Abstract
Two seven-gene phenazine biosynthetic loci were cloned from Pseudomonas aeruginosa PAO1. The operons, designated phzA1B1C1D1E1F1G1 and phzA2B2C2D2E2F2G2, are homologous to previously studied phenazine biosynthetic operons from Pseudomonas fluorescens and Pseudomonas aureofaciens. Functional studies of phenazine-nonproducing strains of fluorescent pseudomonads indicated that each of the biosynthetic operons from P. aeruginosa is sufficient for production of a single compound, phenazine-1-carboxylic acid (PCA). Subsequent conversion of PCA to pyocyanin is mediated in P. aeruginosa by two novel phenazine-modifying genes, phzM and phzS, which encode putative phenazine-specific methyltransferase and flavin-containing monooxygenase, respectively. Expression of phzS alone in Escherichia coli or in enzymes, pyocyanin-nonproducing P. fluorescens resulted in conversion of PCA to 1-hydroxyphenazine. P. aeruginosa with insertionally inactivated phzM or phzS developed pyocyanin-deficient phenotypes. A third phenazine-modifying gene, phzH, which has a homologue in Pseudomonas chlororaphis, also was identified and was shown to control synthesis of phenazine-1-carboxamide from PCA in P. aeruginosa PAO1. Our results suggest that there is a complex pyocyanin biosynthetic pathway in P. aeruginosa consisting of two core loci responsible for synthesis of PCA and three additional genes encoding unique enzymes involved in the conversion of PCA to pyocyanin, 1-hydroxyphenazine, and phenazine-1-carboxamide.
Phenazine compounds produced by fluorescent Pseudomonas species are biologically active metabolites that function in microbial competitiveness (37), the suppression of soilborne plant pathogens (1, 11, 55, 56), and virulence in human and animal hosts (35).
The most widely studied phenazine-producing fluorescent pseudomonad is P. aeruginosa, a gram-negative opportunistic pathogen of animals, insects, nematodes, and plants (30, 33, 35, 46). In humans, P. aeruginosa infects immunocompromised, burned, or injured patients and can cause both acute and chronic lung disease. Strains of P. aeruginosa produce a variety of redox-active phenazine compounds, including pyocyanin, phenazine-1-carboxylic acid (PCA), 1-hydroxyphenazine (1-OH-PHZ), and phenazine-1-carboxamide (PCN) (7, 52, 57).
From 90 to 95% of P. aeruginosa isolates produce pyocyanin (52), and the presence of high concentrations of pyocyanin in the sputum of cystic fibrosis patients has suggested that this compound plays a role in pulmonary tissue damage observed with chronic lung infections (64). This idea is supported by several recent studies which demonstrated that pyocyanin contributes in a variety of ways to the pathophysiological effects observed in airways infected by P. aeruginosa. Pyocyanin interferes with the regulation of ion transport, ciliary beat frequency, and mucus secretion in airway epithelial cells by altering the cytosolic concentration of calcium (15). It may interact with endothelium-derived relaxing factor or with nitric oxide (which plays a central role in the control of blood pressure, blood flow, and immune function) through the formation of a complex, or it may act by inhibition of nitric oxide synthase (29, 58, 59). Phenazines that are produced by P. aeruginosa also can stimulate alveolar macrophages to produce two neutrophil chemotaxins, IL-8 and leukotriene B4, that attract neutrophils into airways, causing an inflammatory response and neutrophil-mediated tissue damage (14, 33).
The unusually broad range of biological activity associated with phenazines is thought to be due to their ability to undergo redox cycling in the presence of various reducing agents and molecular oxygen, which leads to the accumulation of toxic superoxide (O2−) and hydrogen peroxide (H2O2) and eventually to oxidative cell injury or death (6, 25). It also has been shown that pyocyanin can interact synergistically with the siderophore pyochelin and with transferrin cleaved by proteases secreted by both P. aeruginosa and neutrophils in infected lungs to catalyze the formation of the highly cytotoxic hydroxyl radical (·OH), which damages pulmonary endothelial cells (6, 38). In model pathogenesis systems, phenazine synthesis by P. aeruginosa is required for the generation of disease symptoms in plants and for effective killing of the nematode Caenorhabditis elegans and the greater wax moth, Galleria mellonella (30, 35, 46). Phenazine compounds produced in the rhizosphere of plants contribute to the biological control activity of P. aeruginosa against Fusarium wilt of chickpea and Pythium damping-off of bean (1).
