Abstract
A novel archaeal virus, His1, was isolated from hypersaline waters in southeastern Australia. It was lytic, grew only on Haloarcula hispanica (titers of up to 1011 PFU/ml), and displayed a lemon-shaped morphology (74 by 44 nm) previously reported only for a virus of the extreme thermophiles (SSV1). The density of His1 was approximately 1.28 g/ml, similar to that of SSV1 (1.24 g/ml). Purified particles were resistant to low salt concentrations. The genome was linear, double-stranded DNA of 14.9 kb, similar to the genome of SSV1 (15.5 kb). Morphologically, this isolate clearly belongs to the recently proposed Fuselloviridae family of archaeal viruses. It is the first member of this family from the extremely halophilic archaea, and its host, H. hispanica, can be readily manipulated genetically.
Viruses which infect members of each of the three major phenotypic groups of the Archaea—the extreme halophiles, the extreme thermophiles, and the methanogens—have been found (reviewed in references 19 and 30). The viruses described for the extreme halophiles and the methanogens (kingdom Euryarchaeota [29]) are all of the “classical” head-and-tail morphology, a feature that is typical of most eubacterial phages. On the other hand, the extreme thermophiles (kingdom Crenarchaeota [29]) have a number of unusual virus morphotypes including the lemon-shaped SSV1, the rod-shaped TTV1-4, and the icosahedron-headed viruses (19, 30).
The virus SSV1, whose hosts include Sulfolobus shibatae and Sulfolobus isolates P1 and P2 (12), is one of the few well-studied thermophilic viruses (reviewed in reference 31) and the only well-studied lemon-shaped virus. It is temperate and forms stable lysogens by inserting its 15.5-kb circular genome site specifically into the host chromosome. The entire nucleotide sequence of SSV1 has been determined (17), along with the transcriptional map (21), but only four genes have been identified (9, 20).
Of the twelve halophilic viruses that have been reported (2, 13, 19, 28), only two have been studied past basic virological levels. These are ΦH, a temperate but highly mutable virus (due to the transposition of insertion elements) (23; reviewed in reference 31), and the lytic virus HF2 (14, 15). We describe in this report the isolation and initial characterization of a novel virus, His1, which was isolated from hypersaline waters in southeastern Australia and specifically infects Haloarcula hispanica (8).
(The data in this report were first presented at the Australian Society for Biochemistry and Molecular Biology Conference, Canberra, Australia, October 1996.)
Isolation, host range, and plaque morphology.
Hypersaline water samples taken from Avalon saltern, on Corio Bay in Victoria, Australia, were screened for viruses by plating directly on lawns of three halophilic hosts: H. hispanica (ATCC 33960), Haloferax volcanii (NCMB202), and “Haloferax alicantei” (previously Phenon K, Aa2.2 isolate) (7). A plaque was found only on an H. hispanica lawn, and from this a virus, designated His1, was isolated. His1 produced plaques consistently only when plated at 30°C (not at 37°C). The plaques were small (1.5 mm in diameter) and clear, with precise edges. The virus appeared to be lytic, as it gave clear plaques and high titers in liquid cultures.
The host range of His1 was then investigated. Sixteen halobacterial strains from the genera Haloferax, Halorubrum, Halobacterium, the newly proposed genus Haloterrigens (26), and other Haloarcula species—including H. marismortui, H. sinaiiensis, and H. vallismortis—were tested for the ability to act as a host to the newly isolated virus. Apart from the original isolating host, H. hispanica, the halobacteria tested were not susceptible to infection by His1.
For routine use, virus was stored in halophage (HF) diluent or 18% modified growth medium (15) at 4°C, and reserve stocks were kept as isolated plaques on a lawn of H. hispanica, also at 4°C. Small-scale stocks were grown by infection (multiplicity of infection, 0.1) of an early logarithmic H. hispanica culture (50 ml) at 30°C. Liquid cultures (10 to 50 ml) were shaken at 200 rpm and incubated for 1 to 4 days, and plates were incubated for 48 to 72 h, for visible plaques. Virus titers were determined as previously described (15).
