Abstract
Protein folding barriers result from a combination of factors including unavoidable energetic frustration from nonnative interactions, natural variation and selection of the amino acid sequence for function, and/or selection pressure against aggregation. The rate-limiting step for human Pin1 WW domain folding is the formation of the loop 1 substructure. The native conformation of this six-residue loop positions side chains that are important for mediating protein–protein interactions through the binding of Pro-rich sequences. Replacement of the wild-type loop 1 primary structure by shorter sequences with a high propensity to fold into a type-I′ β-turn conformation or the statistically preferred type-I G1 bulge conformation accelerates WW domain folding by almost an order of magnitude and increases thermodynamic stability. However, loop engineering to optimize folding energetics has a significant downside: it effectively eliminates WW domain function according to ligand-binding studies. The energetic contribution of loop 1 to ligand binding appears to have evolved at the expense of fast folding and additional protein stability. Thus, the two-state barrier exhibited by the wild-type human Pin1 WW domain principally results from functional requirements, rather than from physical constraints inherent to even the most efficient loop formation process.
Keywords: β-turn, ligand binding, protein folding, β-sheet, protein function
Globular proteins evolve by mutation and selection. Selection criteria include function, and sufficient thermodynamic stability and folding rate to avoid sustained chaperone binding and proteasome degradation. The selection criteria cannot always be optimized independently over the entire sequence of a protein. For the human Pin1 (hPin1) WW domain (Pin WW hereafter), we have shown that residues important for stability and folding rate are segregated in the sequence (1–4). It is likely that functional selection criteria are predominant once minimal energetic criteria are met. Therefore, sequence evolution to enhance function may lead to a decrease in protein stability and folding rate compared with a sequence optimized for folding energetics.
The hPin1 cell cycle regulatory proline (Pro) cis/trans-isomerase is a two-domain protein (5). In its physiological role, the N-terminal WW domain binds Pro-rich ligands of the consensus sequence (pS/pT)P, whereas the C-terminal domain catalyzes the Pro cis/trans-isomerization at the pS/pT-P peptide bond. NMR solution studies show that the two domains, which are connected by a flexible solvated linker, interact only weakly before ligand binding (6, 7). The structure of the isolated Pin WW domain is virtually superimposable on that of the WW domain in the two-domain hPin1 protein (8). Moreover, Pin WW exhibits sufficient thermodynamic stability for biophysical analysis, folds rapidly, and retains its ligand-binding function (3, 9). These attributes, combined with sequence information on >150 WW domain family members (10, 11), makes Pin WW an excellent small model protein for structure–function–folding studies.
WW domains have been used extensively in experimental and theoretical folding studies (1–4, 12–19). Traditional side-chain and amide-to-ester mutagenesis of Pin WW in conjunction with laser temperature-jump (T-jump) relaxation studies revealed that the folding rate was limited by nucleation at the loop 1 substructure (1–3, 16), similar to the situation observed in SH3 domains (20, 21).
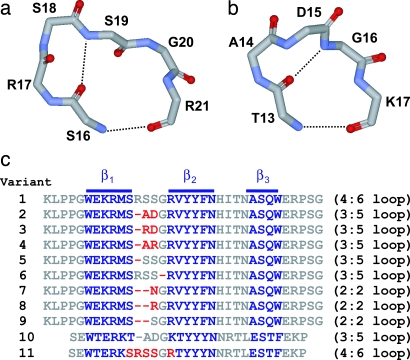
The six-residue loop 1 of Pin WW is unusually long. Sequence alignments of >150 WW domains reveal that a five-residue loop is statistically favored in this protein family, and structural studies on these loops indicate that a type-I G1 bulge conformation seems to be the preferred local structural motif (5, 22–26). An x-ray structure of the full-length hPin1 cis/trans-isomerase revealed that loop 1 in the WW domain adopts an unusual conformation: a four-residue type-II turn intercalated within a larger, six-residue loop (5, 9) (Fig. 1a). NMR solution studies on the isolated Pin WW indicate that loop 1 is intrinsically flexible (8), whereas the topologically equivalent five-residue type-I G1 bulge turn in the FBP28 WW domain (FBP WW) (Fig. 1b) is structurally more ordered (22).
Fig. 1.
Loop structures and sequences of WW domains. (a) Backbone diagram of the loop 1 substructure in WT Pin WW (residues S16–R21) [Protein Data Bank (PDB) ID code 1PIN]. (b) Backbone diagram of the loop 1 substructure in WT FBP WW (residues T13–K17) (PDB ID code 1E01). Backbone H-bonds are indicated by black dotted lines. (c) Aligned sequences of the WT Pin WW domain (variant 1) and loop 1 redesigned variants 2–9 and the redesigned and sequence-minimized FBP WW variants (10 and 11). β-strand residues are colored blue, residues that were mutated or deleted upon loop 1 redesign are in red, and all other residues are in gray.
Mutagenesis studies reveal that the side chain of Arg-17 in loop 1 of Pin WW contributes significantly to ligand-binding energy through recognition of a phosphorylated Ser residue (9). Replacing Arg-17 by Ala results in a 6.3-fold decrease in binding affinity toward the phosphorylated peptide YSPTpSPS, derived from the C-terminal domain of RNA polymerase II, a natural ligand of Pin WW (27). It is therefore possible that the sequence of the loop 1 substructure of Pin WW, with its unusual conformation and intrinsic dynamics, is not optimized for folding kinetics, but instead has been evolutionary optimized for functional duties.
To support this hypothesis, we must demonstrate that significantly faster folding and more stable variants of Pin WW can be made by shortening loop 1 and that such shortened loops result in lower ligand-binding affinity whether Arg-17 is present or not. Structural, thermodynamic, kinetic, and ligand-binding studies on loop 1 Pin WW variants affirm that functional attributes contribute substantially to the selection criteria for this region of Pin WW.
Results
Redesign of the Pin WW Loop 1 Substructure.
In the following, we use the shorthand (X:Y) for β-hairpin classification introduced by Thornton and coworkers (28). The first integer (X) represents the number of residues required for turn formation, and the second integer (Y) identifies the hydrogen (H)-bonding pattern in the closure of the turn. A (2:2) β-hairpin has two residues [(i + 1) and (i + 2) position] in the turn, and loop closure is accomplished by formation of two cross-strand H-bonds. A (3:5) β-hairpin has three turn residues [(i + 1) − (i + 3) position)], but only one cross-strand H-bond is formed upon loop closure.
Loop 1 of WW adopts a rare (4:6) loop conformation, with a type-II four-residue turn (sequence: Ser-16–Arg-17–Ser-18–Ser-19) incorporated into a larger, six-residue (sequence: Ser-16–Arg-17–Ser-18–Ser-19–Gly-20–Arg-21) loop (5, 9) (Fig. 1a). From an evolutionary perspective, the Pin WW loop 1 sequence could have originated from a one-residue insertion into a (3:5) type-I G1 bulge turn, with the additional residue being added N-terminal to the Gly at the bulge position (i + 3) (28, 29). Sequence alignment and structural studies indicate that the type-I G1 bulge turn is the predominant loop 1 conformation in WW domain family members at topologically equivalent positions. The longer loop 1 in Pin WW may be necessary to correctly position the functionally important Arg-17 side chain for optimal ligand binding. To investigate the structure–function–folding relationship in Pin WW, we conducted a loop 1 redesign study, starting with wild-type (WT) Pin WW, referred to as variant 1 in Fig. 1c.
The loop 1 sequence in variant 2 is identical to the sequence in FBP WW, which adopts a five-residue type-I G1 bulge turn conformation (Fig. 1b) (22). Ala, Asp, and Gly occupy the (i + 1) to (i + 3) positions, respectively. Asp exhibits the highest statistical occurrence at the (i + 2) position (30–32), whereas Gly is suited best to adopt the positive ϕ/ψ-dihedral angles at the bulge position (i + 3) in the type-I G1 bulge turn geometry (28, 32–34). The sequences corresponding to variants 3 and 4 were designed to test whether repositioning the functional Arg residue at the (i + 1) or (i + 2) position in a shorter sequence expected to adopt a type-I G1 bulge loop would yield WW domains with ligand-binding affinities comparable with variant 1. Variant 5 was obtained by deletion of the functionally important residue Arg-17 in variant 1. Variant 6 is a negative control in which the (i + 3) bulge position is occupied by a less favorable Ser residue. We predict that variant 6 will be significantly less stable than variants 2–5.
The sequences corresponding to variants 7–9 were envisioned to fold into a (2:2) type-I′ β-hairpin turn. In variant 7, Asn and Gly occupy the (i + 1) and (i + 2) positions, which are particularly well suited to adopt a type-I′ β-turn conformation (35–37). In variant 8 and 9, the (i + 1) positions are occupied by less favorable Arg and Ser residues, respectively (36, 37).
In a reverse control experiment, we replaced the type-I G1 bulge loop in a sequence-minimized version of FBP WW (variant 10) (2) with the longer loop 1 sequence of Pin WW (variant 1), yielding variant 11.
Structural Verification of Pin WW Loop 1 Redesign.
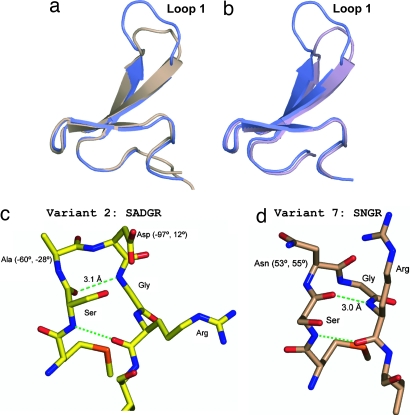
Crystallization of the isolated WW domains was unsuccessful. However, x-ray crystal structures could be obtained for variant 2 (predicted type-I G1 bulge as loop 1) at 1.9-Å resolution and variant 7 (predicted type-I′ β-turn as loop 1) at 1.5-Å resolution in the context of the full-length hPin1 rotamase. An additional Arg-14 → Ala mutation was required for crystallization of variant 2. Details on data collection/refinement statistics and electron density maps of the loop 1 substructure in variants 2 and 7 are provided as, respectively, Table 3 and Fig. 6, which are published as supporting information on the PNAS web site.
Fig. 2 depicts backbone superpositions of variants 1 and 2 (Fig. 2a) and of variants 1 and 7 (Fig. 2b). For variants 2 and 7, loop remodeling causes only local structural rearrangements within the loop 1 substructure of Pin WW. The triple-stranded β-sheet structure, the loop 2 substructure, and the conformation of N- and C-terminal extensions involved in the formation of the hydrophobic stability core are essentially superimposable with those of variant 1 (Fig. 2 a and b). A closer inspection of the loop 1 substructure in variants 2 and 7 confirms the success of our loop 1 redesign. For variant 2 (loop 1 sequence: SADGR), the dihedral angles of Ala (−60°, −28°), Asp (−97°, 12°), and Gly (94.3°, 19°) and two backbone H-bonds between Asn (donor) and Ser (acceptor) (3.1 Å) and Ser (donor) and Arg (acceptor) (2.9 Å) are consistent with a (3:5) type-I G1 bulge turn conformation (29, 33) (Figs. 1b and 2c). For variant 7 (loop 1 sequence: SNGR), the dihedral angles of Asn (53°, 55°) and Gly (70.1°, −2.2°), and two backbone H-bonds between Arg (donor) and Ser (acceptor) (3.0 Å) and Ser (donor) and Arg (acceptor) (2.8 Å) are those expected for a (2:2) type-I′ turn (29, 33) (Fig. 2d).