Although the pathophysiological effects of phenazines produced by P. aeruginosa in host organisms are well studied (6, 14, 15, 33, 34, 38, 64) and pyocyanin-deficient phenotypes frequently have been described (18, 19, 26, 32, 35, 46, 54), the biochemistry and genetics of phenazine synthesis in P. aeruginosa have remained unclear. We describe here cloning and functional analysis of two seven-gene phenazine operons and three phenazine-modifying genes from P. aeruginosa PAO1. Our results show that P. aeruginosa contains a complex phenazine biosynthetic pathway consisting of two homologous core loci (phzA1B1C1D1E1F1G1 and phzA2B2C2D2E2F2G2) responsible for synthesis of PCA and three additional genes (phzM, phzS, and phzH) encoding unique enzymes involved in the conversion of PCA to pyocyanin and PCN. We detected the core biosynthetic operon by Southern hybridization in 21 phenazine-producing pseudomonads, including strains of P. aeruginosa, Pseudomonas fluorescens, Pseudomonas chlororaphis, and Pseudomonas aureofaciens, but not in seven phenazine-producing isolates of Burkholderia cepacia, Burkholderia phenazinium, and Brevibacterium iodinum. Thus, the core biosynthetic pathway is highly conserved in fluorescent Pseudomonas spp. but differs significantly from that in other phenazine-producing bacterial genera.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All of the bacterial strains and plasmids used in this study are described in Table 1. Strains of P. fluorescens, P. aeruginosa, P. chlororaphis, P. aureofaciens, B. cepacia, B. phenazinium, and B. iodinum were grown at 28°C in Luria-Bertani (LB) broth (2). Escherichia coli strains were grown at 28 or 37°C in LB medium. To examine phenazine production, P. aeruginosa strains were grown on PIA plates (Difco Laboratories, Detroit, Mich.) or in PB medium (containing [per liter of distilled water] 20 g of Bacto Peptone [Difco Laboratories], 1.4 g of MgCl2, and 10 g of K2SO4) (18) for 1 to 3 days at 28 or 37°C. Strains of P. fluorescens were tested for phenazine production in LB medium supplemented with 2% glucose. The antibiotics used in this study were gentamicin (300 μg/ml) and carbenicillin (500 μg/ml) in experiments with mutant derivatives of P. aeruginosa; tetracycline (15 to 20 and 100 μg/ml) in experiments with isolates of P. fluorescens and P. aeruginosa, respectively; and ampicillin (80 μg/ml) and tetracycline (12.5 μg/ml) in experiments with E. coli.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description or sequencea | Reference(s) or sourceb |
|---|---|---|
| Brevibacterium iodinum strains | ||
| ATCC 15728 | Phz+, produces iodinin | 57 |
| ATCC 15729 | Phz+, produces iodinin | 57 |
| ATCC 9897 | Phz+, produces iodinin | ATCC |
| Burkholderia cepacia strains | ||
| ATCC 25416 | Phz+, produces 4,9-dihydroxyphenazine-1,6-dicarboxylic acid dimethylester | 31 |
| 1324 | Phz+, biocontrol strain | Lab collection |
| 5.5B#5 | Phz+, biocontrol strain | Lab collection |
| 2257 | Phz(?), biocontrol strain | Lab collection |
| ATCC 52796 | Phz(?), biocontrol strain | 39, 40 |
| Burkholderia phenazinium ATCC 33666 | Phz+, produces iodinin | 49 |
| Pseudomonas aeruginosa strains | ||
| PAO1 | Phz+, produces pyocyanin, PCA, PCN, 1-OH-PHZ | Pseudomonas Genetic Stock Center |
| PAOmxM | Phz+phzM::Gmr | This study |
| PAOmxS | Phz+phzS::Gmr | This study |
| ATCC 23993 | Phz+, produces pyocyanin | ATCC |
| ATCC 25007 | Phz+, produces pyocyanin | 49 |
| ATCC 25011 | Phz+, produces aeruginosins A and B | 65 |
| PAKN 1 | Phz+, produces pyocyanin | Lab collection |
| PAKNP-1 | Phz+, produces pyocyanin | Lab collection |
| PAKNP-2 | