Abstract
The mSin3A corepressor complex contains 7 to 10 tightly associated polypeptides and is utilized by many transcriptional repressors. Much of the corepressor function of mSin3A derives from associations with the histone deacetylases HDAC1 and HDAC2; however, the contributions of the other mSin3A-associated polypeptides remain largely unknown. We have purified an mSin3A complex from K562 erythroleukemia cells and identified three new mSin3A-associated proteins (SAP): SAP180, SAP130, and SAP45. SAP180 is 40% identical to a previously identified mSin3A-associated protein, RBP1. SAP45 is identical to mSDS3, the human ortholog of the SDS3p component of the Saccharomyces cerevisiae Sin3p-Rpd3p corepressor complex. SAP130 does not have detectable homology to other proteins. Coimmunoprecipitation and gel filtration data suggest that the new SAPs are, at the very least, components of the same mSin3A complex. Each new SAP repressed transcription when tethered to DNA. Furthermore, repression correlated with mSin3A binding, suggesting that the new SAPs are components of functional mSin3A corepressor complexes. SAP180 has two repression domains: a C-terminal domain, which interacts with the mSin3A-HDAC complex, and an N-terminal domain, which functions independently of mSin3A-HDAC. SAP130 has a repression domain at its C terminus that interacts with the mSin3A-HDAC complex and an N-terminal domain that probably mediates an interaction with a transcriptional activator. Together, our data suggest that these novel SAPs function in the assembly and/or enzymatic activity of the mSin3A complex or in mediating interactions between the mSin3A complex and other regulatory complexes. Finally, all three SAPs bind to the HDAC-interaction domain (HID) of mSin3A, suggesting that the HID functions as the assembly interface for the mSin3A corepressor complex.
Acetylation, methylation, phosphorylation, and ubiquitination of the amino-terminal tails of the core histones can all contribute to the regulation of transcription (10). Of these modifications, the outcome of histone acetylation is best understood. In general, hyperacetylated histone tails correlates with transcriptional activation and an “open” chromatin conformation whereas hypoacetylated histone tails correlates with transcriptional repression and a “closed“ chromatin conformation (52). Consistent with this paradigm, histone acetyltransferases (HATs) are recruited to promoters via interaction with sequence-specific transcriptional activators and histone deacetylases (HDACs) are recruited to promoters via interactions with sequence-specific transcriptional repressors (6, 21, 29, 44). Both HATs and HDACs are components of large multiprotein complexes (14, 41). The HAT- or HDAC-associated proteins are thought to function in the assembly, targeting, and regulation of these complexes. Ultimately, to understand the function of HAT and HDAC complexes in transcription, it is imperative to identify and characterize each component of these important regulatory assemblies.
Several HDAC-dependent corepressors have been identified in mammalian cells. Among these, the mSin3A and the NuRD/Mi2 complexes contain a module composed of HDAC1, HDAC2, and two histone tail-targeting proteins, RbAp46 and RbAp48 (4, 20, 22, 33, 39, 50, 54, 57, 58). The stoichiometry has not been examined carefully; however, both complexes appear to contain stoichiometric or near-stoichiometric amounts of this HDAC-containing catalytic module. The other identified proteins that associate with mSin3A or NuRD/Mi2 are not related in sequence and are presumably required for functions that are unique to each complex. For example, the NuRD/Mi2 complex contains the CHD3/4 ATPases that impart chromatin remodeling activity to this complex (50, 54, 58) whereas the mSin3A complex lacks intrinsic chromatin-remodeling activity. Interestingly, a fraction of mSin3A appears to interact with the human Swi/Snf chromatin-remodeling complex (30, 47), suggesting that the mSin3A complex may acquire chromatin-remodeling capacity via an indirect mechanism.
mSin3A is a large protein with multiple protein-protein interaction domains and serves as the scaffold on which the corepressor complex assembles (8, 11, 17, 33, 51). The mSin3A corepressor elutes from size exclusion columns over a broad range (30, 47, 54), suggesting that there may be multiple mSin3A complexes within a given cell. However, results from multiple laboratories suggest that HDAC1, HDAC2, RbAp46, RbAp48, SAP30 (32, 59), RBP1 (34), and p33ING1b (30, 48) associate with mSin3A with relatively high stoichiometry and, as such, may represent components of a “core” mSin3A corepressor complex. Other components such as SAP18 appear to associate with mSin3A in a cell-type-specific or perhaps regulated manner (30, 54, 57). The HDAC/RbAp subcomplex is required for much of the transcriptional repression capacity of the mSin3A corepressor (20, 33), whereas the other mSin3A-associated proteins appear to function in the assembly of the complex and the targeting of mSin3A to other proteins or multiprotein complexes. For example, SAP30 is required for repression by some but not all unliganded nuclear hormone receptors (32). It has also been implicated in binding RBP1 and the HDAC/RbAp catalytic module to mSin3A (34, 59). Therefore, SAP30 probably mediates interactions with other corepressor complexes and functions in the assembly of the core complex. In addition, RPB1 functions to tether mSin3A to the tumor suppressor Rb (34). Finally, interactions between the growth suppressor p33ING1b and mSin3A are required for the antiproliferative function of p33ING1b (30, 48); however, the way in which p33ING1b contributes to mSin3A corepressor activity is currently unknown.
mSin3A and mSin3B are close paralogs and were first identified as corepressors required for the transcriptional and growth suppression functions of the Mad1 and Mxi1 proteins (8, 45). Since that time, mSin3A and mSin3B have been implicated as corepressors utilized by a rapidly expanding collection of transcriptional repressors (1, 26). These mSin3A-dependent transcriptional repressors function in diverse cellular processes including proliferation, differentiation, apoptosis, oncogenesis, and cell fate determination. As such, understanding the function of the mSin3A complex will provide insight not only into the mechanism of HDAC-dependent transcriptional repression but also into diverse aspects of cell behavior. Considerable progress has been made in characterizing the core mSin3A complex; however, not all components of the core mSin3A complex have been identified. Here we report the purification of a mSin3A complex from the K562 erythroleukemia cell line that contains many of the known mSin3A-associated proteins and several novel proteins that also associate with mSin3A with high stoichiometry. We have identified three of these novel mSin3A-associated proteins, cloned cDNAs carrying their open reading frames, and provide an initial characterization of their function within the mSin3A complex.
MATERIALS AND METHODS
Purification of a mSin3A complex.
A total of 4.25 × 1010 K562 cells were pelleted in phosphate-buffered saline (PBS) containing 15% glycerol (NIH Cell Culture Center). The pellet was lysed in 2 packed-cell volumes of hypotonic lysis buffer (10 mM KCl, 10 mM HEPES [pH 7.9], 1 mM dithiothreitol [DTT], 1.5 mM MgCl2), and then nuclei were collected by centrifugation. The nuclei were extracted for 1 h at 4°C with 2 packed-cell volumes of extraction buffer (50 mM HEPES [pH 7.9], 0.6 M KCl, 5 mM MgCl2, 20% glycerol, 10% sucrose, 2 mM DTT) containing 1.9 μg of aprotinin per ml, 1 μg of leupeptin per ml, 1 μg of pepstatin per ml, and 50 nM phenylmethylsulfonyl fluoride and dialyzed into 0.1 M KCl column buffer (50 mM HEPES [pH 7.9], 10% glycerol, 1 mM MgCl2, 1 mM DTT) also containing protease inhibitors. The extract was then loaded sequentially onto DEAE-Sepharose and SP-Sepharose columns (Amersham-Pharmacia Biotech). In each case, bound proteins were eluted with a 0.1 to 1.0 M KCl gradient. mSin3A-containing fractions from the DEAE column, determined by Western blot analysis, were pooled, dialyzed into 0.1 M KCl column buffer, and loaded onto the SP-Sepharose column. Following the SP-Sepharose column elution, mSin3A-containing fractions were collected and then divided into two pools. One pool was loaded onto protein A-agarose beads (Sigma) cross-linked to preimmune serum by using dimethyl pimelimidate·2HCl (Pierce), whereas the other pool was loaded onto protein A-agarose cross-linked to anti-GSTA11 (mSin3A) polyclonal antibody (20). The proteins were bound to the antibody columns overnight at 4°C in approximately 0.3 M KCl column buffer. Columns were washed with 10 column volumes of 0.5 M KCl column buffer, eluted with 0.1 M glycine (pH 2.5), and neutralized with 1.5 M Tris (pH 8.7). Fractions containing protein were trichloroacetic acid precipitated with 0.02% deoxycholate as carrier and resuspended in 2× sample buffer (0.15 M Tris [pH 6.8], 20% glycerol, 10% sodium dodecyl sulfate [SDS], 0.02% bromphenol blue). To monitor the integrity of the mSin3A throughout the purification, mSin3A-containing fractions from each ion-exchange column were pooled and run over a Superdex 200 size exclusion column in 25 mM HEPES (pH 7.9)-0.6 M KCl-5% glycerol-0.1% Tween 20-1 mM DTT. Each mSin3A pool eluted with the same profile as mSin3A in the original nuclear extract (data not shown), demonstrating that the complex is quite stable and remained largely intact in the presence of high salt concentrations.
