Abstract
Proteomic approaches require simple and efficient protein purification methodologies that are amenable to high throughput. Biotinylation is an attractive approach for protein complex purification due to the very high affinity of avidin/streptavidin for biotinylated templates. Here, we describe an approach for the single-step purification of transcription factor complex(es) based on specific in vivo biotinylation. We expressed the bacterial BirA biotin ligase in mammalian cells and demonstrated very efficient biotinylation of a hematopoietic transcription factor bearing a small (23-aa) artificial peptide tag. Biotinylation of the tagged transcription factor altered neither the factor's protein interactions or DNA binding properties in vivo nor its subnuclear distribution. Using this approach, we isolated the biotin-tagged transcription factor and at least one other known interacting protein from crude nuclear extracts by direct binding to streptavidin beads. Finally, this method works efficiently in transgenic mice, thus raising the prospect of using biotinylation tagging in protein complex purification directly from animal tissues. Therefore, BirA-mediated biotinylation of tagged proteins provides the basis for the single-step purification of proteins from mammalian cells.
In the postgenome-sequencing era, focus has shifted toward the identification and characterization of the protein complement of cells, the proteome. A crucial aspect of this effort is the utilization of simple and efficient methodologies that are amenable to high-throughput approaches for the purification of proteins and protein complexes (1, 2). As a result, a number of generic affinity-based methodologies have been developed for these purposes, based primarily on the use of specific antibodies or affinity tags that are fused to the protein of interest (3, 4).
Prominent among the affinity-based purification methodologies is the biotin/avidin system. Biotin is a naturally occurring cofactor for metabolic enzymes, which is active only when covalently attached to the enzymes through the action of specific protein–biotin ligases (5). Any biotinylated substrate can be bound very tightly by the proteins avidin and streptavidin. Biotin/avidin binding is the strongest noncovalent interaction known in nature (Kd = 10–15 M), several orders higher than that of commonly used antibodies or other affinity tags. As a result, the biotin/avidin affinity system has numerous applications in modern biological techniques (6). For the purposes of protein purification, in particular, biotinylation offers a number of advantages. For example, the high affinity of biotin for avidin/streptavidin allows purification of the biotinylated protein under high stringency conditions, thus reducing background binding often observed with other affinity tags that elute more easily. In addition, there are very few naturally biotinylated proteins, thus reducing the chance for crossreaction when using biotinylation in protein purification, as opposed to antibodies that may crossreact with several species.
The potential advantages of biotinylation tagging in protein purification have not gone unnoticed (7). The characterization of the minimal amino acid sequence requirements of naturally biotinylated proteins has led to the development of sequence tags that can be biotinylated in bacterial, yeast, insect, and mammalian cells (7–11). Biotinylation can occur either by the cell's endogenous protein–biotin ligases or through the coexpression of an exogenous biotin ligase, in most cases that of the bacterial BirA enzyme. These tags, however, are large in size (at least 63 aa) and may thus affect the structure of the proteins they are fused to. In addition, that these tags can be recognized by endogenous enzymes excludes applications where biotinylation of the tagged protein may need to be regulated. Furthermore, biotinylation using these tags is not very efficient, particularly in mammalian cells (10, 11). Another approach, so far only demonstrated in bacteria (12–15), utilizes small (<23-aa) artificial tags that have been selected through multiple rounds of screening combinatorial peptide libraries for specific biotinylation by BirA biotin ligase (16). These tags have been shown to be biotinylated in vitro with kinetics comparable to those of natural biotin acceptor sequences (17) and may thus serve as excellent substrates for efficient biotinylation in cells by coexpressed biotin ligases.
Simple generic affinity purification methodologies have been increasingly applied for the purposes of large-scale proteomic studies, particularly in yeast (18, 19). However, these often use reagents of variable affinities (e.g., antibodies) that increase background and/or intermediate steps (e.g., prepurification or affinity tag removal) that restrict their simple application in more complex protein sources such as mammalian cells. Considering the advantages of biotinylation, we explored the feasibility of using it for the simple high-affinity one-step purification of tagged proteins from mammalian cells. To these ends, we coexpressed bacterial BirA biotin ligase and hematopoietic transcription factors tagged by an N-terminal fusion of a small artificial peptide previously shown to be biotinylated by BirA (15–17). We show that tagged proteins can be very efficiently and specifically biotinylated in mammalian cells and transgenic mice and can be efficiently purified in a single step by binding to streptavidin beads.
Experimental Procedures
Constructs. The coding region of the Escherichia coli birA biotin–protein ligase gene (20) was cloned from genomic DNA by PCR as an ≈1-kb fragment by using Deep-VENT DNA polymerase (New England Biolabs) and verified by sequencing. The birA gene was recloned into the BglII site of the erythroid expression vector pEV-puromycin, which consists of the human β-globin Locus Control Region (miniLCR), the human β-globin promoter, and the β-globin second intron (21). GATA-1 cDNA cloned in pEV-Neomycin was N-terminally tagged by introducing into the NcoI site at the start codon, an oligonucleotide linker with NcoI overhangs coding for the 23-aa biotinylation tag (16). Tagged erythroid Krüppel-like factor (EKLF) cDNA was constructed in pBluescript by cloning sequentially into the NcoI site start codon firstly an oligonucleotide coding for three copies of the hemagglutinin tag, followed by an oligonucleotide coding for the 23-aa biotinylation tag. Tagged EKLF was then subcloned into pEV-Neomycin.