Fig. 2.
Loop 1 redesign in WT Pin WW. (a and b) Backbone superposition of WT Pin WW (variant 1, 1.3-Å resolution; colored blue) with loop 1 redesigned variant 2 (1.9-Å resolution; brown) (a) and variant 7 (1.5-Å resolution; purple) (b). (c and d) Backbone conformation of loop 1 in variant 2 (c) and loop 1 in variant 7 (d). Backbone H-bonds are indicated by green dashed lines. Turn residues are labeled. See main text for further details.
Structure–Stability Relationship.
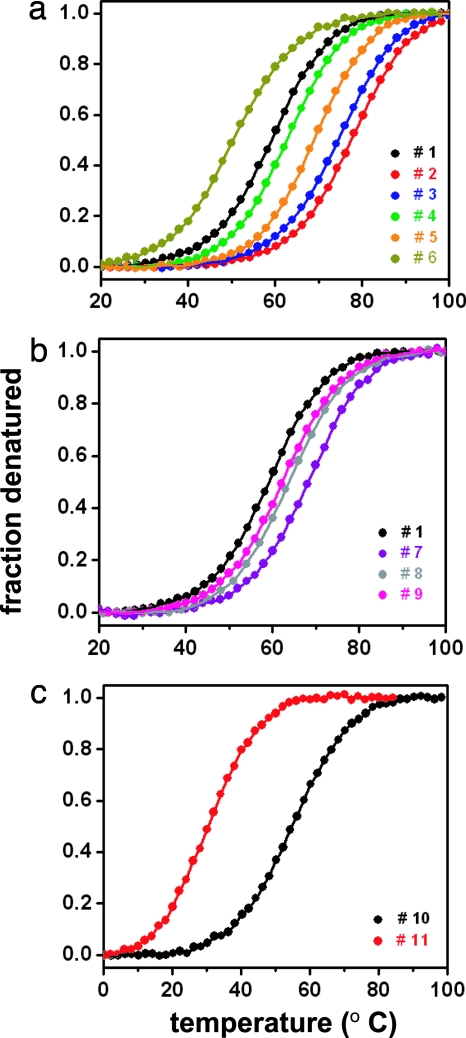
All loop 1 redesigned Pin WW variants are monomeric up to at least 100 μM, as confirmed by analytical gel filtration (data not shown) and unfold cooperatively with essentially identical unfolding cooperativities (Fig. 3a and b). The unfolding transitions were analyzed assuming the validity of a two-state unfolding model (3) (Table 1).
Fig. 3.
Effect of loop 1 redesign on WW domain stability. (a) Normalized equilibrium unfolding transitions for Pin WW (variant 1) and variants 2–6 with either a confirmed (2) or predicted (3–6) (3:5) type-I bulge turn. (b) Normalized equilibrium unfolding transitions for variants 1 and 7–9 with either a confirmed (7) or predicted (8, 9) (2:2) type-I′ β-hairpin turn. (c) Normalized equilibrium unfolding transitions for FBP (WW variant 10) with a confirmed (3:5) type-I G1 bulge turn and variant 11 with a predicted (4:6) loop.
Table 1.
Results from a nonlinear least-squares fitting of the thermal denaturation curves monitored by far-UV CD
| Var. | TM, °C | ΔG1,* kJ·mol−1·K−1 | ΔG2,* kJ·mol−1·K−2 | ΔΔG,† kJ·mol−1 | ΔΔG,‡ kJ·mol−1 |
|---|---|---|---|---|---|
| 1 | 59.0 | 0.3849 (46) | 0.00291 (21) | — | — |
| 2 | 77.5 | 0.4285 (36) | 0.00327 (22) | +7.26 | — |
| 3 | 74.2 | 0.3889 (55) | 0.00451 (26) | +5.61 | — |
| 4 | 62.1 | 0.4076 (58) | 0.00294 (61) | +1.21 | — |
| 5 | 69.2 | 0.4255 (43) | 0.00191 (65) | +4.17 | — |
| 6 | 50.8 | 0.3725 (61) | 0.00006 (77) | −2.89 | — |
| 7 | 68.1 | 0.4215 (39) | 0.00220 (32) | +3.70 | — |
| 8 | 62.6 | 0.3913 (64) | 0.00235 (85) | +1.46 | — |
| 9 | 62.0 | 0.3930 (58) | 0.00228 (75) | +1.21 | — |
| 10 | 54.1 | 0.2904 (150) | 0.00147 (12) | — | — |
| 11 | 30.9 | 0.3438 (33) | −0.00003 (27) | — | −7.11 |
See Fig. 3 for more results. Var., variant no.
*Parameters ΔG1 and ΔG2 are derived from a fit of the equilibrium unfolding curves using a second-order Taylor series expansion of the free energy ΔG(T) about the melting temperature (TM): ΔG(T) = ΔG1(T − TM) + ΔG2(T − TM)2 with ΔG0 = 0. Values in parentheses represent the 1σ confidence intervals.
†ΔΔG = ΔG(Variant 1) − ΔG(Variant i; i = 2–9) at 65°C (average melting temperature for variants 2–9).
‡ΔΔG = ΔG(Variant 10) − ΔG(Variant i; i = 11) at 42°C (average melting temperature for variant 11).