Virus sensitivity to organic solvents, low salt, and temperature.
Diluted His1 was exposed to chloroform and trichlorotrifluoroethane (Arklone; ICI Chemicals and Polymers Runcorn, Cheshire, England) in a volume ratio of 1:4 (solvent to virus) for 2.5 min and 5 min. Incubation was at room temperature with constant agitation, and samples were then removed and diluted. His1 was shown to be sensitive to the organic solvent chloroform, as no viable virus was detected after treatment, but was resistant to a 5-min exposure to trichlorotrifluoroethane (data not shown).
Sensitivity to a low-ionic-strength environment was tested by diluting virus stocks in HF diluent (18% SW) 1,000-fold with pure water, incubating them at room temperature, and determining virus titers at various time intervals. Unlike most other haloviruses, His1 appeared to be resistant to a low concentration of salt (≈0.02%), maintaining a constant titer for at least the 24 h of testing (data not shown).
High-titer virus solutions were also exposed to different temperatures. His1 was able to maintain a stable titer for 3 weeks stored at 4, 20, 30, and 37°C (data not shown).
Virus purification.
To produce high-titer stocks of His1 (>1010 PFU/ml), large (500-ml) cultures of H. hispanica cells in 18% modified growth medium were inoculated with approximately 1011 PFU. The infected cultures were incubated at 30°C in a shaker (200 rpm) for 4 to 7 days, with titers of about 1012 PFU/ml being produced. Although the cultures had not cleared after approximately 4 days, purification proceeded if the titer was determined to be adequately high. The cell debris was pelleted, and virus was precipitated and resuspended as previously described (15), except that polyethylene glycol 8000 (Sigma Chemical Co., St. Louis, Mo.) was used. Resuspended virus was extracted with trichlorotrifluoroethane at a 4:1 volume ratio (sample to trichlorotrifluoroethane) and centrifuged (Sorvall SS34; 6,000 rpm for 10 min at 4°C). The aqueous phase was loaded directly on a CsCl-HF diluent step gradient with the following densities: 1.13, 1.3, and 1.5 g/ml. After centrifugation (Beckman SW28 Ti; 24,000 rpm for 2 h at 10°C) the band(s) visible in the 1.3 g/ml layer was collected and recentrifuged to equilibrium in a CsCl-HF diluent solution of 1.3 g/ml (Beckman 70 Ti; 60,000 rpm for 24 h at 10°C). The resulting band(s) was collected and diluted in HF diluent; the virus was pelleted (Beckman SW55; 35,000 rpm for 75 min at 10°C), resuspended in HF diluent, titrated, and stored at 4°C. The virus particles of His1 had a density of 1.28 g/ml, as determined by equilibrium centrifugation in a CsCl isopynic gradient. This is a relatively low value, but it is similar to that of the thermophilic virus SSV1 (1.24 g/ml) (12).
Electron microscopy.
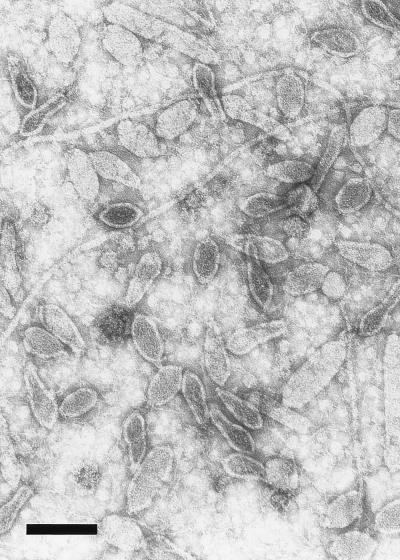
Purified virus preparations were examined by negative-stain electron microscopy (Fig. 1). Virus particles were allowed to adsorb to poly-l-lysine (0.1 mg/ml) (Sigma Chemical Co.)-coated copper grids and were fixed with 1% glutaraldehyde (Merck, Darmstadt, Germany) and negatively stained with 2% uranyl acetate (May & Baker Ltd., Dagenham, England). His1 was found to be similar in morphology to SSV1 of the Fuselloviridae family, i.e., lemon-shaped with a very short tail at one end. Virus particles appeared to be flexible, and some elongated forms were also observed. The regular lemon-shaped particles were about 74 nm long and 44 nm wide with a 7-nm-long tail.
FIG. 1.
Electron micrograph of the halophilic virus His1, taken at a magnification of ×60,000 and negatively stained with uranyl acetate (2% [wt/vol]). Flagella of the host are also visible in this purified preparation. Bar, 100 nm.
Genome of His1.
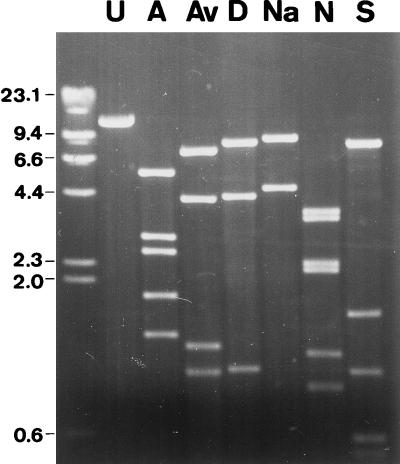
Nucleic acids of purified virus preparations were purified by standard methods with proteinase K (Boehringer, Mannheim, Germany) and 0.1% sodium dodecyl sulfate (BDH Laboratory Supplies, Poole, England), extracted with phenol-chloroform-isoamyl alcohol, and precipitated with ethanol (1). Agarose gel electrophoresis of the isolated viral nucleic acids through 0.8 to 1.0% agarose gels in Tris-acetate-EDTA electrophoresis buffer (1) gave a single, well-resolved band. This, and the ability of a number of nuclease enzymes—including type II restriction enzymes and the exonuclease Bal 31—to digest the genome, indicated that it was double-stranded linear DNA. The estimated size of the His1 genome was 14.9 kb (Table 1; Fig. 2), which is the smallest recorded for any halophilic virus (13) and resembles the size of the SSV1 genome (15.5 kb) (17).
TABLE 1.
Estimated sizes of fragments generated by restriction endonuclease digestion of the His1 genome
| Fragment | Fragment size (kb)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Uncut | AseI | AvaI | DraI | NdeI | NaeI | SphI | SspI | |
| 1 | 14.9a | 5.9b | 8.3 | 9.4 | 3.9 | 10.0 | 10.0 | 8.6 |
| 2 | 3.0 | 4.3 | 4.5 | 3.6 | 4.9 | 4.9 | 1.6 | |
| 3 | 2.7 | 1.3 | 1.0 | 2.5 | 1.0 | |||
| 4 | 1.9b | 1.0 | 2.3 | 0.6 | ||||
| 5 | 1.3 | 1.2 | 0.3 | |||||
| 6 | 0.9 | |||||||
| 7 | 0.3 | |||||||
The restriction digest fragment totals were averaged to estimate the uncut size.
Terminal fragment.
FIG. 2.
Agarose gel electrophoresis of His1 DNA, uncut (U) and cleaved with a number of endonuclease restriction enzymes: AseI (A), AvaI (Av), DraI (D), NaeI (Na), NdeI (N), and SspI (S). DNA size markers (in kilobases) are shown on the left.
The His1 genome is not circularly permuted, as restriction fragments were equimolar and no heterogeneity in the size of the terminal restriction fragments was observed (Fig. 2). Therefore, it is not likely that a headful packaging mechanism is used in the assembly of this virus. The abundance of restriction enzymes able to digest the His1 genome revealed that there was no selection against palindromic sequences, which is a common phage mechanism for resisting host restriction enzymes and which has previously been encountered in the halophilic viruses HF1 and HF2 (14).