Mouse Erythroleukemic (MEL) Cell Transfections. MEL cells (22) were initially electroporated with linearized BirA/pEV, and stable clones were selected under puromycin. Clones were screened for BirA RNA expression by Northern blot analysis. A selected BirA/pEV MEL clone was transfected with tagged GATA-1/pEV, and stable clones were double selected for puromycin and neomycin (Invitrogen).
Nuclear Extract Preparation. MEL cells cultured in DMEM supplemented with 10% FCS at 37°C were induced to differentiate into mature erythroblasts with 2% DMSO for at least 3 days (22). Cells were harvested by centrifugation at 640 × g and washed once with cold PBS. The cell pellet was resuspended in 2.2 M sucrose in 10 mM Hepes, pH 7.9/25 mM KCl/0.15 mM Spermine/0.5 mM Spermidine/1 mM EDTA and incubated for 20 min. Cells were lysed with a blender, and lysis was checked under a light microscope by staining nuclei with Unna (Methylgreen-Pyronin). Nuclei were pelleted by ultracentrifugation at 141,000 × g for 2 h at 4°C, resuspended in lysis buffer (10 mM Hepes, pH 7.9/100 mM KCl/3 mM MgCl2/0.1 mM EDTA/20% glycerol), and extracted by dropwise addition of 3.3 M KCl until the final concentration was ≈400 mM. Insoluble material was removed by ultracentrifugation at 300,000 × g for 1 h at 4°C. Nuclear extracts were aliquoted and stored at –70°C.
Immunoblot Analysis. For analysis of GATA-1 expression and in vivo biotinylation, nuclear extracts (1–2.5 μg/lane) were resolved by SDS/PAGE in an 8% gel and blotted onto ProTran nitrocellulose membrane (Schleicher & Schuell) by using standard procedures. Filters were blocked for 1 h in 5% BSA/1× TBS/0.2% Tween-20 and incubated for 1 h at room temperature with anti-GATA-1 N6 rat monoclonal antibody (Santa Cruz Biotechnology, dilution 1:5,000) or streptavidin–horseradish peroxidase conjugate (HRP) (NEN, dilution 1:10,000). Filters were washed in 1× TBS/0.5M NaCl/0.3% Triton X-100 and incubated in secondary rabbit anti-rat antibody (DAKO, dilution 1:3,000), as above. No secondary antibody step was required after incubating the filters with streptavidin–HRP. Filters were developed by using enhanced chemiluminescence (Amersham Pharmacia).
Binding to Streptavidin Beads. Paramagnetic streptavidin beads [Dynabeads M-280, Dynal (Great Neck, NY)] were blocked by washing three times in TBS with 200 ng/μl purified chicken serum albumin (Sigma-Aldrich). We used ≈20 μl of beads per 1 mg of nuclear extract. Binding was done in 1× TBS/0.3% Nonidet P-40 at 4°C for 1 h to overnight on a rocking platform, followed by six washes in binding solution at room temperature. Bound material was eluted by boiling for 5 min in Laemmli protein sample loading buffer and analyzed by immunoblotting as above.
MS. Proteins eluted from the beads after binding of the BirA nuclear extract were separated by SDS/PAGE electrophoresis on an 8% polyacrylamide gel and stained with Colloidal blue (Invitrogen). The entire lane was cut out and divided into at least 20 gel plugs, which were each further reduced to 1 mm3 gel pieces and dried by using 100% acetonitrile (Fluka) in 60% acetonitrile pretreated tubes (Bioquote, York, U.K.). Proteins were in-gel-digested by using modified trypsin (Roche Diagnostics) in 50 mM ammonium bicarbonate. Digests were analyzed by nanoflow liquid chromatography–tandem MS by using an electrospray ionization quadrupole time-of-flight mass spectrometer (Q-Tof, Micromass, Manchester, U.K.) operating in positive ion mode. A nanoLC system was coupled to the Q-Tof essentially as described (23). Peptide mixtures were delivered to the system by using a Famos autosampler (LC Packings, Amsterdam) at 3 μl/min and trapped on an Aqua C18RP column [Phenomenex (Belmont, CA); column dimensions 1 cm × 100 μm ID, packed in-house]. After flow splitting down to 150–200 nl/min, peptides were transferred to the analytical column (Pep-Map, LC Packings; column dimensions 25 cm × 50 μm ID, packed in-house) in a gradient of acetonitrile (1% per min). Fragmentation of eluting peptides was performed in data-dependent mode, and mass spectra were acquired in full-scan mode. Database searches were performed by using mascot (www.matrixscience.com).