Variants 2 and 3 with a demonstrated and a likely (3:5) type-I G1 bulge turn and with sequence-optimized (i + 2) and (i + 3) turn positions are significantly more stable than WT Pin WW (variant 1) (ΔΔG = 7.26 and 5.61 kJ·mol−1, respectively; see Fig. 3a). The stability of variants 4 and 5 also exceed that of variant 1 (although variant 4 only slightly). The decrease in stability of variants 4 and 5 relative to variants 2 and 3 most likely originates from the replacement of the energetically favorable Asp at the (i + 2) position with a less favorable Arg and Ser residue, respectively (31, 32). As predicted, variant 6, in which a Ser residue replaces the energetically favorable Gly at the (i + 3) position (l-α region of the Ramachandran plot), is the least stable of the single-residue deletion variants and even less stable than variant 1. This result implies that the high stability of variants 2 and 3 cannot be entirely due to a more favorable loop closure entropy but mainly reflects the high propensity of the local sequence to fold into an energetically stable conformation (type-I G1 bulge turn). Generally, shorter loops result in more stable hairpins because of the reduced entropy penalty; however, the conformational preferences of the sequence can contribute substantially as reflected by these results.
Remodeling of Pin WW (variant 1) loop 1 with sequences preferring a (2:2) type-I′ β-turn conformation (variants 7–9) also resulted in a more stable WW domain fold (Fig. 3b). However, even though (2:2) type-I′ turns are statistically more prevalent in β-hairpins, the extent of stabilization was on average lower than that achieved by the five-residue type-I G1 bulge variants, possibly because the latter turns better match the preferred right-handed twist of the WW domain β-sheet. Variant 7 exhibits superior stability in comparison with variants 8 and 9, which might be ascribed to the energetically favorable Asn residue at the (i + 1) position in variant 7 (35, 37).
Substitution of the type-I G1 bulge turn in FBP WW (variant 10) by the longer loop 1 of WT Pin WW (variant 11) leads to a significant destabilization (7.11 kJ·mol−1 at 42°C) (Fig. 3c and Table 1), which is almost identical to the gain in stability obtained at 65°C (7.26 kJ·mol−1) upon engineering of variant 1 into variant 2.
We conclude that non-WT loop 1 substructures increase thermodynamic stability in a nearly independent manner relative to other substructures that are known to be important for thermodynamic stability (1–4, 16). A type-I G1 bulge turn seems to be best suited to connect the twisted β-strands 1 and 2 in the WW domain fold, providing a rationalization for the high statistical preference of this turn type for the loop 1 substructure of the WW domain family.
Structure–Folding Relationship.
The effect of loop 1 remodeling on WW domain folding kinetics was investigated by nanosecond-resolution laser T-jump relaxation. Variants 1, 3, 4, 6, 8, 10, and 11 exhibit single-exponential folding kinetics at all temperatures investigated. Variants 2, 5, 7, and 9 exhibited an additional slower, concentration-dependent relaxation phase, possibly resulting from transient, reversible oligomer formation. For the latter variants, the T-jump-induced relaxation traces were fit with a biexponential decay function, and only the rate of the fast, concentration-independent phase was considered for further analysis (2).
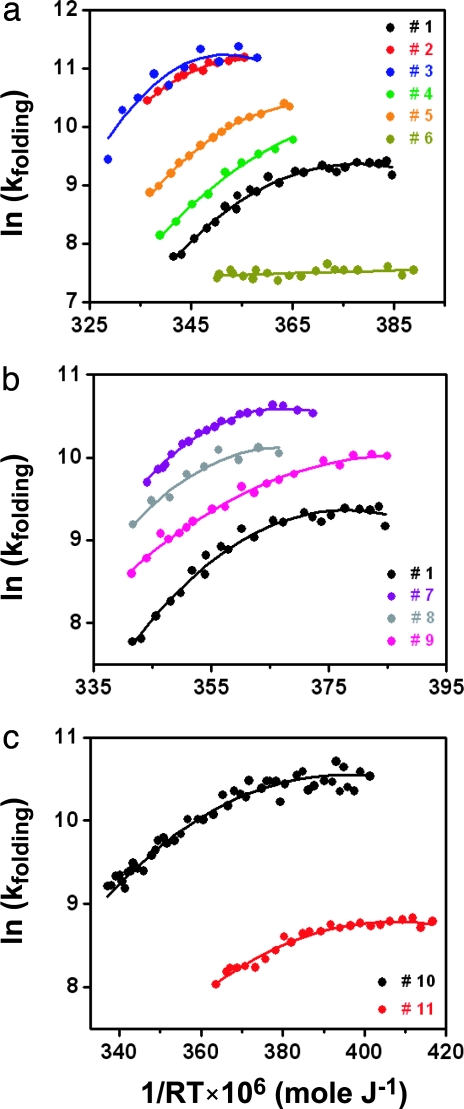
Arrhenius plots of the refolding rates for the redesigned type-I G1 bulge and type-I′ β-turn Pin WW variants are shown in Fig. 4a and b, respectively. A summary of the kinetic parameters is provided in Table 2. Except for the highly destabilized variant 6, all variants fold faster than the WT Pin WW domain and exhibit a pronounced anti-Arrhenius rate turnover at higher temperature. The unfolding rates, which can be computed from Tables 1 and 2, follow a standard Arrhenius-type behavior (see Fig. 7, which is published as supporting information on the PNAS web site). For Pin WW (variants 1–9), both the folding rates under isoenergetic conditions (folding rate at the denaturation midpoint TM) and the rates of maximal folding (Tmax, with Tmax ≈ 7–15°C below TM) correlate reasonably well (correlation coefficients 0.95 and 0.94, respectively) with domain stability (TM) (see Fig. 8, which is published as supporting information on the PNAS web site). Such a strong thermodynamics–kinetics correlation is expected either for a very late transition state or in the absence of a significant folding barrier (16). Loop 1 redesign in FBP WW exhibits the expected effect. Replacing the natural (3:5) type-I G1 bulge loop in variant 10 with the longer and highly destabilizing (4:6) Pin WW loop (variant 11) significantly slows down folding, with rates comparable with Pin WW variant 1 (Table 2). That variant 6 folds slower than all of the other sequences examined implies that the expanded φ/ψ conformational space available to the Gly residue plays an important role in nucleating the WW domain fold.
