Abstract
In investigating the role of metal ions in the pathogenesis of Huntington's disease, we examined the effects of clioquinol, a metal-binding compound currently in clinical trials for Alzheimer's disease treatment, on mutant huntingtin-expressing cells. We found that PC12 cells expressing polyglutamine-expanded huntingtin exon 1 accumulated less mutant protein and showed decreased cell death when treated with clioquinol. This effect was polyglutamine-length-specific and did not alter mRNA levels or protein degradation rates. Clioquinol treatment of transgenic Huntington's mice (R6/2) improved behavioral and pathologic phenotypes, including decreased huntingtin aggregate accumulation, decreased striatal atrophy, improved rotarod performance, reduction of weight loss, normalization of blood glucose and insulin levels, and extension of lifespan. Our results suggest that clioquinol is a candidate therapy for Huntington's disease and other polyglutamine-expansion diseases.
Keywords: polyglutamine, small-molecule therapeutics, R6/2 transgenic mice
Huntington's disease (HD) is an autosomal dominant neurological disorder that causes progressive cognitive, motor, and psychiatric dysfunction over a 10- to 20-year disease course, leading to death (1, 2). HD is caused by CAG-repeat expansion (3) in the 5′ region of the IT15 gene that encodes the ubiquitously expressed 350-kDa protein huntingtin (Htt). The expanded-CAG region encodes polyglutamine (polyQ). When the polyQ region is 40 residues or more, there is virtually 100% penetrance of the disease phenotype (4). The physiologic roles of Htt are not fully understood; however, it was recently found to be important for vesicular transport of brain-derived neurotrophic factor in axons (5).
The toxicity of polyQ-Htt appears to involve a gain-of-function (6) mechanism with possible adjunct involvement of impairment of physiologic Htt function (7). Recent studies suggest that diffusely distributed mono- or oligomeric Htt is the predominant toxic form and not the highly aggregated forms present in inclusion bodies (8–11). New toxic activities may occur because the region containing the expanded polyQ tract binds to itself and other polyQ-containing proteins, especially when cleaved from the full-length protein (12), and may sequester and deactivate transcription factors such as TBP (13), CBP (14), Sp1, and TAFII130 (15). Mutant Htt is also implicated in mitochondrial function defects, including initiation of the mitochondrial permeability transition (16, 17) and loss of trophic support due to decreased production (18, 19) and dominant-negative inhibition of transport of brain-derived neurotrophic factor (5). However, the relative contributions to pathogenesis of the many potentially toxic effects of polyQ expansion of Htt have yet to be determined.
Ongoing synthesis of mutant protein appears necessary to drive the pathogenic process. Yamamoto et al. (20), using a tet-regulatable transgenic system, found that when expression of polyQ-htt exon 1 was inhibited in symptomatic mice, the neuropathologic and behavioral changes were reversed. Similar results have recently been reported in a conditional model of spinocerebellar atrophy type I (SCA1) (21). Finally, specific RNA intererence- (RNAi) mediated inhibition of mutant protein expression reverses symptoms and pathology in animal models of HD (22) and SCA1 (23). These studies suggest that interruption of mutant protein synthesis can abrogate, and even reverse, the pathologic process. However, the practical long-term clinical application of RNAi and other nucleotide-based technologies specifically targeting mutant protein production will require resolution of several problems, including blood–brain barrier penetration and immune responses.
Several small compounds targeting features of polyQ protein metabolism and toxic mechanisms noted above have been found to mitigate symptoms and/or pathology in HD model animals (reviewed in ref. 24), and other compounds have been discovered in screens using in vitro polyQ-expansion disease models that inhibit aggregate formation or cell death (25, 26). To date, however, there has been no report of a small molecule that suppresses expanded CAG-repeat expression.
Several observations suggested the metal-binding compound clioquinol (5-chloro-7-iodo-8-hydroxy-quinoline, CQ) might show efficacy in HD models, including: (i) copper and iron levels are increased in the striata of humans with HD (27); (ii) copper levels are increased in the striatum of rats injected with quinolinic acid (an HD model) (28); (iii) polyQ aggregates recruit Cu/Zn-superoxide dismutase (29); (iv) reactive oxygen species (ROS) have been implicated in HD pathogenesis (30, 31), and metals such as iron, copper, or zinc may participate in the generation or toxic conversion of ROS (reviewed in ref. 32); and (v) CQ is able to penetrate cells and the blood–brain barrier and diminish ROS production (33–36). In addition, although CQ was banned for internal use in the 1970s due to neurotoxicity, recent findings that it enhances Aβ aggregate dissolution, decreases Aβ toxicity, and can be given safely under conditions of controlled dosing and vitamin supplementation have prompted renewed clinical interest. A recent phase II clinical trial showed early positive effects on Alzheimer's disease and no significant CQ toxicity (37).
In this report, we demonstrate that CQ selectively reduces expanded polyQ levels in in vitro HD models. We also show that CQ mitigates pathology and behavioral abnormalities in R6/2 transgenic mice. These data establish CQ as a potential therapeutic agent for HD.
Methods
R6/2 Transgenic and Wild-Type Littermate Mice. A colony of transgenic mice strain R6/2 that is heterozygous for huntingtin exon 1 (containing 145 CAG repeats) was maintained, with founders originating from The Jackson Laboratory. Male transgenic R6/2 mice were bred with background strain (B6CBAF1/J) females. Genotyping was done according to Hockly et al. (38).