Chromatin Pull-Down Assays. MEL cells were treated with 1% formaldehyde in 40 mM Hepes, pH 7.9, at room temperature for 10 min, followed by quenching with 0.125 M glycine. Crosslinked chromatin was fragmented by sonication [10 × 30-s bursts at amplitude 5, followed by 10 × 30-s bursts at amplitude 8 by using a Sanyo Soniprep (Loughborough, U.K.) 150 sonicator] and aliquots corresponding to 4–10 OD260 units were snap-frozen. Pull-downs were carried out by incubating an aliquot of crosslinked chromatin for at least 1 h at 4°C with 20 μl of streptavidin-coated Dynabeads, preblocked with 1 mg/ml BSA and 0.4 mg/ml sonicated salmon sperm DNA. Washes were done according to standard protocols (www.upstate.com/misc/protocols.asp). Elution of bound material and reversal of crosslinks was done in 1% SDS in TE buffer (10 mM Tris/1 mM EDTA, pH 8.0) by overnight incubation at 60°C with shaking. DNA was recovered after deproteinization, and aliquots of pulled-down DNA were assayed by PCR using primers against the erythroid mouse βmaj globin promoter as described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Transgenic Mice. DNA fragments containing the tagged EKLF, and BirA erythroid expression cassettes were released from prokaryotic vector sequences by double digestion with Aat II/Asp 718. The DNA was gel-purified by using Gelase (Epicentre Technologies, Madison, WI) and prepared for microinjection by using EluTip (Schleicher & Schuell) according to the manufacturers' instructions. Microinjection into mouse fertilized eggs was carried out according to standard procedures (24). Screening of mice for transgenics was carried out by Southern blot analysis by using the BirA cDNA and the human β-globin intron II as probes.
Results
Efficient Biotinylation of Tagged GATA-1 in Transfected Cells. The scheme for the specific in vivo biotinylation of tagged proteins in mammalian cells is outlined in Fig. 1A. According to this procedure, a small (23 aa) peptide tag is fused to the protein of interest and coexpressed in cells together with BirA, a bacterial protein–biotin ligase (20). The peptide tag used has been previously isolated from a synthetic peptide library screened for BirA-mediated biotinylation, which occurs specifically at the lysine residue of the tag (16). Protein database searches have identified no naturally occurring proteins that possess a sequence motif similar to that of the peptide tag.
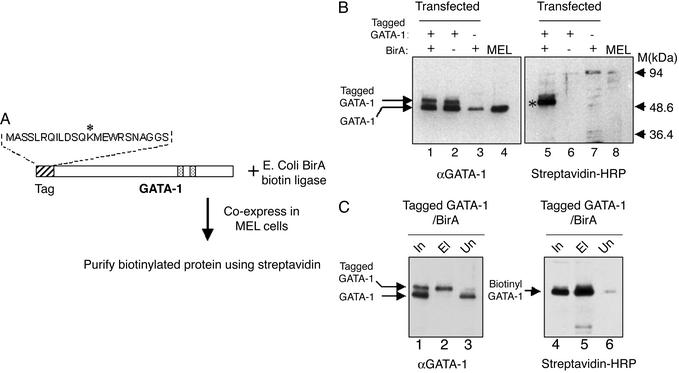
Fig. 1.
(A) Scheme for the specific biotinylation of tagged GATA-1 by BirA biotin ligase in MEL cells. The sequence of the 23-aa peptide tag fused to the N terminus of GATA-1 is shown. The asterisk indicates the lysine residue that becomes specifically biotinylated by BirA. Speckled boxes indicate the positions of the two GATA-1 Zinc-fingers. Tagged GATA-1 and BirA were cloned separately in a mammalian erythroid expression cassette and coexpressed in MEL cells. (B) Biotinylation of tagged GATA-1 in MEL cells. (Left) Western blot with an anti-GATA-1 antibody to detect endogenous and tagged GATA-1 proteins. (Right) Western blot of the same extracts with streptavidin–HRP conjugate to detect biotinylated GATA-1. Nuclear extracts (5 μg per lane) from the double transfectants (lanes 1 and 5) and single transfectants (lanes 2, 3, 6, and 7) for tagged GATA-1 and Bir A were tested. Lanes 4 and 8, nuclear extract from nontransfected MEL cells. Biotinylated GATA-1 (asterisk) is clearly visible in only in the lane of the double transfected cells. Also indicated is the low background detected by streptavidin in MEL nuclear extracts from cells expressing only BirA (Right, lane 7). (C) Efficiency of GATA-1 biotinylation and binding to streptavidin beads. (Left) Western blot using anti-GATA-1 antibody to detect binding of tagged GATA-1 to streptavidin beads (lane 2; starting material for the binding was 2.5 times the amount of nuclear extract shown in the input lane). Input and unbound material are shown in lanes 1 and 3. (Right) The same filter stripped and reprobed with streptavidin–HRP to detect the binding of biotinylated GATA-1 to streptavidin beads (lane 5). Lane 6 shows that very little tagged GATA-1 remains unbound by streptavidin. In this binding experiment, the beads were washed under stringent conditions (0.5 M NaCl/0.3% Triton X-100 in PBS). In, input (nuclear extract); El, eluted material; Un, unbound material.