Fig. 4.
Effect of loop 1 redesign on WW domain folding kinetics. (a) Arrhenius plots of the refolding rate constants (sec−1) for WT Pin WW (variant 1), Pin WW variants 2–5 with either a confirmed (2) or predicted (3–5) (3:5) type-I G1 bulge turn conformation, and Pin WW variant 6 in which the energetically important bulge position is occupied by a less favorable Ser residue. (b) Arrhenius plots of the refolding rate constants for variant 1 and Pin WW variants 7–9 with either a confirmed (7) or predicted (8, 9) (2:2) type-I′ turn conformation. (c) Arrhenius plots of the refolding rate constants for FBP WW variants 10 [confirmed (3:5) type-I G1 bulge turn] and 11 [predicted (4:6) loop]. Color code is the same as in Fig. 3. Solid lines represent fits of the experimental data to a Kramers model by using a temperature-dependent free energy of activation.
Table 2.
Results from a nonlinear least-squares fitting of the kinetic T-jump relaxation data based on fluorescence lifetime changes
| Var. | TM, °C | Tmax,* °C | τ(TM),‡ μs | τ(Tmax),§ μs | ΔG0†,¶ kJ·mol−1 | ΔG1†,¶ kJ·mol−1·K−1 | ΔG2†,¶ kJ·mol−1·K−2 |
|---|---|---|---|---|---|---|---|
| 1 | 59.0 | 45 (1) | 79.2 | 59.4 | 15.00 (7) | 0.2000 (34) | 0.00407 (30) |
| 2 | 77.5 | 64 (1) | 13.3 | 9.8 | 10.65 (3) | 0.2052 (37) | 0.00532 (63) |
| 3 | 74.2 | 67 (1) | 9.8 | 9.1 | 9.65 (20) | 0.147 (26) | 0.0075 (21) |
| 4 | 62.1 | 56‖ | 49.4 | n.d. | 13.82 (12) | 0.233 (23) | 0.0033 (12) |
| 5 | 69.2 | 58 (1) | 33.5 | 20.7 | 13.01 (2) | 0.2305 (29) | 0.00457 (32) |
| 6 | 50.8 | 36‖ | 379.1 | n.d. | 18.83 (7) | 0.1105 (54) | 0.00004 (45) |
| 7 | 68.1 | 55 (1) | 24.4 | 17.3 | 12.07 (2) | 0.2126 (52) | 0.00498 (50) |
| 8 | 62.6 | 58 (1) | 31.0 | 25.5 | 12.54 (14) | 0.152 (28) | 0.0045 (20) |
| 9 | 62.0 | 41‖ | 51.0 | n.d. | 13.92 (5) | 0.1931 (32) | 0.00216 (22) |
| 10 | 54.1 | 31 (1) | 24.6 | 18.1 | 11.74 (6) | 0.1445 (22) | 0.00129 (15) |
| 11 | 30.9 | 23 (1) | 116.0 | 102.0 | 14.58 (4) | 0.1297 (29) | 0.00146 (19) |
See Fig. 4 for more results. Var., variant no.
*Temperature of maximal folding rate. Values in parentheses represent the 1σ confidence intervals.
‡Relaxation time at TM.
§Relaxation time at Tmax. n.d., not determined.
¶The free energy parameters are defined by using a second-order Taylor series approximation about the melting temperature, TM: ΔG† (T) = Δ G0† (TM) + Δ G1†(T − TM) + Δ G2† (T − TM)2. Values in parentheses represent the 1σ confidence intervals.
‖The temperature value is an upper bound, where the actual Tmax is at a lower temperature value.
Folding–Function Relationship.
Our kinetic studies demonstrate that loop 1 substructure formation is rate-limiting in WW domain folding, consistent with previous protein-engineering experiments (1, 3). A more stable loop 1 substructure in Pin WW hastens folding by up to an order of magnitude and stabilizes the domain significantly. To interrogate the influence of loop 1 remodeling on Pin WW function, the binding affinities of the Pin WW variants 1–4, 7, and 8 toward a phosphorylated peptide (sequence: YSPTpSPS), derived from the C-terminal domain of RNA-polymerase II, a natural Pin WW ligand, were determined by isothermal titration calorimetry. Fig. 5a depicts the heat evolved upon titration of a 50 μM solution of variant 1 with a 1 mM stock solution of phosphorylated ligand. Similar to previous binding studies with hYap65 WW (12), the binding isotherm of Pin WW variant 1 lacks a well defined plateau region before reaching the transition area of the titration. An immediate decrease in the amount of heat released in each injection step is evidence that the dissociation constant (Kd) of variant 1 is comparable with (or higher than) the WW domain concentration used (50 μM).
Fig. 5.