Identification and cloning of mSin3A-associated proteins.
The purified mSin3A-associated proteins were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) (4 to 20% polyacrylamide gradient) (Bio-Rad) and stained with Coomassie G-250 (Sigma). Polypeptides were excised, and MALDI-TOF mass spectrometry and microsequencing were carried out by the Rockefeller University Protein/DNA Technology Center. It was found that 33% of the peptide masses and 14 of 14 amino acids of a sequenced peptide from SAP180 matched the 91-kDa RBP1-like protein (AB030181), while 18% of the masses matched the 88-kDa BRCAA1 protein (AF208045). Neither of these clones was full length, and analysis of a genomic sequence (AL356781) suggested that they were alternative splice variants of the same gene. Use of a rat cDNA (AF245512) homologous to RBP1-like and BRCAA1 allowed us to identify a putative 5′ end for SAP180. A full-length SAP180 cDNA was amplified using reverse transcription-PCR with K562 mRNA and matched a revised sequence for RBP1-like (AF214114), subsequently placed into the database. A total of 61% of the original peptide masses matched this full-length clone. The accession number for SAP180 is AY220790.
It was found that 21% of the SAP45 masses and 15 of 15 and 10 of 10 amino acids from two sequenced peptides matched the translation of a partial cDNA clone, FLJ00052 (AK024460). Primers were designed for reverse transcription-PCR of a cDNA fragment that was then used to screen a K562 cell cDNA library. The entire SAP45 open reading frame was isolated in one cDNA clone. A recent publication revealed that SAP45 is identical to mSDS3 (3). Finally, 50% of the SAP130 peptides and 9 of 9 and 6 of 7 amino acids from two sequenced peptides matched a 1,048-amino-acid hypothetical protein, CAB66767. Clone DKFZp434A112Q2 was ordered from the RZPD, Berlin, Germany. The accession number for SAP130 is AY220791.
SAP180, SAP130, and mSDS3 cDNAs were subcloned into pFA-CMV (Stratagene) to generate GAL4 DNA binding domain fusions. SAP180 was subcloned into CMV-Tag2 (Stratagene) to create an N-terminal Flag tag. N-terminal truncations and central regions of SAP180 and SAP130 were generated using PCR, and the amplified cDNA fragments were cloned into the indicated vectors. Truncations of the C termini of SAP180 and SAP130 were made by introducing stop codons at the indicated positions by using the QuikChange XL site-directed mutagenesis kit (Stratagene) and were confirmed by sequencing.
Myc-tagged mSin3A deletion constructs have been described previously (33). mSin3A HID C-terminal truncations were generated by introducing stop codons into pCS2-MT-PAH3+HID by using the QuikChange XL site-directed mutagenesis kit and confirmed by sequencing.
Northern blots.
A cancer cell line blot (Clontech) was incubated for at least 3 h at 42°C in hybridization buffer (5× SSC [1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate], 5× Denhardt's solution, 50% formamide, 1% SDS, 100 μg of sonicated salmon sperm DNA per ml). cDNA fragments from each gene were radiolabeled with [32P]dCTP (Dupont) using the RadPrime DNA-labeling system (Gibco-BRL), boiled for 5 min, and added to the hybridization buffer to 2 × 106 cpm/ml. The blot was hybridized overnight at 42°C and was washed with several volumes of 0.1× SSC-0.1% SDS at 55°C. The blot was exposed to film (Kodak X-OMAT) for 1 to 3 days at −80°C with an intensifying screen (Kodak).
Antibodies.
Anti-GSTA11 (mSin3A) and SAP30 antibodies have been previously described (20, 32). Anti-HDAC1 (Affinity Bioreagents, Inc.), anti-HDAC2 (Zymed), anti-Flag M2 (Sigma), and anti-GAL4 (Santa Cruz Biotech) are available commercially. GST-SAP180 (amino acids 652 to 805), GST-SAP45 (amino acids 83 to 328), and GST-SAP130 (amino acids 301 to 510) were generated by cloning cDNA fragments into pGEX 5×-2 or pGEX4T-1 (Amersham Pharmacia Biotech). Proteins were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in codon (+) Escherichia coli (Stratagene) for 3 to 6 h and purified using glutathione agarose (Amersham Pharmacia). Fractions containing protein were dialyzed into PBS and used to immunize New Zealand White rabbits (Covance Research Products). In preliminary tests, each antibody was able to specifically recognize its cognate antigen under native conditions (data not shown). Anti-rabbit horseradish peroxidase and anti-mouse horseradish peroxidase secondary antibodies were from Amersham Pharmacia Biotech.
Immunoprecipitations.
Nuclear extracts were prepared from 1.0 × 107 K562 cells as described above and immunoprecipitated overnight at 4°C with the indicated antibody or preimmune serum and 10 μl of protein A-agarose (Sigma). The immunoprecipitates were washed four times in 1 ml of L-buffer (PBS, 0.1% NP-40) and eluted from the beads with 2× sample buffer. The eluates were resolved by SDS-PAGE and visualized by immunoblotting with the indicated antibodies. Interactions were detected using enhanced chemiluminescence (Amersham Pharmacia Biotech).
For immunoprecipitations from transfected cells, HEK293 cells were seeded at 1.0 × 106 cells and transfected 16 h later with 5 μg of each expression construct using calcium phosphate (5). At 24 h later, the cells were lysed on the plate in 0.5 ml of L-buffer. Whole-cell extracts were sonicated, cleared, immunoprecipitated, and analyzed as above.
Size exclusion chromatography.
Nuclear extract was prepared from 5.0 × 108 K562 cells as described above and concentrated to 200 μl by centrifugation using a Centricon YM-10 apparatus (Amicon). The concentrated extract was loaded onto a Superose 6 10/30 column (Amersham Pharmacia Biotech) equilibrated with 25 mM HEPES (pH7.9)-0.6 M KCl-5% glycerol-0.1% Tween 20-1 mM DTT. Protein was eluted at 0.3 ml/min, and 0.5-ml fractions were collected over 2 column volumes. Then 36-μl aliquots of fractions were mixed with 5× sample buffer. The remainder of each fraction was immunoprecipiated overnight at 4°C with the indicated antibody and 10 μl of protein A-agarose. Aliquots and immunoprecipitations of the fractions were separated by SDS-PAGE and analyzed by Western blotting with the indicated antibodies.
Luciferase assays.
HEK293 cells were seeded at 2.0 × 105 cells in triplicate 16 h prior to transfection. The cells were transfected with 100 ng of G4-14D luciferase reporter (7, 49), 28 ng of pCMVβ-gal, 100 ng of GAL-fusion cDNA, and sonicated salmon sperm DNA as carrier to 2 μg of DNA per dish using calcium phosphate (5). Trichostatin A (TSA) (BioMol) was added to cells to 100 ng/ml at 8 h after transfection. At 16 h later, cell lysates were prepared in 200 μl of reporter lysis buffer (Promega), and luciferase and β-gal activities were measured as specified by the manufacturers (Promega and Tropix, respectively), using an MLX Microtiter Plate Luminometer (Thermo Labsystems). Results were normalized for transfection efficiency by dividing luciferase relative light units by β-gal relative light units, and averages for the triplicate determinations were taken. Each experiment was repeated at least twice, and the standard error of the mean is reported.