The protein we tagged in testing this system was the essential murine hematopoietic transcription factor GATA-1 (for review, see ref. 25). The tag was fused N-terminally to GATA-1 and expressed under the control of a human β-globin expression cassette in MEL cells, which can be induced to undergo terminal erythroid cell differentiation (22). BirA was also cloned and expressed in MEL cells by using the human globin expression cassette. Clones corresponding to single and double stable transfectants for tagged GATA-1 and BirA were isolated and initially screened for expression of both constructs. In the case of GATA-1, Western blot analysis using a GATA-1 antibody detects the slower-migrating tagged protein as well as the endogenous GATA-1 in nuclear extracts from transfected cells (Fig. 1B, lanes 1 and 2). Expression of BirA was analyzed at the RNA level (data not shown).
We next tested, on selected stable transfectants, whether the tagged GATA-1 protein was biotinylated by BirA. Assaying nuclear extracts from MEL cell clones using a streptavidin–HRP conjugate showed a robust signal corresponding to the tagged GATA-1 protein, detectable only in the lane of the tagged GATA-1/BirA double transfectant (Fig. 1B, lane 5). No biotinylation of tagged GATA-1 is visible in the absence of BirA (Fig. 1B, lane 6). These findings confirm that BirA protein is synthesized in an active form in transfected MEL cells. In addition, very little nonspecific background biotinylation is observed in MEL cell nuclear extracts expressing only BirA (Fig. 1B, lane 7). We therefore conclude that expression of bacterial BirA protein–biotin ligase in MEL cells can specifically biotinylate a mammalian transcription factor bearing a unique peptide tag.
We next tested the efficiency of biotinylation by binding tagged GATA-1 in crude nuclear extracts to streptavidin paramagnetic Dynabeads (Fig. 1C). Analysis of the material eluted from the beads showed that almost all of the tagged GATA-1 protein was bound (compare lane 2 to lanes 1 and 3, Fig. 1C). Reprobing the same filter with streptavidin–HRP shows ≈100% efficiency in the biotinylation and capture of tagged GATA-1 by the beads (Fig. 1C, lanes 4 and 5). In addition, consistent with what was seen in Fig. 1B, there is little background binding of endogenously biotinylated proteins to the beads, as detected by streptavidin–HRP (Fig. 1C, lane 5). These data demonstrate that tagged GATA-1 is very efficiently biotinylated and recovered from extracts by streptavidin binding, with negligible background biotinylation.
Single-Step Purification of Biotinylated GATA-1 from Crude Nuclear Extracts by Streptavidin Binding. We also explored the feasibility of a single-step purification in isolating biotinylated GATA-1 from crude nuclear extracts by directly binding to streptavidin beads under moderate stringency (150 mM NaCl/0.3% Nonidet P-40/200 ng/μl chicken serum albumin). We first tried a control binding from 5 mg of crude nuclear extracts from MEL cells expressing only BirA (Fig. 2, lane 4). The eluted material consisted of approximately five strongly stained bands against a backdrop of much fainter bands (Fig. 2, lane 5). We identified the background binding proteins by excising the whole lane from the gel and analyzing it by liquid chromatography–tandem MS. The proteins thus identified are classified in Table 1 according to biological function and cellular compartment, as defined by the Gene Ontology Consortium (www.geneontology.org). The results showed that the most abundant background proteins identified were the naturally biotinylated carboxylases and associated enzymes, which largely coincided with the intensely staining bands. We also found background binding of abundant nuclear proteins involved in mRNA processing, such as splicing factors, as well as of ribosomal proteins. Together, these three classes of proteins accounted for >80% of background binding under the conditions used (Table 1). It is notable that very few peptides were identified as corresponding to factors involved in transcriptional regulation (Table 1). We conclude that background binding to streptavidin beads is mainly due to endogenous biotinylated proteins as well as abundant nuclear factors involved in mRNA processing and ribosome synthesis and assembly, with very little nonspecific binding of factors involved in transcriptional regulation/activation.
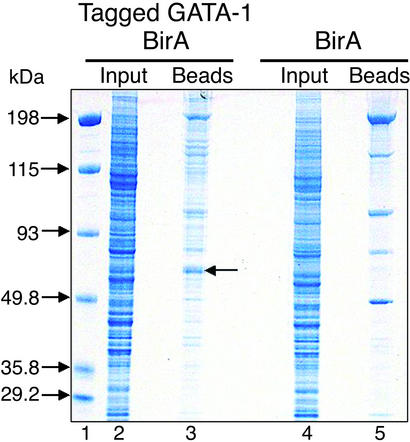
Fig. 2.