Effect of loop 1 redesign in WT Pin WW (variant 1) on peptide-binding activity studied by isothermal titration calorimetry. (a) Raw heat responses upon sequential titration of a solution of variant 1 (50 μM) with 4-μl aliquots of a stock solution of the phosphorylated peptide YSPTpSPS (1 mM; 60 dilution steps in total). (b) Integrated peak areas for variants 1 (black circles) and 3 (blue circles), normalized to moles of phosphorylated peptide ligand added and corrected for dilution effects. The solid black line represents a fit of the data for variant 1 to a single-binding-site model yielding a dissociation constant Kd = 35 ± 25 μM. Analogous to the situation for variant 3, no peptide binding could be detected for WW variants 2, 4, 7, and 8 (see Fig. 9).
Fig. 5b depicts the integrated peak area for variants 1 and 3 after each ligand titration step, normalized to moles of ligand added and corrected for dilution effects. A nonlinear least-squares fit yielded a Kd of 35 ± 25 μM for variant 1, in good agreement with the Kd measured previously by fluorescence polarization (Kd = 34 ± 5.9 μM) (9). Notably, variant 3, which retains the functionally important Arg residue, does not exhibit any measurable heat transfer upon titration with ligand, demonstrating that variant 3, like variants 2, 4, 7, and 8 (see Fig. 9, which is published as supporting information on the PNAS web site), do not bind ligand at the WW domain concentration used (50 μM). No binding data were measured for variants 5, 6, and 9. Variant 6 is less stable and folds significantly slower than variant 1, and therefore any ligand-binding affinity exhibited by variant 6 would not contradict our hypothesis that the loop 1 substructure is optimized for functional duties at the expense of folding energetics. Variants 5 and 9 are closely related to variants 2 and 3 and 7 and 8, respectively, and it is unlikely that variants 5 and 9, which lack the functionally important Arg in loop 1, would exhibit superior binding affinity toward the phosphorylated ligand.
Variant 1, with its thermodynamically and kinetically suboptimal parameters, seems to be the best evolutionary solution for function (YSPTpSPS binding). Deletion of Arg-17 (as in variants 2 and 7), or only its minor repositioning in the shortened but energetically optimized loop 1 context (as in variants 3, 4, and 8), eliminates physiologically meaningful binding. We conclude that the loop 1 sequence and conformation in WT Pin WW (variant 1) is fine-tuned by evolution for optimal recognition and binding affinity to the phosphorylated Ser or Thr residue N-terminal to Pro, at the expense of imparting additional stability and an increased folding rate to the three-stranded β-sheet structure.
Discussion
Evolution optimizes globular proteins for function, as well as for sufficient stability and a high enough folding rate to avoid premature degradation. The solution might be expected to be a compromise. There is no a priori reason why folding and function should both be optimal; they only have to be necessary and sufficient for biological function, no more, no less. Mutations that simultaneously reduce the ability of a protein to function and fold are trivial to find. Our key findings are twofold: (i) mutants exist that exhibit an inverse correlation between increased thermodynamic stability and hastened folding kinetics on the one hand, and (ii) enhanced binding affinity for a natural ligand on the other hand. None of the more stable Pin WW variants with energetically optimized loop 1 sequences (with or without the functionally important Arg residue) can match the ligand-binding affinity of WT Pin WW. Its atypical six-residue loop appears to be the optimal solution for Pin WW function but represents a less than optimal solution with respect to folding rate and fold stability, although no specific in vitro experiment (including binding measurements) can comprehensively evaluate function. Given that Pin WW has the rather unique property of recognizing and binding Ser/Thr phosphorylated side chains through both an Arg side chain and the backbone amide of the loop in question, it is not unexpected that modifications to the loop would result in a substantial reduction in binding free energy.
When the FBP28 WW sequence is used as a host, and its natural loop 1 sequence is replaced by the flexible and longer Pin WW loop 1 sequence, both folding rate and stability decrease. At present, we can only speculate about whether the stable and fast folding loop 1 substructure in FBP28 WW variant 10 coevolved with its in vivo function (26). A recent NMR solution structure of FBP11 WW1 (the first WW domain in human Formin binding protein 11 and a close homologue of FBP WW) in complex with the peptide APPTPPPLPP, suggests that this hypothesis is possible as the loop makes contacts with the ligand (38). Loop 1 (sequence: SPDGR) in FBP11 WW1 adopts a type-I G1 bulge conformation, and its Ser and Pro (Ala in FBP WW) residues provide a second ligand recognition site by interacting with the two methyl groups of L8 in the ligand.
For Pin WW, the nonoptimal folding energetics are related to a specific local structural change at the functional site (loop 1), not merely to overall topological constraints or the lack of need for energetic optimization. The Pin WW loop 1 sequence in either the Pin WW or FBP WW host sequence context slows down folding. This observation is consistent with previous side-chain and amide-to-ester backbone perturbation φ-analyses demonstrating that loop 1 is rate-limiting (φ∼1), whereas loop 2 and the hydrophobic stability core become rate-limiting only after severe mutational destabilization (1, 3, 39). These data are also consistent with molecular dynamics simulations on FBP WW, which revealed that loop 1 nucleates first (14, 15). It will be interesting to investigate whether the optimized loop 1 substructures in Pin WW variants 2 or 7 are still controlling the folding rate or whether another substructure of the WW domain becomes the bottleneck for folding, as elegantly shown by Baker and coworkers (20, 40) for the Protein L–Protein G pair. Our data support the conclusion that for WT Pin WW, evolved functionality is a major contributor to its folding barrier, at least as important as fundamental physical contributions.
Materials and Methods
Pin WW variants were prepared by solid-state peptide synthesis (> 95% purity) as described elsewhere (41). FBP WW variants were expressed recombinantly and purified as described in detail elsewhere (2, 3).
Equilibrium unfolding of Pin and FBP WW variants was monitored by far-UV CD at 229 nm (3). Unfolding transitions were analyzed by using a two-state model, and free energies were extracted as described (3). Normalized unfolding transitions obtained at 10 and 100 μM were superimposable, arguing against irreversible aggregation in the transition region (data not shown).
Relaxation kinetics were acquired with a laser T-jump apparatus (T-jumps between 6 and 15°C) at protein concentrations ranging from 10 to 100 μM. Microscopic rate constants for folding and unfolding were derived from Trp fluorescence decays as described elsewhere (3, 4).
Ligand-binding affinities were measured by isothermal titration calorimetry (Microcal, Amherst, MA) at 25°C (12). A solution of WW domain (50 μM in 20 mM sodium phosphate buffer, pH 7.0) was titrated with a stock solution of a natural Pro-rich ligand (sequence: YpSPTpSPS; 1 mM in 20 mM sodium phosphate buffer, pH 7.0). In total, 50–60 consecutive injections (4 μl per injection) were performed. Five independent titration experiments were conducted to determine the ligand-binding constant of WT Pin WW, and duplicate runs were carried out with the redesigned loop variants. Equilibrium dissociation constants for the Pin WW–ligand complex were determined by fitting the calorimetric data to a single-binding-site model (12).
Crystallization and structure determination of Pin WW loop variants were performed in the context of the two-domain hPin1 cis/trans-isomerase. Pin WW variants were crystallized in 100 mM Hepes buffer (pH 7.5), 1.9–2.2 M ammonium sulfate, and 1% polyethylene glycol (PEG) 400. Diffraction data were collected on a 3 × 3 charge-coupled device detector (Area Detector Systems Corporation, Poway, CA) at beamline 9-1 of the Stanford Synchrotron Radiation Laboratory or 8.2.2. at the Advanced Light Source at −178°C after flash-cooling a crystal cryoprotected with 40% PEG 400. Data were processed and scaled with hkl2000 (42). Structures of the loop 1 variants were solved by molecular replacement (MR) using the structure of WT hPin1 in complex with the peptide YSPTpSPS (see above) (Protein Data Bank ID code 1f8a) as an initial searching model (9). Refinement was carried out by using the program cns (43). Model building was performed in o (44) by using a combination of shake–omit 2Fo − Fc and Fo − Fc density maps. A test set of 5% of reflections was omitted from each data set for the calculation of Rfree (45).
Supplementary Material
Acknowledgments
M.G. and H.N. were supported by National Science Foundation Grant MCB 0316925. J.W.K., M.D., and M.J. were supported by National Institutes of Health Grant GM 051105, The Skaggs Institute of Chemical Biology, and the Lita Annenberg Hazen Foundation. M.J. was supported by postdoctoral fellowships from the Deutsche Forschungsgemeinschaft (DFG) and the La Jolla Interfaces in Science program. Y.Z. was supported by National Institutes of Health Grant CA 054418. J.P.N. is a Howard Hughes Medical Institute Investigator.
Abbreviations
- hPin1
human Pin1
- Pin WW
hPin1 WW domain
- FBP WW
FBP28 WW domain
- T-jump
temperature-jump.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1zcn and 2f21).
References
- 1.Deechongkit S., Nguyen H., Powers E. T., Dawson P. E., Gruebele M., Kelly J. W. Nature. 2004;430:101–105. doi: 10.1038/nature02611. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen H., Jager M., Moretto A., Gruebele M., Kelly J. W. Proc. Natl. Acad. Sci. USA. 2003;100:3948–3953. doi: 10.1073/pnas.0538054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jager M., Nguyen H., Crane J. C., Kelly J. W., Gruebele M. J. Mol. Biol. 2001;311:373–393. doi: 10.1006/jmbi.2001.4873. [DOI] [PubMed] [Google Scholar]
- 4.Crane J. C., Koepf E. K., Kelly J. W., Gruebele M. J. Mol. Biol. 2000;298:283–292. doi: 10.1006/jmbi.2000.3665. [DOI] [PubMed] [Google Scholar]
- 5.Ranganathan R., Lu K. P., Hunter T., Noel J. P. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 6.Bayer E., Goettsch S., Mueller J. W., Griewel B., Guiberman E., Mayr L. M., Bayer P. J. Biol. Chem. 2003;278:26183–26193. doi: 10.1074/jbc.M300721200. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs D. M., Saxena K., Vogtherr M., Bernado P., Pons M., Fiebig K. M. J. Biol. Chem. 2003;278:26174–26182. doi: 10.1074/jbc.M300796200. [DOI] [PubMed] [Google Scholar]
- 8.Kowalski J. A., Liu K., Kelly J. W. Biopolymers. 2002;63:111–121. doi: 10.1002/bip.10020. [DOI] [PubMed] [Google Scholar]
- 9.Verdecia M. A., Bowman M. E., Lu K. P., Hunter T., Noel J. P. Nat. Struct. Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 10.Sudol M. Prog. Biophys. Mol. Biol. 1996;65:113–132. doi: 10.1016/s0079-6107(96)00008-9. [DOI] [PubMed] [Google Scholar]
- 11.Sudol M., Hunter T. Cell. 2000;103:1001–1004. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 12.Koepf E. K., Petrassi H. M., Ratnaswamy G., Huff M. E., Sudol M., Kelly J. W. Biochemistry. 1999;38:14338–14351. doi: 10.1021/bi991105l. [DOI] [PubMed] [Google Scholar]