Several linear double-stranded DNA phages have a terminal protein linked to the 5′ ends of their DNA (22). His1 DNA was isolated with and without proteinase K being present and then digested with AseI and separated on a 1% agarose gel. The untreated samples had the same restriction profile as the proteinase K-treated samples, indicating that His1 does not possess terminal proteins, which would have caused retardation of the terminal fragments in the gel (data not shown).
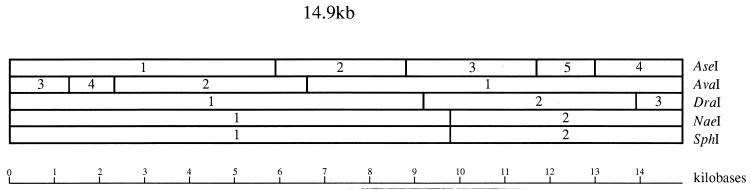
Exonuclease treatment with Bal 31 (New England Biolabs) over various periods of time, prior to digestion with the endonuclease restriction enzyme AseI, was the initial procedure used to map the genome. The results of these experiments enabled the terminal AseI fragments to be established (Fig. 3), as well as the order in which the AseI fragments are arranged within the genome. With a basic AseI map established, double digests with AseI and different restriction enzymes were able to establish the positions of additional restriction sites (Fig. 3).
FIG. 3.
Physical map of the genome of the virus His1. The map was constructed with five endonuclease restriction enzymes: AseI, AvaI, DraI, NaeI, and SphI. The numbers within the map correspond to fragment numbers in Table 1, and a scale is shown beneath the map.
Viral structural proteins.
Attempts were made to study the proteins of purified virus particles. Despite numerous attempts with conventional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10) or the protocols described for analyzing the small and highly hydrophobic SSV1 proteins (20), no well-resolved bands were observed.
Persistent infection state.
Although His1 appeared to be lytic, due to the clear plaques and high titers, liquid cultures of virus did not clear (even at multiplicities of infection as high as 5), and host cells which appeared to be persistently infected were isolated. Attempts to produce single-step growth curves also were not successful, as virus appeared to be liberated from cells continuously after a 4- to 6-h eclipse. This may have been due to the host continually extruding virus particles with little or no cell death, a phenomenon previously observed with SSV1 (11). Alternatively, His1 may have been able to form a stable state with the host.
Southern blot hybridization of the host chromosome from a persistently infected culture was therefore performed to determine if His1 is temperate or capable of producing a carrier state. DNAs were isolated from H. hispanica cultures that appeared to be persistently infected; i.e., cells extruded virus, as shown by plaques around the streaked bacteria on an uninfected indicator lawn. Chromosomal DNA was purified as previously described by Dyall-Smith and Doolittle (5), and plasmid extraction was performed by the alkaline lysis miniprep protocol of Ausubel et al. (1) with two modifications: (i) halobacterial cells were centrifuged for 2 min at 13,000 rpm, and (ii) 1 M NaCl–5 mM Tris (pH 7.5) was used to resuspend the cell pellet before proceeding with cell lysis.
Southern blot hybridization showed that no plasmid-form or chromosomally integrated virus DNA existed in these host cells (data not shown). Linear His1 DNA was detected in very small amounts, probably indicating that not every cell in the culture contained virus DNA. The evidence indicated that the persistent infection was not of a lysogenic nature but was a carrier state, as reported for many other halophilic viruses (18, 24, 25, 27). In this respect, His1 differs from the other known lemon-shaped virus, SSV1, which forms lysogens (11, 31).