Deacetylase assays.
HEK293 cells were transfected with 5 μg of expression construct and immunoprecipitated as above. After being washed three times with L-buffer, beads were incubated with 50 μM of Fluor de Lys acetylated substrate (BioMol) for 1 h at 37°C. Reaction products were developed and assayed as specified by the manufacturer, except that TSA and developer were used at 1:1200 dilutions. Fluorescence was excited and measured using a Bio-Tek FL600 Microplate fluorescence reader. Experiments were carried out in duplicate, and the standard deviation was calculated.
RESULTS
Purification of a novel mSin3A complex.
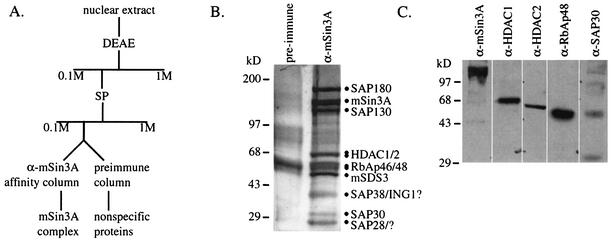
Immunoprecipitations of mSin3A from metabolically labeled U937, Jurkat, and K562 cells consistently reveal multiple mSin3A-associated proteins, several of which have been identified by our laboratory and others (4, 20, 22, 30, 32-34, 39, 48, 57, 59). To identify additional novel mSin3A-associated proteins (SAPs), a combination of ion-exchange and affinity chromatography was used to purify an mSin3A complex from K562 cells (Fig. 1A). To control for nonspecific interactions with the beads, a parallel sample was run on a “preimmune” affinity column.
FIG. 1.
Purification of an mSin3A complex from K562 cells. (A) Purification scheme used to purify an mSin3A complex from K562 nuclear extracts. (B) A silver-stained SDS-PAGE gel showing proteins from the mSin3A and preimmune affinity columns. Proteins that were retained specifically on the anti-mSin3A column are indicated. Question marks indicate that the identities of the proteins were not confirmed. (C) A Western blot of eluates from the mSin3A affinity column, demonstrating that the known components of the mSin3A complex copurified over two ion-exchange columns and were retained by the affinity matrix.
Several polypeptides from the mSin3A antibody affinity column migrated with molecular masses corresponding to mSin3A itself and to previously identified mSin3A-associated proteins (Fig. 1B). Western blotting (Fig. 1C) and MALDI-TOF (data not shown) revealed that mSin3A, HDAC1, HDAC2, RbAp48, and SAP30 were retained on the anti-mSin3A affinity column but not on the preimmune column (data not shown). Several additional polypeptides bound the anti-mSin3A column but not the preimmune column, suggesting that they also associated specifically with mSin3A (Fig. 1B). These mSin3A-associated proteins had apparent molecular masses of 180, 130, 45, 38, and 28 kDa. We present our identification and characterization of SAP180, SAP130, and SAP45 below; however, we have not yet been able to identify SAP38 or SAP28. The apparent molecular mass of SAP38 is consistent with its being a member of the ING family. These SAPs are not strongly represented in previously purified mSin3A complexes, suggesting that we have identified new mSin3A-associated proteins. We have also purified a similar mSin3A complex from U937 cells (data not shown), suggesting that it is not restricted to K562 cells.
Identification of new mSin3A-associated proteins.
To identify the novel polypeptides associated with mSin3A, both peptide mass and sequence information were used (Table 1). The tryptic peptides for the 180-kDa polypeptide band had masses that matched the retinoblastoma binding protein 1 (RBP1) (18), previously identified as an mSin3A-associated protein (29), and a second unique but similar protein, RBP1-like. In addition, the sequence from one peptide derived from the 180-kDa polypeptide was a 100% match to RBP1-like (Table 1). Therefore, the mSin3A-associated polypeptide of 180 kDa is probably a mixture of RBP1 and RBP1-like.
TABLE 1.
Data used to identify unknown mSin3A-associated proteins
| Protein | MALDI-TOF massesa | Sequenceb | ||||
|---|---|---|---|---|---|---|
| SAP180 | 1,164.72 | 1,399.62 | 1,492.76 | 1,558.84 | 1,678.85 | TGFYSGFSEVAEKR |
| 1,165.68 | 1,430.71 | 1,503.82 | 1,561.82 | 1,800.01 | ||
| 1,182.62 | 1,434.83 | 1,522.73 | 1,590.83 | 2,064.05 | ||
| 1,347.67 | 1,477.76 | 1,527.80 | 1,634.92 | 2,098.05 | ||
| 1,368.74 | ||||||
| Rbp1 | 1,165.68 | 1,399.62 | 1,549.81 | 1,707.80 | 1,923.98 | None |
| 1,202.63 | 1,503.82 | 1,561.82 | 1,800.01 | 2,027.01 | ||
| 1,383.84 | 1,527.80 | |||||
| SAP130 | 1,066.53 | 1,331.78 | 1,677.13 | 2,136.13 | PQQITHTSP | |
| 1,225.77 | 1,362.79 | 1,720.93 | LLVKAE(K) | |||
| SAP45 | 703.38 | 1,186.59 | 1,398.79 | 1,558.79 | 1,989.09 | QLQQLQEGTLQEYQK |
| 856.39 | 1,242.64 | 1,402.69 | 1,818.04 | 2,087.19 | ENLIAELEEK | |
| 989.50 | 1,292.74 | 1,468.81 | 1,833.08 | |||
Only masses used to identify the indicated protein are listed.
Sequences match the indicated protein 100% except for amino acids in parentheses.
At the time we identified RBP1-like as an mSin3A-associated protein, a full-length cDNA did not exist. A cDNA encoding full-length SAP180 was obtained from a K562 cDNA library. This cDNA encodes a protein with a predicted mass of 148 kDa that migrates at approximately 180 kDa when in vitro transcribed and translated (data not shown). Our SAP180 cDNA is nearly identical to RBP1-like, with differences probably resulting from sequencing ambiguity or polymorphisms. Overall, SAP180 is 40% identical to RBP1, with the majority of the homology arising from four domains: a Tudor domain (38), an AT-rich interacting domain (ARID) (27), a chromodomain (24) and a C-terminal region that is unique to SAP180 and RBP1 (Fig. 2A). The Tudor domain and the chromodomain appear to function primarily as protein-protein interaction domains, whereas ARIDs bind DNA. Therefore, these motifs suggest possible roles for SAP180 in targeting the mSin3A complex to DNA, stabilization of the complex via multiple interaction domains, or recruitment of additional factors that may contribute to repression. For RBP1, the conserved C-terminal domain binds mSin3A indirectly through interactions with SAP30 (34). We show below that the C-terminal domain of SAP180 also binds the mSin3A complex.
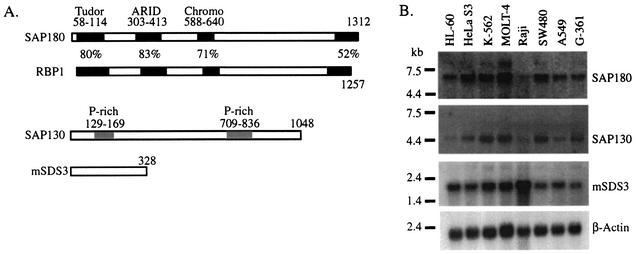
FIG. 2.
Identification and expression of new mSin3A-associated proteins. (A) Diagrams of the predicted open reading frames for each newly identified mSin3A-associated protein. Putative structural domains are boxed. Because of the high similarity between the two proteins, SAP180 is compared to RBP1, with regions of 50% identity or higher shown in black boxes. (B) A multiple cancer cell line blot probed with cDNA fragments encoding each of the SAPs and a β-actin loading control.