Colloidal blue-stained gel of a binding experiment of crude nuclear extracts to streptavidin beads. Lane 1, marker (M). Lane 2, input nuclear extract from tagged GATA-1/BirA double transfected cells (≈12 μg). Lane 3, proteins eluted after direct binding to streptavidin beads of ≈5 mg of crude nuclear extracts from tagged GATA-1/BirA transfected cells. Lane 4, input nuclear extract from tagged GATA-1/BirA transfected cells. Lane 5, proteins eluted after binding to streptavidin beads ≈5 mg of nuclear extract from BirA transfected cells. Arrow in lane 3 indicates protein band containing purified biotinylated GATA-1, as determined by MS.
Table 1. Summary of background binding proteins.
| Biological process | Cellular component | Total no. of peptides | Remarks |
|---|---|---|---|
| Metabolism | Mitochondrion | >180 | Carboxylases, acyltransferases, etc. |
| mRNA processing | Nucleus | 120 | Splicing factors, hnRNPs, ATP-dependent RNA helicases, etc. |
| Protein biosynthesis | Cytosol | 93 | Ribosomal proteins, etc. |
| Receptor activity? | Unknown | 17 | Single protein: thyroid hormone receptor-associated protein |
| RNA processing? | Unknown | 16 | Four proteins with RNA-binding motifs |
| Cytoskeleton | 13 | Actin, lamin | |
| Chromatin assembly | Nucleosome | 13 | Histones |
| Apoptotic program | Nucleolus | 10 | Single protein: apoptotic chromatin condensation inducer in the nucleus |
| No information | Unknown | 10 | Bcl-2-associated transcription factor |
| Ribosome biogenesis | Nucleolus | 9 | SnoRNA-binding proteins, fibrillarin |
| Protein targeting | Nucleus | 5 | Single protein: cdc5-like |
| Electron transport | Microsomes | 5 | Single protein: oxidoreductase |
| No information | Nucleolar | 4 | myb-binding protein 1a |
| Chromatin | Nucleus | 3 | Single protein: SAP 18 |
| Modification | |||
| DNA recombination | Nucleus | 2 | Two proteins: pontin and reptin (RuvB-like) |
| Transcription regulation | Nucleus | 2 | Two proteins: Y box transcription factor 1 (1 peptide); thymocyte selection associated HMG box (1 peptide) |
| Total no. of peptides | >500 | ||
| Gel slices | ∼21 | ||
| Protein molecular mass range, kDa | 15-274 |
Proteins identified by MS were classified according to biological function and cellular localization, with the criteria used by the Gene Ontology Consortium. Also shown is the number of peptides identified for each class of proteins.
We next tried binding a similar amount of crude nuclear extract (5 mg) from tagged GATA-1/BirA double transfectants, under the same conditions (Fig. 2, lane 2). The staining pattern of the lane with the eluted material (Fig. 2, lane 3) was significantly different to that observed with the background binding in lane 5, indicating a significant enrichment in proteins coeluting with tagged GATA-1 (Fig. 2, lane 3 vs. lane 5). Biotinylated GATA-1 and coeluting proteins may dilute out or compete for binding with the nonspecific proteins observed in lane 5. The protein enrichment visible in lane 3 may thus correspond to copurified GATA-1-interacting proteins. In the eluted material, we also observed a strongly stained band migrating with a size similar to that expected for tagged GATA-1 (Fig. 2, arrow, lane 3). We confirmed the presence of GATA-1 in this band by gel excision and MS. The detailed analysis of all proteins copurifying with tagged GATA-1 will be published elsewhere (P.R., F.G., and J.S., unpublished results). Taken together, these data demonstrate the quantitative biotinylation of tagged GATA-1 in MEL cells and its efficient purification from crude protein extracts in a single-step procedure by direct binding to streptavidin beads, with few background binding bands corresponding primarily to endogenously biotinylated proteins and easily identifiable abundant nuclear/nucleolar proteins.