CQ in water was orally administered to mice at 30 mg/kg per day starting at 3 weeks of age and continued until the mice were deceased or killed for tissues. Just before administration, CQ (≈6 mg) was mixed with 1 ml of drinking water in a 1.5-ml microcentrifuge tube. The tube was mixed by agitation, 0.1 ml of the dispersed CQ solution taken up into a 1-cc syringe attached to a feeding needle, and the dose delivered by oral gavage (similar to ref. 39). Littermate R6/2 mice were gavaged with water vehicle alone. At the end of CQ treatment, mice were anesthetized and transcardially perfused with PBS followed by 4% paraformaldehyde. Blood glucose levels were determined after at least 8 h of fasting in 11-week-old mice (n = 6 for wild type, n = 4 for vehicle-treated R6/2, n = 6 for CQ-treated R6/2 littermates) using a 2300 STAT Plus glucose analyzer (Yellow Springs Instruments). Fasting blood insulin levels in animals fasted for 8 h were measured by using an Ultra Sensitive Rat Insulin ELISA Kit (Crystal Chem, Downers Grove, IL) according to the manufacturer's instructions. All procedures were performed in accordance with protocols approved by the San Francisco Veterans Administration Medical Center Animal Care Committee.
Cell Culture, Transfection, and CQ Treatment. polyQ-htt-GFP fusion constructs, in which the polyQ region is encoded by CAA- and CAG-containing tandem repeats (CAACAGCAACAACAGCAG)n, were obtained from the Hereditary Disease Foundation, Santa Monica, CA. PC12 and HEK293 cells were maintained in MEM supplemented with 10% FBS, switched to Opti-MEM without serum for 6 h during transfection using Lipofectamine 2000 (Invitrogen), and returned to serum-containing medium thereafter (up to 48 h). Cells were exposed to CQ (Sigma) at 0.5–8 μM for 30 min before and after (but not during) transfection. Control cultures received equivalent amounts of DMSO-containing vehicle (≤0.1% DMSO, final concentration).
Western Blotting. Cells were lysed in PBS buffer containing 1% Triton X-100 and protease inhibitor mixture (Roche Applied Science, Indianapolis) and passed through a 28-gauge needle to ensure the release of Htt inclusions from the nuclei. Whole mouse brains were homogenized in 50 mM Tris·HCl (pH 7.4)/150 mM NaCl/1% Triton X-100/complete protease inhibitor mixture. Brain homogenate was precleared by centrifuging at 7,500 × g for 90 s at 4°C. The supernatant was passed through a 28-gauge needle and boiled in SDS sample buffer for 10 min. Fifty micrograms of total protein was loaded onto 10% SDS gel containing a 4% stacking gel. Monoclonal antibodies anti-polyQ (1:2,000), anti-GAPDH (1:10,000), anti-GFP (1:4,000), and anti-Htt (EM48) in 1:1,000 dilution were used for immunoblotting. All Western blot experiments were performed at least three times, and band intensities were averaged.
Northern Analysis. TRIzol reagent (Invitrogen) was used to isolate total RNA. Ten micrograms of total RNA was electrophoresed through 1.25% agarose gel and transferred onto Hybond n + membrane. The membrane was baked in an 80°C oven for 15 min and UV-crosslinked. Exon 1 of htt containing 103 CAA/CAG repeats was used as a probe for Northern blot detection according to the AlkPhos Direct Labeling and Detection System (Amersham Pharmacia Biosciences). One hundred nanograms of the probe was hybridized to the membrane in a 60°C rotating hybridization oven overnight. The membrane was scanned by using the Typhoon 9410 digital scanner (Amersham Pharmacia Biosciences) to yield the signal image. The experiment was performed three times, and band intensities were averaged.
Cell Viability and Section Measurement. Plasma membrane integrity was evaluated by using propidium iodide at 20 μg/ml final concentration. Approximately 200 cells transfected with the Httexon1-Q103-egfp plasmid were counted for each treatment group 48 h after transfection. Cells were analyzed at that time, because expanded protein accumulation had reached a plateau, and 90% of cell death occurred by this time point. The experiment was repeated twice, and counts were averaged. Counts of transfected cells were averaged from three independent randomly selected fields. imagej 1.32j (National Institutes of Health, Bethesda) was used to measure the size of the lateral ventricles (n ≥ 6 for each group).
Pulse–Chase Assay. HEK293 cells at 90% confluence on 100-mm2 plates coated with poly(d-lysine) (0.1 μg/μl) were transfected for 4 h in Opti-MEM medium before replacement of the medium with MEM supplemented with 10% FBS. After another 5-h incubation, cells were trypsinized and replated in two wells of a six-well plate, then incubated for another 14 h at 37°C. The medium was replaced with methionine-free MEM medium supplemented with 5% FBS and cells incubated for 1.5 h with 4 μM of CQ or DMSO vehicle added during the last 30 min of incubation. One mCi (1 Ci = 37 GBq) of [35S]methionine (Amersham Pharmacia Biosciences) in 12 ml of methionine-free MEM was prepared as a pulse-stock medium. Cells were pulsed for 10 min by replacing the medium with 1 ml of the pulse-stock medium. At different time points, cells were washed twice with ice-cold PBS and lysed in 1 ml of immunoprecipitation lysis buffer (50 mM Tris·HCl, pH 7.5/150 mM NaCl/1% Nonidet P-40/0.5% sodium deoxycholate/one tablet minicomplete protease inhibitor/10 ml of buffer). Cells were harvested by scraping, placed in a 1.5-ml microcentrifuge tube, and lysed with constant agitation at 4°C for at least 30 min before immunoprecipitation of the Httexon1-Q103-egfp protein using anti-polyQ antibody and protein G agarose beads (Roche Applied Science). Assays were performed at least three times.