A database search using SAP130 mass and sequence data (Table 1) resulted in the identification of a hypothetical protein with a molecular mass of 110 kDa, whose existence was predicted by an uncharacterized expressed sequence tag. When the cDNA was subcloned and in vitro transcribed and translated, a protein migrating at roughly 130 kDa was observed (data not shown). The only recognizable motifs in the SAP130 protein are two proline-rich regions (Fig. 2A). In several instances, proline-rich regions are known to mediate protein-protein interactions (25), but their presence in SAP130 provides little insight into its function within the mSin3A complex.
We identified SAP45 peptides in a partial expressed sequence tag and used this sequence information to obtain a cDNA encoding full-length SAP45. SAP45 does not contain any recognizable sequence motifs; however, an iterative BLAST search using PSI-BLAST revealed homology to S. cerevisiae Sds3p, a protein previously identified as a component of the S. cerevisiae Sin3p complex (16). While this paper was in preparation, the mammalian SDS3 protein (mSDS3) was identified (3) and is identical to SAP45; therefore, we have adopted this nomenclature.
To determine the expression patterns for each novel SAP, a cancer cell line Northern blot was probed with cDNAs encoding SAP180, SAP130, or mSDS3 (Fig. 2B). Transcripts for each SAP were detected in each cell line; however, there was some variation in their expression levels between different cell lines. For example, Sap180 and Sap130 are less abundant in the B-cell line Raji while mSDS3 levels are highest in this cell line. Therefore, the novel SAPs are broadly expressed, but these data suggest that the mSin3A complex may exhibit cell type heterogeneity.
Newly identified proteins associate with an HDAC-containing mSin3A complex.
To confirm that the novel SAPs are components of an mSin3A complex, we generated polyclonal antibodies against SAP180, SAP130, and mSDS3. In preliminary tests, these antibodies recognized their respective in vitro-transcribed and -translated protein specifically (data not shown). Nuclear extracts from K562 cells were immunoprecipitated with these antibodies and one against mSin3A, and the associated proteins were detected by Western blotting. Each antibody was able to immunoprecipitate mSin3A, HDAC1, HDAC2, and mSDS3, suggesting that these proteins are components of a single complex. None of these proteins were immunoprecipitated with preimmune serum, demonstrating that the observed associations were specific (Fig. 3A). Finally, mSin3A has been reported to interact with members of the Swi/Snf complex, and the HDAC/RbA module also associates with the NURD/MI2 complex (54, 58); however, no members of either complex were detected in SAP180 immunoprecipitates (data not shown).
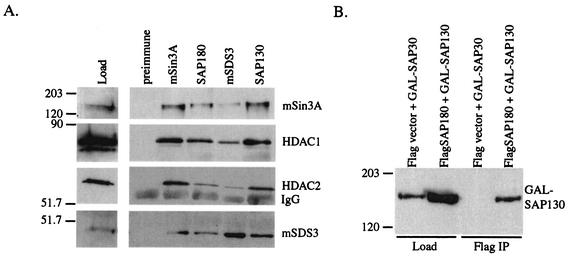
FIG. 3.
SAP180, SAP130, and mSDS3 are components of an HDAC-containing mSin3A complex. (A) mSin3A, HDAC1, HDAC2, and mSDS3 Western blots of preimmune, mSin3A, SAP180, mSDS3, and SAP130 immunoprecipitates from K562 cell nuclear extract. “Load” represents 1/50 of the nuclear extract used for the immunoprecipitations. IgG, immunoglobulin G. (B) GAL4 Western blot of Flag immunoprecipitates (IP) of whole-cell lysates from HEK293 cells transiently transfected with the indicated constructs. An aliquot (1/50) of the lysate used in the immunopreciptitation was also probed with GAL4 antibody.
SAP180 and SAP130 can both associate with mSin3A, HDAC1, HDAC2, and mSDS3; however, neither SAP180 nor SAP130 antibodies work well for Western blotting (data not shown). Therefore, it was not possible to determine whether SAP180 and SAP130 are components of the same mSin3A complex or associate independently with distinct pools of mSin3A. To test these different possibilities, we transfected HEK293 cells with Flag-tagged SAP180 (Flag-SAP180) and with a fusion between SAP130 and the DNA binding domain of GAL4 (GAL-SAP130). Anti-Flag immunoprecipitates contained both Flag-SAP180 (data not shown) and GAL-SAP130 (Fig. 3B), suggesting that they can occupy the same mSin3A complex.
To further characterize this new mSin3A complex, we fractionated K562 nuclear extracts over a gel filtration column. To reduce nonspecific protein interactions, the column was run in the presence of 0.6 M KCl and 0.1% Tween. Consistent with previous results, mSin3A eluted as a broad high-molecular-mass peak between the void volume and approximately 500 kDa. Consistent with their inclusion in an mSin3A complex, HDAC1, HDAC2, RbAp48, and mSDS3 also eluted in a similar high-molecular-mass range (Fig. 4A). Interestingly, mSDS3 eluted in a second peak between approximately 500 kDa and 43 kDa, demonstrating that a significant fraction of the cellular mSDS3 is not associated with mSin3A, but may be a subunit of other multiprotein complexes.
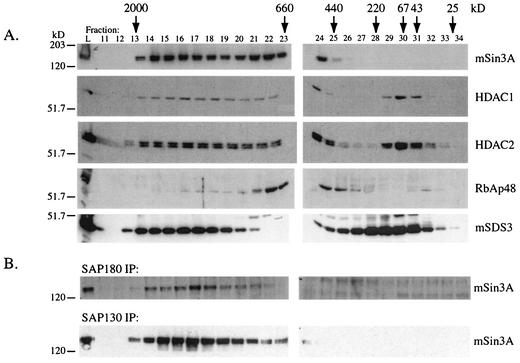
FIG. 4.
Components of the mSin3A corepressor complex coelute from a size exclusion column. (A) mSin3A, HDAC1, HDAC2, RbAp48, and mSDS3 Western blots of aliquots (1/50 of total) of fractions collected from a Superose 6 column. “L” indicates 1/20 of the amount of protein loaded onto the column. Arrows indicate where size standards eluted from the column. (B) mSin3A Western blots of SAP180 and SAP130 immunoprecipitates (IP) of fractions eluted from the Superose 6 column.
To detect the presence of SAP180 and SAP130 in the column fractions, we performed immunoprecipitations with each antibody and assayed for the presence of mSin3A by Western blotting. Both antibodies were able to immunoprecipitate mSin3A from the fractions that contained mSin3A, providing evidence that both proteins were present in these fractions and associated with mSin3A (Fig. 4B). However, since we cannot detect SAP180 or SAP130 by Western blotting, we cannot rule out the possibility that they are also components of other complexes that migrate at lower molecular mass, nor can we determine whether they are present in excess of mSin3A. Together, our purification data (Fig. 1), our coprecipitation data (Fig. 3), and the fact that mSin3A, HDAC1, HDAC2, mSDS3, SAP180, and SAP130 all coelute over the same size range (Fig. 4) provide compelling evidence for the existence of a single multiprotein complex containing each of these proteins.
SAP180, SAP130, and mSDS3 bind to the HDAC-interacting domain of mSin3A.