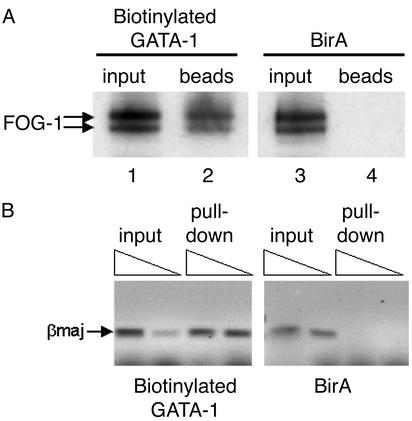
Biotinylation Does Not Affect the Protein-Interacting or DNA-Binding Properties of GATA-1. Because it is possible that addition of the peptide tag and/or biotinylation may affect the properties of the tagged protein, we tested whether biotinylated GATA-1 could still undergo protein–protein interactions with a known GATA-1 partner, such as FOG-1 (26), and whether it could bind in vivo to known GATA-1 gene targets such as the mouse βmaj globin promoter. We carried out a pull-down experiment of biotinylated GATA-1 by binding nuclear extracts to streptavidin beads and tested whether FOG-1 was also copurified. We found FOG-1 to be pulled down from extracts expressing biotinylated GATA-1 but not from extracts expressing BirA only (Fig. 3A, lanes 2 and 4). We have also found FOG-1 to be copurifying with biotinylated GATA-1 by MS. We also carried out a chromatin pull-down (ChIP) experiment in which sonicated chromatin from formaldehyde-crosslinked MEL cells was incubated with streptavidin beads, followed by elution of the bound material and recovery of the pulled-down DNA. Using primers specific for the βmaj promoter, we found enrichment for these sequences in the DNA pulled down from the chromatin of cells expressing biotinylated GATA-1 but not from cells expressing BirA (Fig. 3B). We have also found that biotinylation does not affect the subnuclear distribution normally observed with GATA-1 (Fig. 5, which is published as supporting information on the PNAS web site; ref. 27) and that biotinylated GATA-1 displays an identical biochemical fractionation profile as endogenous GATA-1 in MEL cell nuclear extracts (data not shown). Taken together, these results provide strong evidence that the properties of GATA-1 are not affected by biotinylation tagging. In addition, these data also demonstrate the application of biotinylation tagging as an alternative to antibodies in methods involving an affinity purification step, such as protein pull-downs or a ChIP assay.
Fig. 3.
(A) Binding biotinylated GATA-1 to streptavidin beads specifically pulls down FOG-1, as detected by Western blotting by using a FOG-1 antibody. By contrast, FOG-1 cannot be pulled down by streptavidin in nuclear extracts expressing biotinylated expressing BirA only (Right). FOG-1 is detected as a doublet (26). (B) Streptavidin pull-down of βmaj globin promoter sequences from crosslinked chromatin from MEL cells expressing biotinylated GATA-1 (Left) or BirA only (Right). Triangles indicate increasing amounts of pulled-down crosslinked chromatin used as template in PCR reactions in detecting amplification of the βmaj sequences. Specific enrichment for βmaj sequences is observed in pulled-down chromatin from cells expressing biotinylated GATA-1 but not from cells expressing BirA only.
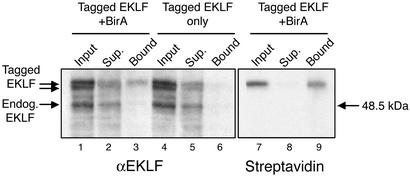
Biotinylation Tagging in Transgenic Mice. The ability to directly isolate the biotin-tagged protein from crude extracts in a single step raises the prospect of using this approach in the purification of tagged proteins from limiting sources such as mouse tissues. We therefore tested whether BirA-mediated biotinylation tagging would also work in vivo in transgenic mice. Because GATA-1 overexpression leads to embryonic lethality in mice (28), we tested this approach by tagging the essential erythropoietic transcription factor EKLF (29). Mouse EKLF cDNA was tagged with the biotinylation tag and a double hemagglutinin epitope and microinjected in mouse eggs to establish transgenic mouse lines. Similarly, transgenic mouse lines were also established by microinjecting the BirA/erythroid expression cassette construct. Transgenic mouse lines with detectable expression of tagged EKLF and BirA in erythroid cells were selected and crossbred. In vivo biotinylation of tagged EKLF was assessed in nuclear extracts prepared from the fetal livers of 13.5-day postcoitum embryos. Western blot analysis with an anti-EKLF antibody detected endogenous EKLF as well as tagged EKLF, which was visualized as a doublet (Fig. 4, lane 1). Binding of the fetal liver nuclear extracts to streptavidin beads shows that only the top band in the doublet is retained, suggesting that it is biotinylated (Fig. 4, lanes 2 and 3). This observation is confirmed by probing the same blot with streptavidin–HRP, which detects only a single band (Fig. 4, lanes 7 and 9). The doublet detected by the EKLF antibody is most likely due to the differential utilization of translation initiation codons at the N-terminally fused biotinylation tag (top band) and at the double hemagglutinin epitope present immediately downstream, which also contains an initiation codon (bottom band). As a result, only the top band bearing the biotinylation tag serves as a substrate for BirA, thus further demonstrating the in vivo specificity of this approach. The binding of nuclear extracts to streptavidin beads also showed that a significant proportion (≈50%) of tagged EKLF is biotinylated in the fetal livers of transgenic mice. We speculate that the difference in biotinylation efficiencies between transfected cells and fetal livers (apart from the use of different fusion proteins) may reflect an in vivo limitation in the availability of biotin (biotin is abundant in the FCS used to supplement cell culture media) or a difference in BirA expression levels. These results demonstrate that the specific biotinylation of tagged EKLF by bacterial BirA can also be achieved with high efficiency in vivo in transgenic mice.
Fig. 4.
