68:87–112. doi:10.1006/bijl.1999.0332 [Google Scholar]
- Ho S.Y, Larson G. Molecular clocks: when times are a-changin'. Trends Genet. 2006;22:79–83. doi: 10.1016/j.tig.2005.11.006. doi:10.1016/j.tig.2005.11.006 [DOI] [PubMed] [Google Scholar]
- Larson G, et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science. 2005;307:1618–1621. doi: 10.1126/science.1106927. doi:10.1126/science.1106927 [DOI] [PubMed] [Google Scholar]
- Loftus R.T, MacHugh D.E, Bradley D.G, Sharp P.M, Cunningham P. Evidence for two independent domestications of cattle. Proc. Natl Acad. Sci. USA. 1994;91:2757–2761. doi: 10.1073/pnas.91.7.2757. doi:10.1073/pnas.91.7.2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacHugh D.E, Edwards C.J, Bailey J.F, Bancroft D.R, Bradley D.G. The extraction and analysis of ancient DNA from bone and teeth: a survey of current methodologies. Anc. Biomol. 2000;3:81–102. [Google Scholar]
- Mannen H, Tsuji S, Loftus R.T, Bradley D.G. Mitochondrial DNA variation and evolution of Japanese black cattle (Bos taurus) Genetics. 1998;150:1169–1175. doi: 10.1093/genetics/150.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannen H, et al. Independent mitochondrial origin and historical genetic differentiation in north eastern Asian cattle. Mol. Phylogenet. Evol. 2004;32:539–544. doi: 10.1016/j.ympev.2004.01.010. doi:10.1016/j.ympev.2004.01.010 [DOI] [PubMed] [Google Scholar]
- Noe-Nygaard N, Price T, Hede S. Diet of aurochs and early cattle in southern Scandinavia: evidence from 13C and 15N stable isotopes. J. Archaeol. Sci. 2005;32:855–871. doi:10.1016/j.jas.2005.01.004 [Google Scholar]
- Poplin F. Origine du mouflon de Corse dans une nouvelle perspective paléontologique: par marronnage. Annales Génétique et Sélection Animale. 1979;11:133–143. doi: 10.1186/1297-9686-11-2-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall K.A. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. doi:10.1093/bioinformatics/14.9.817 [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond A. University of Oxford; Oxford, UK: 2004. Tracer—MCMC trace analysis tool. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shapiro B, et al. Rise and fall of the Beringian steppe bison. Science. 2004;306:1561–1565. doi: 10.1126/science.1101074. doi:10.1126/science.1101074 [DOI] [PubMed] [Google Scholar]
- Taberlet P, Fumagalli L, Wust-Saucy A.G, Cosson J.F. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. doi:10.1046/j.1365-294x.1998.00289.x [DOI] [PubMed] [Google Scholar]
- Troy C.S, MacHugh D.E, Bailey J.F, Magee D.A, Loftus R.T, Cunningham P, Chamberlain A.T, Sykes B.C, Bradley D.G. Genetic evidence for Near-Eastern origins of European cattle. Nature. 2001;410:1088–1091. doi: 10.1038/35074088. doi:10.1038/35074088 [DOI] [PubMed] [Google Scholar]
- van Andel T.H. Late Quaternary sea-level changes and archaeology. Antiquity. 1989;63:733–745. [Google Scholar]
- Vigne J.-D, Helmer D. Nouvelles analyses sur les débuts de l'élevage dans le centre et l'ouest méditerranéen. In: Vaquer J, editor. Le Néolithique du nord-ouest Méditerranéen, Actes XXIVe Congr. Préhist. Fr. Soc. Préhist. Fr; Paris, France: 1999. pp. 129–146. [Google Scholar]
- Vigne, J.-D., Carrère, I., Saliège, J.-F., Person, A., Bocherens, H., Guilaine, J. & Briois, F. 2000 Predomestic cattle, sheep, goat and pig during the late 9th and the 8th millenniun cal. BC on Cyprus: preliminary results of Shillourokambos (Perkklisha, Limassol). In Archaeozoology of the Near East IV, Proc. 4th Int. Symp. Archaeozoology of Southwestern Asia and Adjacent Areas, vol. 32 (eds M. Mashkour, A. M. Choyke, H. Buitenhuis & F. Poplin), pp. 52–75. Groningen, The Netherlands: Archaeological Research and Consultancy.
- Vigne, J.-D., Carrère, I. & Guilaine, J. 2003 Unstable status of early domestic ungulates in the Near East: the example of Shillourokambos (Cyprus, IX-VIIIth millennia cal. BC). In Le Néolithique de Chypre, vol. 43 (eds J. Guilaine & A. Le Brun), pp. 239–251. Greece: Bull. Corr. Héllenique.
- Yang D.Y, Eng B, Waye J.S, Dudar J.C, Saunders S.R. Technical note: improved DNA extraction from ancient bones using silica-based spin columns. Am. J. Phys. Anthropol. 1998;105:539–543. doi: 10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1. doi:10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Discussion of the T haplotypes in morphological aurochs specimens. Figure 4. Primer strategy view for the amplification of the control region. Table S1. Table of archaeological Bos samples studied, with associated codes, skeletal element used and extra information supplied. Table S2. The variable positions in control region sequences of the aurochsen consensus haplotype (P), novel aurochs haplotype (E). Table S3. Assessment of individuals whose haplotype did not match with the archaeozoological assessment of the remains. Table S4. Genetic distance matrix generated by DNADIST to determine the position of the novel E haplotype. Table S5. Radiocarbon dates conducted as part of this study, with associated collagen values. Table S6. Osteological measurement of the distal breadth of the radius CPC-04