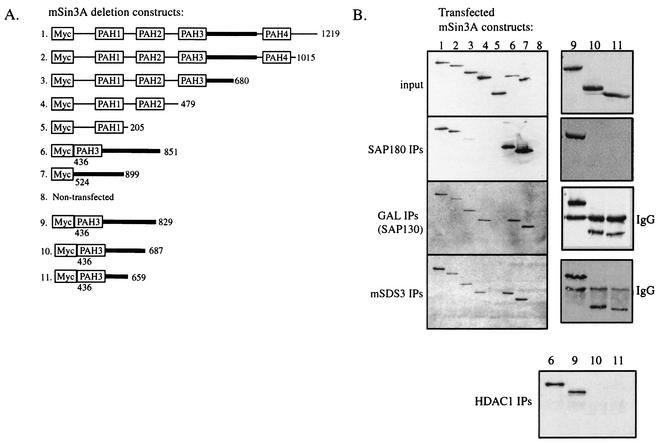
To determine the regions of mSin3A required for interaction with SAP180, SAP130, and mSDS3, HEK293 cells were transfected with a series of Myc-tagged mSin3A deletion constructs (Fig. 5A) and immunoprecipitations were performed with antibodies specific for SAP180 and mSDS3. To assay for interactions between SAP130 and mSin3A, lysates from HEK293 cells transfected with GAL-SAP130 and the different mSin3A constructs were immunoprecipitated with anti-GAL4. Western blotting with an anti-Myc monoclonal antibody showed that all of the Myc-tagged mSin3A constructs expressed to similar levels (input, Fig. 5B). SAP180, SAP130, and mSDS3 all bound an mSin3A fragment containing amino acids 524 to 899 (Fig. 5B, lanes 7). Furthermore, each SAP also bound a fragment of mSin3A that terminated at amino acid 851 (lanes 6), establishing the C-terminal boundary of the binding domain. The region between 524 and 851 is sufficient for HDAC binding and hence is termed the HDAC-interacting domain (HID) (33). Interestingly, both SAP130 and mSDS3 also coimmunoprecipitated mSin3A fragments lacking the HID (lanes 4), suggesting the presence of a second binding site for these proteins in the N-terminal half of mSin3A.
FIG. 5.
SAP180, SAP130, and mSDS3 bind to the mSin3A HDAC-interaction domain (A) Diagram of Myc-tagged mSin3A deletion constructs (constructs 1 to 11). (B) Myc Western blots of cell lysates and immunoprecipitates (IPs) of HEK293 cells transfected with Myc-tagged mSin3A deletion constructs (constructs 1 to 11). Input represents 1/50 of total cell lysates used in the immunoprecipitations. The antibodies used in the immunoprecipitations are indicated. IgG, immunoglobulin G.
The HID is relatively large and could contain multiple protein binding sites; therefore, to more precisely determine the binding site for SAP180, SAP130, and mSDS3 within the HID, we made three additional mSin3A deletion constructs that terminated at amino acids 829, 687, and 659 (Fig. 5A, constructs 9 to 11). Since the binding site for HDAC1 within the HID is not well defined, it was also included in this analysis. SAP180 and HDAC1 interacted with only the largest PAH3+HID fragment that terminated at amino acid 829 (Fig. 5B, lanes 9), suggesting that their binding site(s) encompasses amino acids 687 to 829 of mSin3A. By contrast, both SAP130 and mSDS3 coimmunoprecipitated each of the PAH3+HID truncations (lanes 9 to 11), delineating a binding site(s) between amino acids 524 and 659. These data suggest that at least two regions of the HID mediate protein-protein interactions between mSin3A and multiple mSin3A-associated proteins.
SAP180, SAP130, and mSDS3 repress transcription when targeted to DNA.
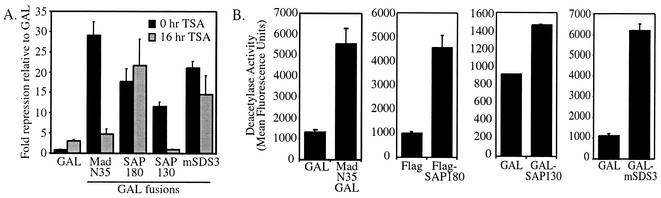
SAP130 and mSDS3 lack recognizable DNA binding domains, and although SAP180 contains an ARID, a specific binding site for SAP180 has not been determined. Therefore, to test the intrinsic transcriptional activity of the new SAPs and to demonstrate that they are components of functional mSin3A corepressor complexes, GAL-SAP180, GAL-SAP130, and GAL-mSDS3 chimeras were tested for the ability to repress transcription from a GAL-dependent luciferase reporter gene. The dependence of transcriptional repression on HDAC activity was examined by performing these experiments in the absence or presence of the deacetylase inhibitor TSA. As previously reported, recruitment of the mSin3A complex to DNA via the MadN35GAL fusion resulted in TSA-sensitive, and therefore HDAC-dependent, transcriptional repression (7, 20). Similarly, GAL-SAP180, GAL-SAP130, and GAL-mSDS3 all repressed transcription (Fig. 6A). As predicted, TSA blocked repression by GAL-SAP130 but, interestingly, had little or no effect on transcriptional repression by GAL-SAP180 or GAL-mSDS3 (Fig. 6A). One potential explanation for this lack of response to TSA is that GAL-SAP180 and GAL-mSDS3 can recruit HDAC-independent corepressors in addition to the HDAC-dependent mSin3A complex. To confirm that the GAL fusions interact with active HDACs, they were tested for their ability to immunoprecipitate HDAC activity. Like MadN35GAL, each SAP immunoprecipitated HDAC activity above background levels (Fig. 6B). Together, the transcription and deacetylase assays suggest that GAL-SAP180, GAL-SAP130, and GAL-mSDS3 can recruit an HDAC-dependent mSin3A complex to DNA, resulting in transcriptional repression. Furthermore, GAL-SAP180 and GAL-mSDS3 may also recruit HDAC-independent corepressors to DNA.
FIG. 6.
New SAPs repress transcription and immunoprecipitate deacetylase activity. (A) Repression of a Gal4-dependent luciferase reporter gene relative to GAL by GAL fusions to SAP180, SAP130, mSDS3, and the N-terminal 35 amino acids of Mad1. Black bars represent data from cells that received no TSA, and gray bars represent data from cells treated with 100 ng of TSA per ml for 16 h prior to the assay. Error bars represent the standard error of the mean. (B) Deacetylase activity (measured as fluorescence) in MadN35GAL, Flag-SAP180, GAL-SAP130, and GAL-mSDS3 immunoprecipitates. The mean activity from duplicate samples is plotted, with error bars showing the standard deviation.
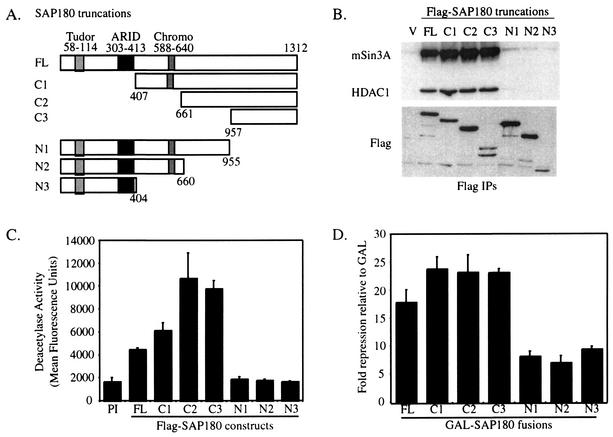
To determine which regions of SAP180 bound mSin3A and repressed transcription, N-terminal and C-terminal deletion mutants of SAP180 were made (Fig. 7A). Initially, the region of SAP180 that interacted with mSin3A-HDAC was determined. Flag-SAP180 deletion constructs were transfected into HEK293 cells, and the truncated SAP180 proteins were immunoprecipitated with FlagM2-agarose. Western blotting showed that both mSin3A and HDAC1 bound strongly to the region at the C terminus of SAP180 that is conserved with RBP1 (Fig. 7B, construct C3). Western blotting also revealed that each SAP180 deletion construct was expressed in excess (Fig. 7B). The HDAC activity in SAP180 immunoprecipitates was also measured. As expected, only the SAP180 fragments containing the C-terminal mSin3A-HDAC binding domain immunoprecipitated HDAC activity above background (Fig. 7C). Together, these data suggest that an enzymatically active mSin3A complex interacts with the C terminus of SAP180.
FIG. 7.
SAP180 contains HDAC1-dependent and -independent repression domains. (A) Diagram of SAP180 deletion constructs. (B) mSin3A, HDAC1, and Flag Western blots of FlagM2 immunoprecipitates (IPs) from whole-cell lysates from HEK293 cells transfected with Flag-tagged SAP180 deletion constructs. (C) Deacetylase activity in FlagM2 immunoprecipitates from cells transfected with Flag-tagged SAP180 truncations. Assays were done with duplicate samples, and the standard deviation was calculated. (D) Transcriptional repression of a Gal4-dependent luciferase reporter gene by GAL-SAP180 deletion constructs relative to GAL. Error bars show the standard errors of the means.
To determine which regions of SAP180 contribute to transcriptional repression, the same deletion constructs used above were fused to GAL and tested for their ability to repress transcription. As predicted, each GAL-SAP180 fusion that interacted with mSin3A-HDAC1 (C1, C2, and C3) repressed transcription, approximately 23-fold (Fig. 7D). In addition, transcriptional repression by the smallest fragment that bound mSin3A and HDAC, C3, was partially sensitive to TSA (data not shown), further suggesting a functional interaction between mSin3A and SAP180. Interestingly, the other truncations, N1, N2, and N3, also repressed transcription, but in this case repression was more modest, being six- to eightfold. Since HDAC activity was not detected in association with the N-terminal fragments, the observed repression may be independent of HDAC activity, consistent with the observation that full-length SAP180 is not TSA sensitive (Fig. 6A). Together, these results suggest that SAP180 has at least two domains that interact independently with distinct corepressor complexes.
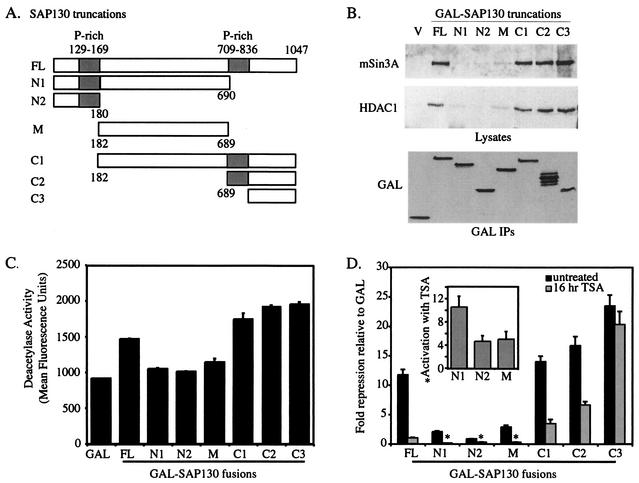
Similar analyses were performed to investigate the function of SAP130. A collection of SAP130 deletion constructs were made and fused to GAL (Fig. 8A). To map the regions of SAP130 that were required for interaction with mSin3A and HDAC1, each GAL-SAP130 construct was transfected into HEK293 cells and anti-GAL immunoprecipitates were probed for mSin3A and HDAC1 by Western blotting (Fig. 8B). Both mSin3A and HDAC1 bound to the C terminus of SAP130 between amino acids 836 and 1047, defining their binding site on SAP130. Western blots of cell lysates verified that all GAL-SAP130 fusions were expressed to similar levels (Fig. 8B). Each GAL-SAP130 construct that bound mSin3A and HDAC1 also immunoprecipitated HDAC enzymatic activity (Fig. 8C) and repressed transcription (Fig. 8D), suggesting that repression by GAL-SAP130 depends on association with the mSin3A-HDAC complex.
FIG. 8.
SAP130 contains HDAC-dependent and -independent repression domains (A) Diagram of deletion constructs for SAP130. (B) mSin3A and HDAC1 Western blots of anti-GAL immunoprecipitates (IPs) of HEK293 cells transfected with GAL-SAP130 fusions. GAL Western blots were done on whole-cell lysates using 1/25 of the amount used for the immunoprecipitations. (C) Deacetylase activity in immunoprecipitates of each GAL-SAP130 deletion construct done in duplicate, with the standard error between samples calculated. (D) Transcriptional repression of a Gal4-dependent luciferase reporter gene by each of the GAL-SAP130 truncations relative to GAL, with error bars displaying the standard error of the mean. Black bars represent data from cells that received no TSA, and gray bars represent data from cells treated with 100 ng of TSA per ml for 16 h prior to the assay. The inset shows the fold activation relative to GAL of GAL-SAP130 truncations N1, N2, and M when treated with 100 ng of TSA per ml for 16 h.
We have also used this collection of GAL-SAP130 fusion proteins to determine which region was responsible for the TSA sensitivity observed for full-length SAP130 (Fig. 6A). Construct C3, at the C terminus of SAP130, was able to bind mSin3A and HDAC and repressed transcription slightly better than did full-length SAP130, but, unexpectedly, this repression was not sensitive to TSA, suggesting that this C-terminal domain can interact with an HDAC-dependent mSin3A complex and an HDAC-independent corepressor. By contrast, transcriptional repression by each of the other constructs was TSA sensitive. Interestingly, GAL fusions to fragments N1, N2, and M, actually activated transcription following TSA treatment (Fig. 8D, inset), perhaps revealing the presence of binding sites for a transcriptional coactivator in the first 689 amino acids of SAP130.
DISCUSSION
Here we describe the identification and initial characterization of three novel proteins, SAP45/mSDS3, SAP130, and SAP180, which associate with the mSin3A corepressor with high stoichiometry. Each of these proteins could immunoprecipitate known components of the mSin3A complex as well as one another. In addition, each of the putative mSin3A-associated proteins, HDAC1, HDAC2, RbAp48, and mSDS3, coeluted with mSin3A from gel filtration columns. Finally, SAP130 and SAP180 associated with mSin3A in the high-molecular-mass fractions from the gel filtration columns. Therefore, together with the mSin3A affinity purification, which identified the majority of known components of the mSin3A complex, and previous data showing that SAP30 associates with mSin3A with high stoichiometry (32, 59), these experiments provide evidence for the existence of a multiprotein complex that contains, at the very least, mSin3A, SAP180, SAP130, HDAC1, HDAC2, RbpA48, mSDS3, and SAP30. We could not definitively identify SAP38, but based on its molecular mass, it is probably a member of the p33ING family (30, 48). We are still trying to determine the identity of SAP28. Finally, in contrast to the results of others (57), we did not detect SAP18 in our mSin3A purification, suggesting that its association with mSin3A may be cell type specific or regulated. Because this mSin3A complex was identified by affinity purification and it is highly salt stable, it probably represents the major mSin3A-containing complex in K562 cells.
We detected peptide masses for both SAP180 and RBP1 within the 180-kDa mSin3A-associated polypeptide band, suggesting that it was a mixture of SAP180 and RBP1. We are currently investigating whether SAP180 and RBP1 are members of the same mSin3A complex or whether they are components of distinct mSin3A complexes. RBP1 was originally identified as a binding partner for the Rb transcriptional corepressor and was subsequently shown to interact with mSin3A (18, 34). An LXCXE motif in RBP1 binds the Rb pocket (35); however, there is no easily identifiable LXCXE motif in SAP180 (data not shown), suggesting that it cannot bind Rb. Therefore, RBP1-containing and SAP180-containing complexes may differentially target Rb.
mSDS3, SAP130, and SAP180 all repressed transcription when targeted to a reporter via a heterologous DNA binding domain, and repression by SAP130 (Fig. 8), SAP180 (Fig. 7), and mSDS3 (data not shown) correlated with mSin3A binding, suggesting that each of these proteins can interact with functional mSin3A corepressor complexes. At present it is unclear why transcriptional repression by SAP130 is sensitive to TSA whereas transcriptional repression by mSDS3 and SAP180 is completely insensitive. It may relate to the GAL tethering assay used to measure their activity. We detected mSDS3, SAP130, and SAP180 in association with each other and with mSin3A; therefore, we think that this assay must reflect recruitment of other, currently unknown, activities to the mSin3A complex by the new SAP proteins. At this point, we cannot rule out the formal possibility that this assay also measures the recruitment of other transcription complexes, which lack mSin3A, by mSDS3, SAP130, or SAP180. Because we observed mSDS3 in high-molecular-mass complexes that lack mSin3A (Fig. 4), this argument may be particularly relevant for this protein. Regardless of the precise mechanism, our data indicate that mSDS3, SAP130, and SAP180 interact with both mSin3A and other regulatory activities. As discussed below, mSDS3 is probably required for the assembly and/or stability of the mSin3A-HDAC corepressor whereas SAP180 is probably involved in stabilizing the mSin3A complex on DNA and in mediating interactions between mSin3A and HDAC-independent corepressors. The function of SAP130 is more complex, but our data suggest that, like SAP180, it can interact with mSin3A- and HDAC-independent corepressors; however, unlike SAP180, SAP130 also appears to interact with a transcriptional coactivator(s).
In addition to recruiting HDACs to repress transcription, mSin3A can repress transcription by HDAC-independent mechanisms (20, 33); however, the identity of this HDAC-independent corepressor is not yet known. The N terminus of SAP180, the C terminus of SAP130, and mSDS3 can all repress transcription by HDAC-independent mechanisms, suggesting that they may recruit one or multiple HDAC-independent corepressors to mSin3A. TLE, the mammalian homolog of Drosophila groucho, appears to repress transcription by both HDAC-dependent and -independent mechanisms (15) and interacts with mSin3A (13, 55, 56), suggesting that it may account for some fraction of the HDAC-independent corepressor activity of mSin3A. We are currently testing whether TLE can interact with mSDS3, SAP130, or SAP180.
The N terminus of SAP180 may mediate an interaction with HDAC-independent corepressor complexes. Furthermore, a repression domain has also been mapped to a similar region of the related RBP1 (35), suggesting that this function may be conserved. The N terminus of SAP180 contains a Tudor domain and a chromodomain that probably serve as protein-protein interaction domains and are candidate binding sites for the putative HDAC-independent corepressor. Tudor domains have been identified in a number of proteins that are components of ribonucleoprotein complexes and have been suggested to function as protein-protein interaction domains (12, 43, 46). Chromodomains are found in a number of proteins involved in chromatin organization and modification (24) and function as interaction interfaces that can associate with protein or nucleic acid (2, 9, 31, 40). The solution structure of the chromodomain of HP1 bound to histone H3 methylated at lysine 9 revealed the residues required for the folding of chromodomain and binding to this methylated lysine (23, 42). Mapping of the primary amino acid sequence of the chromodomain of SAP180 to this structure suggests that the chromodomain of SAP180 forms the canonical chromodomain fold and probably binds protein rather than RNA (data not shown). Furthermore, this analysis also suggests that the chromodomain of SAP180 is unlikely to bind methylated histone H3, raising the possibility that it may recognize other methylated histones or perhaps methylated nonhistone proteins (53). Therefore, given the demonstrated roles of Tudor domains and chromodomains in mediating protein-protein interactions, it seems most likely that the Tudor domains and chromodomains of SAP180 mediate interactions between the mSin3A complex and other regulatory complexes and/or modified histone tails.
In contrast, the ARID domain of SAP180 probably mediates protein-DNA interactions. The ARID domain was first characterized in the Drosophila transcription factor Dead-ringer and was shown to bind AT-rich sequences (19). However, subsequent characterization of other ARIDs showed that they do not bind AT-rich sequences exclusively and, in several cases, bind DNA nonspecifically (27). We were unable to identify a high-affinity consensus binding site by using the ARID of SAP180 in a binding-site selection experiment (data not shown). As such, it is likely that the ARID of SAP180 binds DNA nonspecifically or with low affinity. Because the ARIDs of SAP180 and RBP1 are 91% similar, it is likely that they will bind DNA similarly. Therefore, we propose that the ARID of SAP180 and/or RBP1 may function to stabilize the mSin3A-HDAC complex on DNA via interactions that are sequence independent. mSin3A-HDAC complexes can be targeted to DNA via interactions with sequence-specific repressors like Mad1 (8, 26) and by an unidentified nonspecific constitutive targeting mechanism (37). It is conceivable that the ARID of SAP180 and/or RBP1 could stabilize mSin3A-HDAC complexes under either of these circumstances.
Transcriptional assays carried out in the presence of TSA revealed that the C terminus of SAP130 may interact with both HDAC-dependent and -independent corepressors. Paradoxically, the N terminus of SAP130, which had only weak repression activity on it own, activated transcription approximately sixfold in the presence of TSA (Fig. 8). We have yet to investigate this interesting finding; however, because the activity of many transcription factors is regulated by their acetylation and deacetylation state (28), one potential model is that acetylation-deacetylation of SAP130 itself may regulate the binding of a coactivator. Consistent with this model, we have identified potentially acetylated peptides from SAP130 by MALDI-TOF mass spectrometry (data not shown). Together, these data suggest that the N terminus of SAP130 has the capacity to interact with a transcriptional coactivator. As such, SAP130 might recruit this activity to mSin3A, modulating its transcriptional repression activity. The relatively low level of HDAC activity found in association with SAP130 may also reflect such a regulatory mechanism.
SAP45 is identical to mSDS3 and appears to be the mammalian homolog of S. cerevisiae Sds3p. Sds3p interacts with Sin3p and is required for the integrity and the enzymatic activity of the S. cerevisiae Sin3p-Rpd3p complex (16, 36). In mammalian cells, reduction of the levels of mSDS3 by RNAi resulted in a decrease in HDAC1 activity (3), suggesting that like SDS3p, mSDS3 may contribute to the stability and/or the activity of the mSin3A-HDAC corepressor. Our gel filtration experiment shows that perhaps as much as 50% of the cellular mSDS3 is in high-molecular-mass complexes that lack mSin3A, suggesting that it has functions outside of its association with mSin3A. A percentage of HDAC1 and HDAC2 are also found in these fractions. It will be of interest to establish whether mSDS3 functions in the preassembly of an HDAC1/HDAC2/RbAp46/RbpAp48 complex, as suggested from the experiments with yeast (36), or whether it is involved in the assembly and stabilization of novel complexes.
Finally, our data suggest that the HID is the focal point for the assembly for the mSin3A complex. It lies between PAH3 and PAH4 of mSin3A, is highly conserved, and was originally identified as the binding site for HDAC1 and HDAC2 (33). Here we show that the HID can also interact with mSDS3, SAP130, and SAP180. Furthermore, we have shown that the HID can interact with members of the mortality factor family (56) and the TLE/groucho corepressor (unpublished data). Therefore, the HID probably has dual function: functioning as a binding pocket that mediates the assembly of the “core” mSin3A complex and also in mediating interactions with other regulatory proteins.
Acknowledgments
We thank Laura Palanker for making the GST-SAP45 fusion protein and for performing preliminary testing of the SAP45 antibody, R. Eisenman for providing Myc-tagged mSin3A constructs, and I.-C. Tsai for making the mSin3A-HID truncations. We thank Bradley Cairns for advice in protein purification and David Virshup for a critical review of the manuscript.
DNA sequencing and oligonucleotide synthesis were supported by Cancer Center Support Grant 2P30 CA42014. This work was supported by NIH grant GM55668, American Cancer Society Research Scholar grant RSG-01-049-01-GMC, and funds from the Huntsman Cancer Foundation. D.E.A. is a scholar of the Leukemia and Lymphoma Society.
REFERENCES
- 1.Ahringer, J. 2000. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 16:351-356. [DOI] [PubMed] [Google Scholar]
- 2.Akhtar, A., D. Zink, and P. B. Becker. 2000. Chromodomains are protein-RNA interaction modules. Nature 407:405-409. [DOI] [PubMed] [Google Scholar]
- 3.Alland, L., G. David, H. Shen-Li, J. Potes, R. Muhle, H. C. Lee, H. Hou, Jr., K. Chen, and R. A. DePinho. 2002. Identification of mammalian Sds3 as an integral component of the Sin3/histone deacetylase corepressor complex. Mol. Cell. Biol. 22:2743-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alland, L., R. Muhle, H. Hou, Jr., J. Potes, L. Chin, N. Schreiber-Agus, and R. A. DePinho. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387:49-55. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 6.Ayer, D. E. 1999. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 9:193-198. [DOI] [PubMed] [Google Scholar]
- 7.Ayer, D. E., C. D. Laherty, Q. A. Lawrence, A. Armstrong, and R. N. Eisenman. 1996. Mad proteins contain a dominant transcription repression domain. Mol. Cell. Biol. 16:5772-5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayer, D. E., Q. A. Lawrence, and R. N. Eisenman. 1995. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80:767-776. [DOI] [PubMed] [Google Scholar]
- 9.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 10.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 11.Brubaker, K., S. M. Cowley, K. Huang, L. Loo, G. S. Yochum, D. E., Ayer, R. N. Eisenman, and I. Radhakrishnan. 2000. Solution structure of the interaction domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex. Cell 103:655-665. [DOI] [PubMed] [Google Scholar]
- 12.Buhler, D., V. Raker, R. Luhrmann, and U. Fischer. 1999. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum. Mol. Genet. 8:2351-2357. [DOI] [PubMed] [Google Scholar]
- 13.Choi, C. Y., Y. H. Kim, H. J. Kwon, and Y. Kim. 1999. The homeodomain protein NK-3 recruits groucho and a histone deacetylase complex to repress transcription. J. Biol. Chem. 274:33194-33197. [DOI] [PubMed] [Google Scholar]
- 14.Cosma, M. P. 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10:227-236. [DOI] [PubMed] [Google Scholar]
- 15.Courey, A. J., and S. Jia. 2001. Transcriptional repression: the long and the short of it. Genes Dev. 15:2786-2796. [DOI] [PubMed] [Google Scholar]
- 16.Dorland, S., M. L. Deegenaars, and D. J. Stillman. 2000. Roles for the Saccharomyces cerevisiae SDS3, CBK1 and HYM1 genes in transcriptional repression by SIN3. Genetics 154:573-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eilers, A. L., A. N. Billin, J. Liu, and D. E. Ayer. 1999. A 13-amino acid amphipathic alpha-helix is required for the functional interaction between the transcriptional repressor Mad1 and mSin3A. J. Biol. Chem. 274:32750-32756. [DOI] [PubMed] [Google Scholar]
- 18.Fattaey, A. R., K. Helin, M. S. Dembski, N. Dyson, E. Harlow, G. A. Vuocolo, M. G. Hanobik, K. M. Haskell, A. Oliff, D. Defeo-Jones, et al. 1993. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene 8:3149-3156. [PubMed] [Google Scholar]
- 19.Gregory, S. L., R. D. Kortschak, B. Kalionis, and R. Saint. 1996. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol. Cell. Biol. 16:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 21.Hassig, C. A., and S. L. Schreiber. 1997. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr. Opin. Chem. Biol. 1:300-308. [DOI] [PubMed] [Google Scholar]
- 22.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs, S. A., and S. Khorasanizadeh. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295:2080-2083. [DOI] [PubMed] [Google Scholar]
- 24.Jones, D. O., I. G. Cowell, and P. B. Singh. 2000. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays 22:124-137. [DOI] [PubMed] [Google Scholar]
- 25.Kay, B. K., M. P. Williamson, and M. Sudol. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14:231-241. [PubMed] [Google Scholar]
- 26.Knoepfler, P. S., and R. N. Eisenman. 1999. Sin meets NuRD and other tails of repression. Cell 99:447-450. [DOI] [PubMed] [Google Scholar]
- 27.Kortschak, R. D., P. W. Tucker, and R. Saint. 2000. ARID proteins come in from the desert. Trends Biochem. Sci. 25:294-299. [DOI] [PubMed] [Google Scholar]
- 28.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 30.Kuzmichev, A., Y. Zhang, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol. Cell. Biol. 22:835-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 32.Laherty, C. D., A. N. Billin, R. M. Lavinsky, G. S. Yochum, A. C. Bush, J. M. Sun, T. M. Mullen, J. R. Davie, D. W. Rose, C. K. Glass, M. G. Rosenfeld, D. E. Ayer, and R. N. Eisenman. 1998. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell 2:33-42. [DOI] [PubMed] [Google Scholar]
- 33.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 34.Lai, A., B. K. Kennedy, D. A. Barbie, N. R. Bertos, X. J. Yang, M. C. Theberge, S. C. Tsai, E. Seto, Y. Zhang, A. Kuzmichev, W. S. Lane, D. Reinberg, E. Harlow, and P. E. Branton. 2001. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol. Cell. Biol. 21:2918-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai, A., J. M. Lee, W. M. Yang, J. A. DeCaprio, W. G. Kaelin, Jr., E. Seto, and P. E. Branton. 1999. RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol. Cell. Biol. 19:6632-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lechner, T., M. J. Carrozza, Y. Yu, P. A. Grant, A. Eberharter, D. Vannier, G. Brosch, D. J. Stillman, D. Shore, and J. L. Workman. 2000. Sds3 (suppressor of defective silencing 3) is an integral component of the yeast Sin3-Rpd3 histone deacetylase complex and is required for histone deacetylase activity. J. Biol. Chem. 275:40961-40966. [DOI] [PubMed] [Google Scholar]
- 37.Li, J., Q. Lin, W. Wang, P. Wade, and J. Wong. 2002. Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev. 16:687-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacKenzie, A. E., and N. H. Gendron. 2001. Tudor reign. Nat. Struct. Biol. 8:13-15. [DOI] [PubMed] [Google Scholar]
- 39.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 40.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 41.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen, P. R., D. Nietlispach, H. R. Mott, J. Callaghan, A. Bannister, T. Kouzarides, A. G. Murzin, N. V. Murzina, and E. D. Laue. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416:103-107. [DOI] [PubMed] [Google Scholar]
- 43.Ponting, C. P. 1997. Tudor domains in proteins that interact with RNA. Trends Biochem. Sci. 22:51-52. [DOI] [PubMed] [Google Scholar]
- 44.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 45.Schreiber-Agus, N., L. Chin, K. Chen, R. Torres, G. Rao, P. Guida, A. I. Skoultchi, and R. A. DePinho. 1995. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell 80:777-786. [DOI] [PubMed] [Google Scholar]
- 46.Selenko, P., R. Sprangers, G. Stier, D. Buhler, U. Fischer, and M. Sattler. 2001. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 8:27-31. [DOI] [PubMed] [Google Scholar]
- 47.Sif, S., A. J. Saurin, A. N. Imbalzano, and R. E. Kingston. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skowyra, D., M. Zeremski, N. Neznanov, M. Li, Y. Choi, M. Uesugi, C. A. Hauser, W. Gu, A. V. Gudkov, and J. Qin. 2001. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J. Biol. Chem. 276:8734-8739. [DOI] [PubMed] [Google Scholar]
- 49.Sterneck, E., C. Muller, S. Katz, and A. Leutz. 1992. Autocrine growth induced by kinase type oncogenes in myeloid cells requires AP-1 and NF-M, a myeloid specific, C/EBP-like factor. EMBO J. 11:115-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong, J. K., C. A. Hassig, G. R. Schnitzler, R. E. Kingston, and S. L. Schreiber. 1998. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395:917-921. [DOI] [PubMed] [Google Scholar]
- 51.Wang, H., I. Clark, P. R. Nicholson, I. Herskowitz, and D. J. Stillman. 1990. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol. Cell. Biol. 10:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, J., and M. Grunstein. 2000. 25 years after the nucleosome model: chromatin modifications. Trends Biochem. Sci. 25:619-623. [DOI] [PubMed] [Google Scholar]
- 53.Xu, W., H. Chen, K. Du, H. Asahara, M. Tini, B. M. Emerson, M. Montminy, and R. M. Evans. 2001. A transcriptional switch mediated by cofactor methylation. Science 294:2507-2511. [DOI] [PubMed] [Google Scholar]
- 54.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]
- 55.Yochum, G. S., and D. E. Ayer. 2001. Pf1, a novel PHD zinc finger protein that links the TLE corepressor to the mSin3A-histone deacetylase complex. Mol. Cell. Biol. 21:4110-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yochum, G. S., and D. E. Ayer. 2002. A role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and TLE. Mol. Cell. Biol. 22:7868-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357-364. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279-289. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, Y., Z. W. Sun, R. Iratni, H. Erdjument-Bromage, P. Tempst, M. Hampsey, and D. Reinberg. 1998. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell 1:1021-1031. [DOI] [PubMed] [Google Scholar]