- 13.Koepf E. K., Petrassi H. M., Sudol M., Kelly J. W. Protein Sci. 1999;8:841–853. doi: 10.1110/ps.8.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson N., Johnson C. M., Macias M., Oschkinat H., Fersht A. Proc. Natl. Acad. Sci. USA. 2001;98:13002–13007. doi: 10.1073/pnas.221467198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferguson N., Pires J. R., Toepert F., Johnson C. M., Pan Y. P., Volkmer-Engert R., Schneider-Mergener J., Daggett V., Oschkinat H., Fersht A. Proc. Natl. Acad. Sci. USA. 2001;98:13008–13013. doi: 10.1073/pnas.221467398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen H., Jaeger M., Kelly J. W., Gruebele M. J. Phys. Chem. B. 2005;109:15182–15186. doi: 10.1021/jp052373y. [DOI] [PubMed] [Google Scholar]
- 17.Karanicolas J., Brooks C. L., III Proc. Natl. Acad. Sci. USA. 2004;101:3432–3437. doi: 10.1073/pnas.0304825101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russ W. P., Lowery D. M., Mishra P., Yaffe M. B., Ranganathan R. Nature. 2005;437:579–583. doi: 10.1038/nature03990. [DOI] [PubMed] [Google Scholar]
- 19.Socolich M., Lockless S. W., Russ W. P., Lee H., Gardner K. H., Ranganathan R. Nature. 2005;437:512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- 20.Grantcharova V. P., Riddle D. S., Santiago J. V., Baker D. Nat. Struct. Biol. 1998;5:714–720. doi: 10.1038/1412. [DOI] [PubMed] [Google Scholar]
- 21.Martinez J. C., Pisabarro M. T., Serrano L. Nat. Struct. Biol. 1998;5:721–729. doi: 10.1038/1418. [DOI] [PubMed] [Google Scholar]
- 22.Macias M. J., Gervais V., Civera C., Oschkinat H. Nat. Struct. Biol. 2000;7:375–379. doi: 10.1038/75144. [DOI] [PubMed] [Google Scholar]
- 23.Huang X., Poy F., Zhang R., Joachimiak A., Sudol M., Eck M. J. Nat. Struct. Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 24.Kanelis V., Rotin D., Forman-Kay J. D. Nat. Struct. Biol. 2001;8:407–412. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- 25.Wiesner S., Stier G., Sattler M., Macias M. J. J. Mol. Biol. 2002;324:807–822. doi: 10.1016/s0022-2836(02)01145-2. [DOI] [PubMed] [Google Scholar]
- 26.Otte L., Wiedemann U., Schlegel B., Pires J. R., Beyermann M., Schmieder P., Krause G., Volkmer-Engert R., Schneider-Mergener J., Oschkinat H. Protein Sci. 2003;12:491–500. doi: 10.1110/ps.0233203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meinhart A., Kamenski T., Hoeppner S., Baumli S., Cramer P. Genes Dev. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- 28.Sibanda B. L., Blundell T. L., Thornton J. M. J. Mol. Biol. 1989;206:759–777. doi: 10.1016/0022-2836(89)90583-4. [DOI] [PubMed] [Google Scholar]
- 29.Sibanda B. L., Thornton J. M. J. Mol. Biol. 1993;229:428–447. doi: 10.1006/jmbi.1993.1044. [DOI] [PubMed] [Google Scholar]
- 30.Jourdan M., Griffiths-Jones S. R., Searle M. S. Eur. J. Biochem. 2000;267:3539–3548. doi: 10.1046/j.1432-1327.2000.01381.x. [DOI] [PubMed] [Google Scholar]
- 31.Zerella R., Chen P. Y., Evans P. A., Raine A., Williams D. H. Protein Sci. 2000;9:2142–2150. doi: 10.1110/ps.9.11.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blandl T., Cochran A. G., Skelton N. J. Protein Sci. 2003;12:237–247. doi: 10.1110/ps.0228603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milner-White E. J. Biochim. Biophys. Acta. 1987;911:261–265. doi: 10.1016/0167-4838(87)90017-3. [DOI] [PubMed] [Google Scholar]
- 34.Hutchinson E. G., Thornton J. M. Protein Sci. 1994;3:2207–2216. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramirez-Alvarado M., Blanco F. J., Niemann H., Serrano L. J. Mol. Biol. 1997;273:898–912. doi: 10.1006/jmbi.1997.1347. [DOI] [PubMed] [Google Scholar]
- 36.Ramirez-Alvarado M., Kortemme T., Blanco F. J., Serrano L. Bioorg. Med. Chem. 1999;7:93–103. doi: 10.1016/s0968-0896(98)00215-6. [DOI] [PubMed] [Google Scholar]
- 37.Stanger H. E., Gellman S. H. J. Am. Chem. Soc. 1998;120:4236–4237. [Google Scholar]
- 38.Pires J. R., Parthier C., Aido-Machado R., Wiedemann U., Otte L., Bohm G., Rudolph R., Oschkinat H. J. Mol. Biol. 2005;348:399–408. doi: 10.1016/j.jmb.2005.02.056. [DOI] [PubMed] [Google Scholar]
- 39.Fersht A. R., Matouschek A., Serrano L. J. Mol. Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 40.Nauli S., Kuhlman B., Baker D. Nat. Struct. Biol. 2001;8:602–605. doi: 10.1038/89638. [DOI] [PubMed] [Google Scholar]
- 41.Kaul R., Angeles A. R., Jager M., Powers E. T., Kelly J. W. J. Am. Chem. Soc. 2001;123:5206–5212. doi: 10.1021/ja0102890. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z., Minor W. Methods Enzymol. 1997;276:307326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J.-S., Kuszewski J., Nilges M., Pannu N. S., et al. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Jones T. A., Zou J. Y., S. W. C., Kjeldgaard M. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 45.Brunger A. T. Acta Crystallogr. D. 1993;49:24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.