In summary, His1 is a novel halophilic virus of the Archaea which plaques only on the isolating host H. hispanica. The typical lemon-shaped morphology of His1 indicates it should be classified within the family Fuselloviridae, the type species of which is the Sulfolobus virus SSV1 (12). His1 is the first halophilic member of this virus family and expands the range of hosts to include organisms in two kingdoms of the Archaea. This morphotype has not been previously found among the Bacteria or Eukarya and would appear to be restricted to the Archaea, perhaps reflecting a deep evolutionary history within this domain. His1 is also the first “culturable” halophilic virus of this morphotype which appears to be geographically widespread, as lemon-shaped virus-like particles have been observed by electron microscopy in hypersaline water taken from the Dead Sea (16) and in two Spanish solar salterns (6). Since genetic manipulation within the Archaea is currently more advanced in halobacteria (3, 4), the isolation of His1 offers the prospect of study of this novel virus group at a much finer level than previously possible.
Acknowledgments
We gratefully acknowledge the excellent electron microscopy carried out by Jocelyn Carpenter, and we thank Lisette Moran for technical assistance.
These studies were supported by a grant from the Australian Research Council. C. Bath was supported by a Melbourne University postgraduate scholarship.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley; 1987. [Google Scholar]
- 2.Brown J W, Daniels C J, Reeve J N. Gene structure, organization, and expression in archaebacteria. Crit Rev Microbiol. 1989;16:287–338. doi: 10.3109/10408418909105479. [DOI] [PubMed] [Google Scholar]
- 3.Doolittle W F, Lam W L, Schalkwyk L C, Charlebois R L, Cline S W, Cohen A. Progress in developing the genetics of the halobacteria. Biochem Soc Symp. 1992;58:73–78. [PubMed] [Google Scholar]
- 4.Dyall-Smith, M. Unpublished data.
- 5.Dyall-Smith M L, Doolittle W F. Construction of composite transposons for halophilic Archaea. Can J Microbiol. 1994;40:922–929. doi: 10.1139/m94-148. [DOI] [PubMed] [Google Scholar]
- 6.Gúixa-Boixareu N, Calderón-Paz J I, Heldal M, Bratbak G, Pedrós-Alió C. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat Microb Ecol. 1996;11:215–227. [Google Scholar]
- 7.Holmes M L, Dyall-Smith M L. A plasmid vector with a selectable marker for halophilic archaebacteria. J Bacteriol. 1990;172:756–761. doi: 10.1128/jb.172.2.756-761.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Juez G, Rodriguez-Valera F, Ventosa A, Kushner D J. Haloarcula hispanica spec. nov. and Haloferax gibbonsii spec. nov., two new species of extremely halophilic archaebacteria. Syst Appl Microbiol. 1986;8:75–79. [Google Scholar]
- 9.Koonin E V. Archaebacterial virus SSV1 encodes a putative DnaA-like protein. Nucleic Acids Res. 1992;20:1143. doi: 10.1093/nar/20.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 11.Martin A, Yeats S, Janekovic D, Reiter W-D, Aicher W, Zillig W. SAV1, a temperate u.v.-inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldaricus isolate B12. EMBO J. 1984;3:2165–2168. doi: 10.1002/j.1460-2075.1984.tb02107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy: classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. [Google Scholar]
- 13.Nuttall S D. Ph.D. thesis. Parkville, Australia: University of Melbourne; 1994. [Google Scholar]