Specific biotinylation of tagged EKLF in transgenic mouse embryos. Nuclear extracts from the fetal liver of 13.5-days postcoitum embryos from a tagged EKLF/BirA double transgenic line (lanes 1–3 and 7–9) and from a tagged EKLF transgenic line (lanes 4–6) were bound to streptavidin beads. Tagged and biotinylated EKLF in input nuclear extract, unbound material (sup., supernatant), and bound material was detected by an EKLF antibody (Left) or by streptavidin–HRP (Right). EKLF biotinylation and binding to the beads is detected only in extracts from double transgenic embryos.
Discussion
In this paper, we have demonstrated that expression of the bacterial BirA biotin ligase in mammalian cells and transgenic mice leads to the quantitative biotinylation of specific transcription factors bearing a small artificial peptide tag. We also showed that, at least for GATA-1, fusion of the peptide tag and specific biotinylation do not interfere with the protein's properties. We demonstrated that biotinylation can be effectively used for the single-step affinity purification of the tagged protein by binding to streptavidin beads.
Small (<23-aa) biotinylation tags have been previously obtained through multiple rounds of screening combinatorial peptide libraries for specific biotinylation by the BirA biotin ligase (16) and have been shown to be biotinylated at rates similar to those of naturally occurring substrates (17). Such peptide tags have been subsequently used for the specific biotinylation of fusion proteins in E. coli (12–15). Our work shows the utility of such a small tag for the efficient BirA-mediated biotinylation of specific fusion proteins in mammalian cells. Larger (>63-aa) tags derived from biotin acceptor domains present in naturally biotinylated proteins have been previously used in biotinylating fusion proteins in mammalian cells (10, 11). However, there are obvious advantages in using smaller artificial tags. First, small tags are much less likely to affect the structure and thus the properties of the fusion protein in vivo. It should be noted that tag size can be reduced even further to ≈14 aa without compromising biotinylation efficiencies (17). Second, the use of small tags avoids the extra complication of tag removal by proteolytic cleavage that is often necessary with larger tags (e.g., TAP tag, ref. 4). Third, artificial tags are unlikely to be recognized and biotinylated by endogenous biotin ligases. Indeed, in our assays, we observed no biotinylation (in nuclear extracts) of the fusion proteins in the absence of BirA. It is also important to note that expression of BirA in mammalian cells did not lead to an increase in nonspecific background biotinylation in nuclear extracts. We conclude that, using this approach, biotinylation of tagged proteins in mammalian cells is a highly specific tightly regulated process that occurs only in the presence of BirA.
The demonstration that a small peptide tag can be efficiently biotinylated in mammalian cells provides a very useful tool with a number of advantages for protein purification. First, we showed that the biotinylated protein can be efficiently purified directly from a crude extract in a single-step procedure, whereas most commonly used affinity tags require a number of purification steps before the affinity-binding step. Second, there is low specific background binding primarily due to the small number of endogenous naturally biotinylated proteins. Under the mild conditions we used in our purification procedure, we observed five strongly staining background protein bands binding to streptavidin that corresponded to naturally biotinylated proteins, such as carboxylases, as well binding by abundant nuclear proteins such as splicing factors. Third, the very strong binding of biotin to avidin/streptavidin offers another advantage in that it allows increasingly higher stringencies to be used during purification without fear of early elution of the tagged protein. Not only may this higher stringency further reduce nonspecific background binding (e.g., by splicing factors), but it can also serve as a measure of the strength of interactions of proteins copurifying with the biotin-tagged protein. Indeed, a systematic comparison of different tags for purifying a rat neurotensin receptor in E. coli showed biotinylation to be the best approach in terms of efficiency and purity (12). Furthermore, the inclusion of specific protease cleavage sites downstream of the biotinylation tag may be used to specifically cleave and elute the tagged protein complex from streptavidin, while leaving the endogenous biotinylated background proteins still bound. Fourth, another advantage is the availability of modified forms of avidin with lower binding affinities for biotin (e.g., monomeric avidin with a Kd of ≈5 × 10–8 M), which allow elution of bound biotinylated proteins under native conditions. This option offers the possibilities of purifying active tagged protein complexes as well as the better resolution and identification of purified proteins by 2D gel electrophoresis. Finally, it is very significant that the BirA-mediated specific biotinylation can also be efficiently achieved in transgenic animals, thus raising the prospect of using limiting animal tissues for protein purification by using high-affinity interaction with streptavidin. This is, in principle, a major advantage over other tagging approaches, such as the highly efficient TAP tag approach (4), which also does not require prepurification steps. The TAP tag includes two Ig-binding domains from the Stahylococcus aureus protein A, which may be a problem when expressed in transgenic animals that naturally express antibodies.
In conclusion, the efficient biotinylation of specific proteins in mammalian cells demonstrated here raises the prospect of applying the advantages and flexibility of the well-developed biotin/avidin technology in the single-step purification and characterization of mammalian protein complexes.
Supplementary Material
Acknowledgments
We are grateful to Dr. Stuart Orkin (Harvard Medical School) for the FOG-1 antibody, Dr. Mariette van de Corput for help with immunofluorescence, and John Kong-a-San for mouse transgenesis. This work was supported by the European Union (to J.S., P.R., and E.K.) and the Netherlands Organization for Scientific Research (NWO).