Brain Cryosectioning and Immunohistochemistry. Perfused mouse brains were fixed in 4% paraformaldehyde for 1 h at room temperature and stored in 30% sucrose overnight at 4°C. Brain tissue was frozen at -70°C for at least 30 min before cryosectioning at 40-μm thickness by using a Leica CM1850 cryostat (McBain Instruments, Chatsworth, CA). Eleven-week-old mouse brains (n > 5 for each treatment group) sections were incubated at room temperature for 1 h in blocking buffer containing 2% goat serum, 0.2% Triton X-100, and 0.1% BSA in PBS. Then, EM48 anti-Htt monoclonal antibody (Chemicon) at a 1:100 dilution was incubated with the tissue at 4°C overnight. Colorimetric signal detection was performed by using diaminobenzidine substrate and the ABC kit (Vector Laboratories), according to manufacturer's protocol.
Behavioral Tests. Rotarod performance (n ≥ 6 for each treatment group) was assessed as described by Hockly et al. (38) with the modification that the SDI Rota-Rod (San Diego Instruments, San Diego) was set to linearly increase in speed to 40 rpm in 4-min ramp time. For testing clasping, 10-week-old mice (n = 8 for each treatment group) were suspended by the tail for 30 s, and the foot-clasping time was scored such that a 0- to 5-s clasping duration was given a score of 1, 5–10 s a score of 2, and >10 s a score of 3.
Results
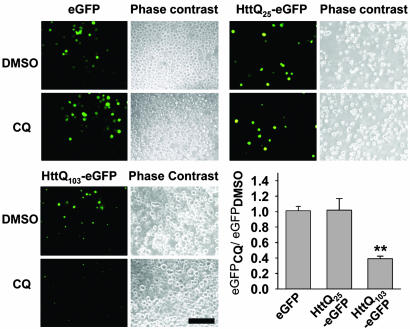
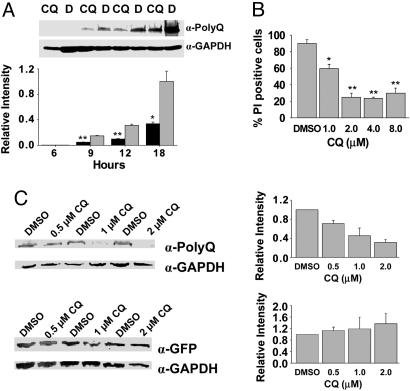
CQ Selectively Down-Regulates polyQ Protein Levels and Relieves Cell Death in Vitro. Transiently transfected PC12 cells expressing Htt exon-1 containing a 103-residue polyQ-encoding region fused to enhanced GFP gene (Httexon1-Q103-egfp) produced a less fluorescent product when treated with CQ at 4 μM, whereas Httexon1-Q25–egfp and egfp alone were unaffected at 48 h posttransfection (Fig. 1). Similar results were obtained with HEK293 cells. Differential expression of Httexon1-Q103-egfp signal could be detected visually at 6 h posttransfection (not shown) and was demonstrable on Western blotting of HEK293 cell extracts beginning at 9 h posttransfection (Fig. 2A). Concordant with this reduction in mutant protein levels, cell survival was improved by CQ in a dose-dependent fashion with a half-maximal effect at 1–2 μM (Fig. 2B). Cell death began to increase at 8 μM, likely due to dose-dependent toxic effects of CQ. Western blotting of cell extracts at 48 h showed a dose-dependent decrease in Httexon1-Q103-egfp levels beginning at 0.5 μM CQ and reaching 3.2-fold at 2 μM, which was not observed with egfp alone (Fig. 2C). The lack of effects on Httiexon1-Q25–egfp and egfp alone suggested that CQ was not causing a generalized decrease in protein synthesis or a reduction in transfection efficiency or affecting GFP maturation.
Fig. 1.
CQ down-regulates expanded polyQ-egfp fluorescence in vitro. Bars and error bars represent mean + SEM. n ≥ 3 for each data set. Fluorescence microscopy and quantitation of EGFP-, Httexon1-Q25-egfp-, and Httexon1-Q103-egfp-expressing cells. (Bar, 100 μm.) **, P < 0.03.
Fig. 2.
CQ down-regulates polyQ protein expression and decreases cell death in vitro. Bars and error bars represent mean + SEM. n ≥ 3 for each data set. Western data are normalized to the GAPDH signal. (A) Western blotting and quantitation of Httexon1-Q103-egfp (polyQ Ab) in HEK293 cell lysates; times posttransfection as indicated. (B) Viability (propidium iodide staining) of Httexon1-Q103-egfp cells treated with CQ. *, P ≤ 0.03; **, P < 0.002 vs. vehicle control. (C) Western blotting and quantitation of Httexon1-Q103-egfp (polyQ Ab) and EGFP (GFP Ab) in cell lysates. ***, P = 0.02.