- 14.Nuttall S D, Dyall-Smith M L. Halophage HF2: genome organization and replication strategy. J Virol. 1995;69:2322–2327. doi: 10.1128/jvi.69.4.2322-2327.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nuttall S D, Dyall-Smith M L. HF1 and HF2: novel bacteriophages of halophilic archaea. Virology. 1993;197:678–684. doi: 10.1006/viro.1993.1643. [DOI] [PubMed] [Google Scholar]
- 16.Oren A, Bratbak G, Heldal M. Occurrence of virus-like particles in the Dead Sea. Extremophiles. 1997;1:143–149. doi: 10.1007/s007920050027. [DOI] [PubMed] [Google Scholar]
- 17.Palm P, Schleper C, Grampp B, Yeats S, McWilliam P, Reiter W D, Zillig W. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shiibatae. Virology. 1991;185:242–250. doi: 10.1016/0042-6822(91)90771-3. [DOI] [PubMed] [Google Scholar]
- 18.Pauling C. Bacteriophages of Halobacterium halobium: isolation from fermented fish sauce and primary characterization. Can J Microbiol. 1982;28:916–921. doi: 10.1139/m82-138. [DOI] [PubMed] [Google Scholar]
- 19.Reiter W, Zillig W, Palm P. Archaebacterial viruses. Adv Virus Res. 1988;34:143–188. doi: 10.1016/s0065-3527(08)60517-5. [DOI] [PubMed] [Google Scholar]
- 20.Reiter W-D, Palm P, Henschen A, Lottspeich F, Zillig W, Grampp B. Identification and characterization of the genes encoding three structural proteins of the Sulfolobus virus-like particle SSV1. Mol Gen Genet. 1987;206:144–153. [Google Scholar]
- 21.Reiter W-D, Palm P, Yeats S, Zillig W. Gene expression in archaebacteria: physical mapping of constitutive and UV-inducible transcripts from the Sulfolobus virus-like particle SSV1. Mol Gen Genet. 1987;209:270–275. doi: 10.1007/BF00329653. [DOI] [PubMed] [Google Scholar]
- 22.Salas M. Phages with protein attached to the DNA ends. In: Calendar R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Press; 1988. pp. 169–191. [Google Scholar]
- 23.Stolt P, Zillig W. Gene regulation in halophage ΦH: more than promoters. Syst Appl Microbiol. 1994;16:591–596. [Google Scholar]
- 24.Torsvik T, Dundas I D. Halophilic phage specific for Halobacterium salinarium str.1. In: Caplan S R, Ginzburg M, editors. Energetics and structure of halophilic microorganisms. Amsterdam, The Netherlands: Elsevier/North-Holland Biomedical Press; 1978. pp. 609–614. [Google Scholar]
- 25.Torsvik T, Dundas I D. Persisting phage infection in Halobacterium salinarium str.1. J Gen Virol. 1980;47:29–36. [Google Scholar]
- 26.Ventosa, A., M. C. Gutierrez, M. Kamekura, and M. L. Dyall-Smith. Proposal for the transfer of Halococcus turkmenicus, “Halobacterium trapanicum” JCM9743, and strain GSL-11 to Haloterrigens turkmenicus gen. nov., comb. nov. Int. J. Syst. Bacteriol., in press. [DOI] [PubMed]
- 27.Wais A C, Kon M, MacDonald R E, Stollar B D. Salt-dependent bacteriophage infecting Halobacterium cutirubrum and Halobacterium halobium. Nature. 1975;256:314–315. doi: 10.1038/256314a0. [DOI] [PubMed] [Google Scholar]
- 28.Witte A, Baranyi U, Klein R, Sulzner M, Luo C, Wanner G, Krüger D H, Lubitz W. Characterization of Natronobacterium magadii phage ΦCh1, a unique archaeal phage containing DNA and RNA. Mol Microbiol. 1997;23:603–616. doi: 10.1046/j.1365-2958.1997.d01-1879.x. [DOI] [PubMed] [Google Scholar]
- 29.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zillig W, Gropp F, Henschen A, Neumann H, Palm P, Reiter W D, Rettenberger M, Schnabel H, Yeats S. Archaebacterial virus host systems. Syst Appl Microbiol. 1986;7:58–66. [Google Scholar]
- 31.Zillig W, Reiter W-D, Palm P, Gropp F, Neumann H, Rettenberger M. Viruses of archaebacteria. In: Calendar R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Press; 1988. pp. 517–555. [Google Scholar]