Abbreviations: MEL, mouse erythroleukemic; HRP, horseradish peroxidase; EKLF, erythroid Krüppel-like factor.
References
- 1.Lamond, A. I. & Mann, M. (1997) Trends Cell Biol. 7, 139–142. [DOI] [PubMed] [Google Scholar]
- 2.Kumar, A. & Snyder, M. (2002) Nature 415, 123–124. [DOI] [PubMed] [Google Scholar]
- 3.Jarvik, J. W. & Telmer, C. A. (1998) Annu. Rev. Genet. 32, 601–618. [DOI] [PubMed] [Google Scholar]
- 4.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M. & Seraphin, B. (1999) Nat. Biotechnol. 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- 5.Chapman-Smith, A. & Cronan, J. E., Jr. (1999) Trends Biochem. Sci. 24, 359–363. [DOI] [PubMed] [Google Scholar]
- 6.Savage, M. D., Mattson, G., Desai, S., Nielander, G. W., Morgensen, S. & Conklin, E. J. (1992) Avidin-Biotin Chemistry: A Handbook (Pierce Chemical Company, Rockford, IL).
- 7.Cronan, J. E., Jr. (1990) J. Biol. Chem. 265, 10327–10333. [PubMed] [Google Scholar]
- 8.Duffy, S., Tsao, K. L. & Waugh, D. S. (1998) Anal. Biochem. 262, 122–1228. [DOI] [PubMed] [Google Scholar]
- 9.Cronan, J. E., Jr., & Reed, K. E. (2000) Methods Enzymol. 326, 440–458. [DOI] [PubMed] [Google Scholar]
- 10.Parrott, M. B. & Barry, M. A. (2000) Mol. Ther. 1, 96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parrott, M. B. & Barry, M. A. (2001) Biochem. Biophys. Res. Commun. 281, 993–1000. [DOI] [PubMed] [Google Scholar]
- 12.Tucker, J. & Grisshammer, R. (1996) Biochem. J. 317, 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatsumi, H., Fukuda, S., Kikuchi, M. & Koyama, Y. (1996) Anal. Biochem. 243, 176–180. [DOI] [PubMed] [Google Scholar]
- 14.Tsao, K. L., DeBarbieri, B., Michel, H. & Waugh, D. S. (1996) Gene 169, 59–64. [DOI] [PubMed] [Google Scholar]
- 15.Smith, P. A., Tripp, B. C., DiBlasio-Smith, E. A., Lu, Z., LaVallie, E. R. & McCoy, J. M. (1998) Nucleic Acids Res. 26, 1414–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schatz, P. J. (1993) Biotechnology 11, 1138–1143. [DOI] [PubMed] [Google Scholar]
- 17.Beckett, D., Kovaleva, E. & Schatz, P. J. (1999) Protein Sci. 8, 921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, et al. (2002) Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- 19.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. (2002) Nature, 415, 180–183. [DOI] [PubMed] [Google Scholar]
- 20.Howard, P. K., Shaw, J. & Otsuka, A. J. (1985) Gene 35, 321–331. [DOI] [PubMed] [Google Scholar]
- 21.Needham, M., Gooding, C., Hudson, K., Antoniou, M., Grosveld, F. & Hollis, M. (1992) Nucleic Acids Res., 20, 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antoniou, M. (1991) in Methods in Molecular Biology: Gene Transfer and Expression Protocols, ed. Murray E. J. (Humana, Clifton, NJ), Vol. 7, pp. 421–434. [DOI] [PubMed] [Google Scholar]
- 23.Meiring, H. D., van der Heeft, E., ten Hove, G. J. & de Jong, A. P. J. M. (2002) J. Sep. Sci. 25, 557–568. [Google Scholar]
- 24.Hogan, B., Beddington, R., Costantini, F. & Lacey, E. (1994) Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plain-view, NY).
- 25.Cantor, A. B. & Orkin, S. H. (2002) Oncogene 21, 3368–3376. [DOI] [PubMed] [Google Scholar]
- 26.Tsang, A. P., Visvader, J. E., Turner, C. A., Fujiwara, Y., Yu, C., Weiss, M. J., Crossley, M. & Orkin, S. H. (1997) Cell 90, 109–119. [DOI] [PubMed] [Google Scholar]
- 27.Elefanty, A. G., Antoniou, M., Custodio, N., Carmo-Fonseca, M. & Grosveld, F. G. (1996) EMBO J. 15, 319–333. [PMC free article] [PubMed] [Google Scholar]
- 28.Whyatt, D., Lindeboom, F., Karis, A., Ferreira, R., Milot, E., Hendriks, R., de Bruijn, M., Langeveld, A., Gribnau, J., Grosveld, F. & Philipsen, S. (2000) Nature 406, 519–524. [DOI] [PubMed] [Google Scholar]
- 29.Bieker, J. J. (2001) J. Biol. Chem. 276, 34355–34358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.