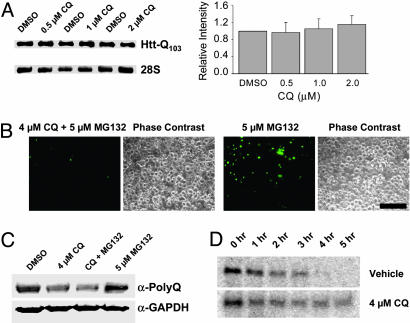
CQ Does Not Affect polyQ mRNA Abundance or Protein Degradation Rate. In examining potential mechanisms for the effects of CQ on Htt accumulation, we considered the possibilities that CQ might act to inhibit Htt aggregation or directly influence Htt synthesis or degradation. Because the early accumulation of Htt was inhibited by CQ, we focused on the aspects of Htt synthesis and turnover. To determine whether CQ might down-regulate polyQ-Htt expression by reducing mRNA levels, we examined the levels of Httexon1-Q103-egfp mRNA present 48 h posttransfection by Northern analysis (Fig. 3A). CQ had no significant effect on mRNA levels at concentrations that markedly diminished protein levels (Fig. 2B). In addition, no differences in mRNA content were observed by using RT-PCR at earlier time points (6, 9, 12, and 18 h; data not shown). Because Htt-exon 1 is subject to ubiquitination and proteasomal degradation (40, 41), we examined the possibility that CQ might work by increasing proteasome-mediated turnover. If this were so, we would expect that the effects of CQ on Httexon1-Q103-egfp expression might be reversed by a proteasome inhibitor. Httexon1-Q103-egfp-transfected HEK293 cells were treated with the panproteasome inhibitor MG132 in the presence or absence of CQ, visualized (compare Figs. 1 and 3B) and blotted (Fig. 3C). Suppression of Httexon1-Q103-egfp protein levels by CQ was unaffected by MG132 treatment. To further support this result and assess the possibility degradation through alternative paths, pulse–chase experiments were performed. No significant differences in the rates of Httexon1-Q103-egfp degradation were observed in the presence of CQ (Fig. 3D), although there may have been a small trend toward decreased degradation at longer time points in CQ-treated cells.
Fig. 3.
CQ does not affect polyQ mRNA levels or protein degradation. Bars and error bars represent mean + SEM. n ≥ 3 for each data set. (A) Northern blotting and quantitation of Httexon1-Q103-egfp transcripts. (B) Fluorescence microscopy of mutant protein accumulation in Httexon1-Q103-egfp-expressing cells in presence of MG132 ± CQ. (Bar, 100 μm.) (C) Western analysis of cells treated with MG132 and CQ. (D) Pulse–chase analysis of Httexon1-Q103-egfp turnover in the presence and absence of CQ.
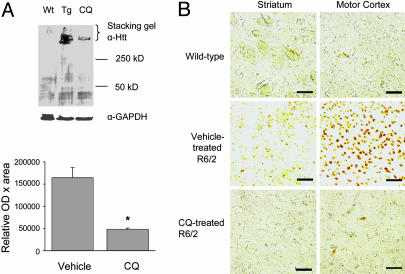
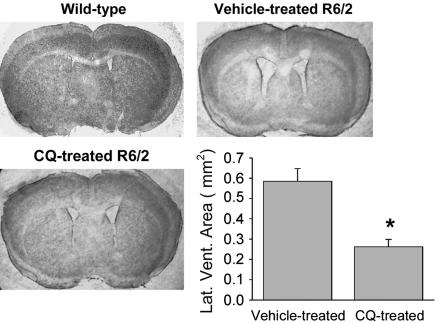
CQ Inhibits Accumulation of Aggregated Htt in Vivo. We next asked whether CQ would also reduce mutant Htt accumulation in vivo by using the HD transgenic mouse model R6/2. After 8 weeks of daily oral CQ treatment, R6/2 mice had a 3.9-fold reduction in the quantity of aggregated Htt, as indicated by Western blotting of whole-brain extracts (Fig. 4A). To examine the anatomical distribution of the changes in expression, immunohistochemistry with the anti-Htt aggregate antibody EM48 was performed, revealing that CQ treatment inhibited the accumulation of aggregated Htt in both the striatum and motor cortex (Fig. 4B).
Fig. 4.
CQ inhibits mutant Htt aggregate accumulation in vivo. Bars and error bars represent mean + SEM. (A) Representative Western blotting and quantitation of high molecular weight mutant Htt in whole-brain homogenates from 11-week-old R6/2 transgenic mice. Wt, wild-type; Tg, vehicle-treated R6/2; CQ, CQ-treated R6/2. n = 3. *, P = 0.01. (B) Inhibition of neuronal intranuclear inclusion formation by treatment with CQ. Representative images of striatum and cortex from 11-week-old mice; wild-type, R6/2-vehicle treated, or R6/2-CQ treated, as indicated. Htt aggregates were visualized by using EM48 Ab. Results were similar for six or more animals in each treatment group. (Bar, 50 μm.)
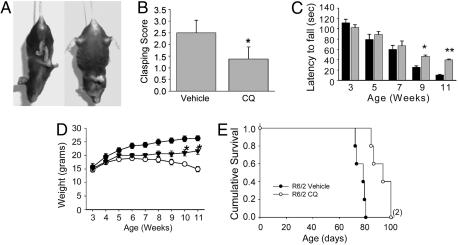
CQ Treatment Improves HD Pathology and Enhances Motor Function and Survival in R6/2 Mice. The major sites of cell loss in HD brain are the striatum and the cortex (42, 43) and in R6/2 mice, striatal atrophy is reflected by expansion of the size of the lateral ventricles. Compared with the sham-treatment group, the treated group had a 55% reduction of lateral ventricle area in standardized sections (Fig. 5). Foot clasping, a dystonic posturing of the hind limbs on suspension by the tail (Fig. 6A), is a characteristic behavior of the later stages of the disease in R6/2. CQ treatment decreased the average clasping score at 10 weeks from 2.5 (of 3 maximum) to 1.3, a 48% reduction (Fig. 6B). The treated group also scored significantly better on rotarod performance, a test of agility and muscle coordination, between 9 and 11 weeks of age (Fig. 6C). The initial trend to a therapeutic effect of CQ on rotarod performance in R6/2 mice began when symptoms such as tremors and weight loss developed. With respect to weight changes, without CQ treatment, weight gain in R6/2 diverged from the wild-type curve and reached a plateau between the fifth and seventh weeks, which was followed by a steady weight loss (Fig. 6D). With CQ treatment, weight gain in R6/2 was comparable to that of the vehicle-treated group up to the seventh week of age but trended toward a gain thereafter, with weights significantly greater than untreated animals observed in the tenth and eleventh weeks (Fig. 6D). The average lifespan of untreated animals in this series was ≈76 d, within the originally described range (10–13 weeks) of R6/2 lifespan (44). CQ treatment extended the lifespan to an average of 92 d, a 20% increase in longevity over the littermate control group (Fig. 6E). This falls within the range of survival improvement (7–31%) seen with several therapeutic agents tested in this mouse model [reviewed in ref. 24).
Fig. 5.
CQ decreases polyQ-mediated pathology in vivo. Bars and error bars represent mean + SEM. Representative images of the cerebrum of 11-week-old mice and quantitation of lateral ventricle areas, showing decreased striatal atrophy in 11-week-old CQ-treated R6/2 mice. n ≥ 6 for each treatment group. *, P < 0.001 vs. vehicle-treated mice.
Fig. 6.
CQ positively effects behavior, weight, and survival in vivo. Bars and error bars represent mean + SEM. (A) Illustration of clasping behavior. (Left) Wild-type mouse. (Right) Ten-week-old R6/2 mouse exhibiting clasping. (B) Clasping score at 10 weeks of age in CQ and vehicle-treated R6/2. n = 8. *, P < 0.0004 vs. vehicle. (C) Rotarod testing of CQ and vehicle-treated R6/2. Bars and error bars represent mean + SEM. Black bars, vehicle-treated; gray bars, CQ-treated. n ≥ 6 for each group. *, P < 0.03; **, P < 0.001 vs. vehicle-treated animals. (D) Body weight over time of wild-type (filled circles), vehicle-treated R6/2 (open circles), and CQ-treated R6/2 (filled triangles); symbols and bars represent mean ± SEM. n ≥ 6 for each group, *, P < 0.01 vs. vehicle-treated animals by Student's t test; also, P < 0.0001 overall, for CQ-treated vs. vehicle-treated animals by ANOVA with post hoc Bonferroni/Dunn testing. (E) Kaplan–Meier analysis of R6/2 lifespan, vehicle treated (closed circles) vs. CQ treated (open circles). n = 5 per group. P = 0.0018 by log-rank Mantel–Cox test.
CQ Mitigates HD of the Endocrine Pancreas. R6/2 mice develop diabetes mellitus due to dysregulation of insulin gene expression associated with sequestration of transcription factors by mutant Htt, which has suggested a mechanism for the elevated incidence of diabetes in individuals with HD (45–47). We found that after 8 weeks of CQ treatment, 11-week-old R6/2 mice had normal fasting blood glucose levels (4.1 ± 0.3 mM), comparable to that of wild-type littermate controls (5.2 ± 0.8 mM). R6/2 sham-treated animals had significantly higher fasting blood glucose levels (7.6 ± 0.4), characteristic of diabetes (Table 1). Consistent with the blood glucose level was a reciprocal change in blood insulin levels. The sham group had a significantly lower fasting blood insulin level (166 ± 5.13 pg/ml) than the treated group (415 ± 54.60 pg/ml) (Table 1), which was similar to that of wild-type mice in these experiments and in those reported previously (24).
Table 1. Fasting plasma glucose and insulin levels in 11-week-old mice.
| Wild type | Vehicle-treated R6/2 | CQ-treated R6/2 | |
|---|---|---|---|
| Glucose, mM | 5.2 ± 0.8 | 7.6 ± 0.4 | 4.1 ± 0.3* |
| Insulin, pg/ml | 403 ± 6.4 | 166 ± 5.1 | 415 ± 54.6** |
Data are mean ± SEM. n ≤ 4 for each treatment group. *, P < 0.0004 for glucose; **, P = 0.0065 for insulin levels comparing vehicle and CQ-treated groups.
Discussion
We have found that CQ decreases polyQ-expanded protein accumulation and promotes cell survival in an in vitro model of HD and decreases symptoms and increases lifespan in HD model animals. To begin to elucidate the mechanism of action of CQ in these models, we investigated the metabolism of the mutant protein by examining changes in mRNA and protein levels and degradation on exposure to the drug. Northern analysis suggested that mRNA levels were not affected by CQ. Early rates of mutant protein accumulation were reduced, whereas inhibition of proteasomal degradation had little effect; moreover, pulse–chase experiments suggested that overall rates of degradation were unchanged or slowed. These findings are consistent with models in which CQ interacts directly or indirectly with mRNA, RNA-binding proteins, or the translation apparatus, but they do not completely rule out other effects on RNA metabolism or posttranslational events, such as aggregation. Moreover, the relevance of the in vitro findings to the effects of CQ in transgenic mice remains unclear, and other compounds such as creatine and caspase inhibitors (reviewed in ref. 24) decrease aggregate accumulation in such models as well.
As a metal-binding compound, CQ may affect numerous cellular processes. Iron, zinc, copper, manganese, magnesium, and other metals play important roles in the structure and function of DNA, RNA, and some proteins. Of note, CQ is a relatively weak chelator and will not significantly recruit tightly bound metals (48), but under various pathologic conditions, metals may be released, or their complexes loosened, allowing effects of even low-affinity chelators. Chelation of iron has been implicated as a mechanism of CQ inhibition of MPTP toxicity (49) and has been suggested to be broadly neuroprotective (36). Interestingly, Htt may be indirectly transcriptionally regulated by iron or the redox state (50), although no such regulatory element sequences are present in the constructs used in our in vitro experiments. Copper chelation can reduce the production of hydrogen peroxide formed by copper–Aβ peptide and perhaps other complexes (34), and accumulation of copper is a feature of the affected regions in HD (27); however, its form and reactivity have not been characterized. Zinc is a structural component of numerous proteins involved in RNA and protein synthesis, and its binding is regulated by levels of ROS in the cell (51). Thus, through indirect antioxidant effects or direct effects on zinc trafficking, CQ may have significant effects on cellular metabolism. In addition, independent of chelation, quinols similar to CQ have been found to bind to RNA and modulate RNA–protein interactions (52). Thus, CQ might influence mutant protein accumulation and cell survival through chelation of iron, copper, zinc, or other ion or through direct interactions with RNA or protein, by stabilizing secondary structures or interfering with RNA–protein interactions or by other effects.
Conclusion
This work suggests that CQ may have therapeutic value for HD and raises the possibility that the elucidation of its mechanism of action will identify new specific targets for the mitigation of expanded polyQ toxicity.
Acknowledgments
This work was funded by a Veterans Administration Merit Review grant (to S.M.M.).
Author contributions: T.N., A.H., and S.M.M. designed research; T.N., A.H., and S.M.M. performed research; T.N., A.H., and S.M.M. analyzed data; and T.N. and S.M.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HD, Huntington's disease; Htt, huntingtin; CQ, clioquinol; polyQ, polyglutamine.
References
- 1.Bates, G. P., Harper, P. S. & Jones, L. (2002) Huntington's Disease (Oxford Univ. Press, Oxford, U.K.).
- 2.Young, A. B. (2003) J. Clin. Invest. 111, 299-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snell, R. G., MacMillan, J. C., Cheadle, J. P., Fenton, I., Lazarou, L. P., Davies, P., MacDonald, M. E., Gusella, J. F., Harper, P. S. & Shaw, D. J. (1993) Nat. Genet. 4, 393-397. [DOI] [PubMed] [Google Scholar]
- 4.Ambrose, C. M., Duyao, M. P., Barnes, G., Bates, G. P., Lin, C. S., Srinidhi, J., Baxendale, S., Hummerich, H., Lehrach, H., Altherr, M., et al. (1994) Somat. Cell Mol. Genet. 20, 27-38. [DOI] [PubMed] [Google Scholar]
- 5.Gauthier, L. R., Charrin, B. C., Borrell-Pages, M., Dompierre, J. P., Rangone, H., Cordelieres, F. P., De Mey, J., MacDonald, M. E., Lessmann, V., Humbert, S., et al. (2004) Cell 118, 127-138. [DOI] [PubMed] [Google Scholar]
- 6.Schilling, G., Becher, M. W., Sharp, A. H., Jinnah, H. A., Duan, K., Kotzuk, J. A., Slunt, H. H., Ratovitski, T., Cooper, J. K., Jenkins, N. A., et al. (1999) Hum. Mol. Genet. 8, 397-407. [DOI] [PubMed] [Google Scholar]
- 7.Gunawardena, S., Her, L. S., Brusch, R. G., Laymon, R. A., Niesman, I. R., Gordesky-Gold, B., Sintasath, L., Bonini, N. M. & Goldstein, L. S. (2003) Neuron 40, 25-40. [DOI] [PubMed] [Google Scholar]
- 8.Arrasate, M., Mitra, S., Schweitzer, E. S., Segal, M. R. & Finkbeiner, S. (2004) Nature 431, 805-810. [DOI] [PubMed] [Google Scholar]
- 9.Cummings, C. J., Reinstein, E., Sun, Y., Antalffy, B., Jiang, Y., Ciechanover, A., Orr, H. T., Beaudet, A. L. & Zoghbi, H. Y. (1999) Neuron 24, 879-892. [DOI] [PubMed] [Google Scholar]
- 10.Saudou, F., Finkbeiner, S., Devys, D. & Greenberg, M. E. (1998) Cell 95, 55-66. [DOI] [PubMed] [Google Scholar]
- 11.Watase, K., Weeber, E. J., Xu, B., Antalffy, B., Yuva-Paylor, L., Hashimoto, K., Kano, M., Atkinson, R., Sun, Y., Armstrong, D. L., et al. (2002) Neuron 34, 905-919. [DOI] [PubMed] [Google Scholar]
- 12.Gafni, J., Hermel, E., Young, J. E., Wellington, C. L., Hayden, M. R. & Ellerby, L. M. (2004) J. Biol. Chem. 279, 20211-20220. [DOI] [PubMed] [Google Scholar]
- 13.Schaffar, G., Breuer, P., Boteva, R., Behrends, C., Tzvetkov, N., Strippel, N., Sakahira, H., Siegers, K., Hayer-Hartl, M. & Hartl, F. U. (2004) Mol. Cell 15, 95-105. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, H., Nucifora, F. C., Jr., Ross, C. A. & DeFranco, D. B. (2003) Hum. Mol. Genet. 12, 1-12. [DOI] [PubMed] [Google Scholar]
- 15.Dunah, A. W., Jeong, H., Griffin, A., Kim, Y. M., Standaert, D. G., Hersch, S. M., Mouradian, M. M., Young, A. B., Tanese, N. & Krainc, D. (2002) Science 296, 2238-2243. [DOI] [PubMed] [Google Scholar]
- 16.Choo, Y. S., Johnson, G. V., MacDonald, M., Detloff, P. J. & Lesort, M. (2004) Hum. Mol. Genet. 13, 1407-1420. [DOI] [PubMed] [Google Scholar]
- 17.Ruan, Q., Lesort, M., MacDonald, M. E. & Johnson, G. V. (2004) Hum. Mol. Genet. 13, 669-681. [DOI] [PubMed] [Google Scholar]
- 18.Zuccato, C., Tartari, M., Crotti, A., Goffredo, D., Valenza, M., Conti, L., Cataudella, T., Leavitt, B. R., Hayden, M. R., Timmusk, T., et al. (2003) Nat. Genet. 35, 76-83. [DOI] [PubMed] [Google Scholar]
- 19.Zuccato, C., Ciammola, A., Rigamonti, D., Leavitt, B. R., Goffredo, D., Conti, L., MacDonald, M. E., Friedlander, R. M., Silani, V., Hayden, M. R., et al. (2001) Science 293, 493-498. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto, A., Lucas, J. J. & Hen, R. (2000) Cell 101, 57-66. [DOI] [PubMed] [Google Scholar]
- 21.Zu, T., Duvick, L. A., Kaytor, M. D., Berlinger, M. S., Zoghbi, H. Y., Clark, H. B. & Orr, H. T. (2004) J. Neurosci. 24, 8853-8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper, S. Q., Staber, P. D., He, X., Eliason, S. L., Martins, I. H., Mao, Q., Yang, L., Kotin, R. M., Paulson, H. L. & Davidson, B. L. (2005) Proc. Natl. Acad. Sci. USA 102, 5820-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia, H., Mao, Q., Eliason, S. L., Harper, S. Q., Martins, I. H., Orr, H. T., Paulson, H. L., Yang, L., Kotin, R. M. & Davidson, B. L. (2004) Nat. Med. 10, 816-820. [DOI] [PubMed] [Google Scholar]
- 24.Beal, M. F. & Ferrante, R. J. (2004) Nat. Rev. Neurosci. 5, 373-384. [DOI] [PubMed] [Google Scholar]
- 25.Heiser, V., Engemann, S., Brocker, W., Dunkel, I., Boeddrich, A., Waelter, S., Nordhoff, E., Lurz, R., Schugardt, N., Rautenberg, S., et al. (2002) Proc. Natl. Acad. Sci. USA 99, Suppl. 4, 16400-16406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollitt, S. K., Pallos, J., Shao, J., Desai, U. A., Ma, A. A., Thompson, L. M., Marsh, J. L. & Diamond, M. I. (2003) Neuron 40, 685-694. [DOI] [PubMed] [Google Scholar]
- 27.Dexter, D. T., Carayon, A., Javoy-Agid, F., Agid, Y., Wells, F. R., Daniel, S. E., Lees, A. J., Jenner, P. & Marsden, C. D. (1991) Brain 114, 1953-1975. [DOI] [PubMed] [Google Scholar]
- 28.Perez, P., Flores, A., Santamaria, A., Rios, C. & Galvan-Arzate, S. (1996) Arch. Med. Res. 27, 449-452. [PubMed] [Google Scholar]
- 29.Kim, S. J., Kim, T. S., Kim, I. Y., Hong, S., Rhim, H. & Kang, S. (2003) Biochem. Biophys. Res. Commun. 307, 660-665. [DOI] [PubMed] [Google Scholar]
- 30.Browne, S. E., Ferrante, R. J. & Beal, M. F. (1999) Brain Pathol. 9, 147-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maksimovic, I. D., Jovanovic, M. D., Colic, M., Mihajlovic, R., Micic, D., Selakovic, V., Ninkovic, M., Malicevic, Z., Rusic-Stojiljkovic, M. & Jovicic, A. (2001) Vojnosanit Pregl. 58, 237-242. [PubMed] [Google Scholar]
- 32.Barnham, K. J., Masters, C. L. & Bush, A. I. (2004) Nat. Rev. Drug Discov. 3, 205-214. [DOI] [PubMed] [Google Scholar]
- 33.Ogata, M., Watanabe, S., Tateishi, J., Kuroda, S. & Otsuki, S. (1973) Lancet 1, 1248-1249. [DOI] [PubMed] [Google Scholar]
- 34.Opazo, C., Huang, X., Cherny, R. A., Moir, R. D., Roher, A. E., White, A. R., Cappai, R., Masters, C. L., Tanzi, R. E., Inestrosa, N. C., et al. (2002) J. Biol. Chem. 277, 40302-40308. [DOI] [PubMed] [Google Scholar]
- 35.Milton, N. G. (2004) Drugs Aging 21, 81-100. [DOI] [PubMed] [Google Scholar]
- 36.Mattson, M. P. (2004) Ann. N. Y. Acad Sci. 1012, 37-50. [DOI] [PubMed] [Google Scholar]
- 37.Ritchie, C. W., Bush, A. I., Mackinnon, A., Macfarlane, S., Mastwyk, M., MacGregor, L., Kiers, L., Cherny, R., Li, Q. X., Tammer, A., et al. (2003) Arch. Neurol. 60, 1685-1691. [DOI] [PubMed] [Google Scholar]
- 38.Hockly, E., Woodman, B., Mahal, A., Lewis, C. M. & Bates, G. (2003) Brain Res. Bull. 61, 469-479. [DOI] [PubMed] [Google Scholar]
- 39.Cherny, R. A., Atwood, C. S., Xilinas, M. E., Gray, D. N., Jones, W. D., McLean, C. A., Barnham, K. J., Volitakis, I., Fraser, F. W., Kim, Y., et al. (2001) Neuron 30, 665-676. [DOI] [PubMed] [Google Scholar]
- 40.Davies, S. W., Turmaine, M., Cozens, B. A., DiFiglia, M., Sharp, A. H., Ross, C. A., Scherzinger, E., Wanker, E. E., Mangiarini, L. & Bates, G. P. (1997) Cell 90, 537-548. [DOI] [PubMed] [Google Scholar]
- 41.Holmberg, C. I., Staniszewski, K. E., Mensah, K. N., Matouschek, A. & Morimoto, R. I. (2004) EMBO J. 23, 4307-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Storey, E., Kowall, N. W., Finn, S. F., Mazurek, M. F. & Beal, M. F. (1992) Ann. Neurol. 32, 526-534. [DOI] [PubMed] [Google Scholar]
- 43.Meade, C. A., Deng, Y. P., Fusco, F. R., Del Mar, N., Hersch, S., Goldowitz, D. & Reiner, A. (2002) J. Comp. Neurol. 449, 241-269. [DOI] [PubMed] [Google Scholar]
- 44.Mangiarini, L., Sathasivam, K., Seller, M., Cozens, B., Harper, A., Hetherington, C., Lawton, M., Trottier, Y., Lehrach, H., Davies, S. W., et al. (1996) Cell 87, 493-506. [DOI] [PubMed] [Google Scholar]
- 45.Andreassen, O. A., Dedeoglu, A., Stanojevic, V., Hughes, D. B., Browne, S. E., Leech, C. A., Ferrante, R. J., Habener, J. F., Beal, M. F. & Thomas, M. K. (2002) Neurobiol. Dis. 11, 410-424. [DOI] [PubMed] [Google Scholar]
- 46.Farrer, L. A. (1985) Clin. Genet. 27, 62-67. [DOI] [PubMed] [Google Scholar]
- 47.Hurlbert, M. S., Zhou, W., Wasmeier, C., Kaddis, F. G., Hutton, J. C. & Freed, C. R. (1999) Diabetes 48, 649-651. [DOI] [PubMed] [Google Scholar]
- 48.Padmanabhan, G., Becue, I. & Smith, J. A. (1989) in Analytical Profiles of Drug Substances, ed. Florey, K. (Academic, New York), pp. 57-90.
- 49.Kaur, D., Yantiri, F., Rajagopalan, S., Kumar, J., Mo, J. Q., Boonplueang, R., Viswanath, V., Jacobs, R., Yang, L., Beal, M. F., et al. (2003) Neuron 37, 899-909. [DOI] [PubMed] [Google Scholar]
- 50.Hilditch-Maguire, P., Trettel, F., Passani, L. A., Auerbach, A., Persichetti, F. & MacDonald, M. E. (2000) Hum. Mol. Genet. 9, 2789-2797. [DOI] [PubMed] [Google Scholar]
- 51.Larabee, J. L., Hocker, J. R. & Hanas, J. S. (2005) Arch. Biochem. Biophys. 434, 139-149. [DOI] [PubMed] [Google Scholar]
- 52.Richter, S., Parolin, C., Gatto, B., Del Vecchio, C., Brocca-Cofano, E., Fravolini, A., Palu, G. & Palumbo, M. (2004) Antimicrob. Agents Chemother. 48, 1895-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]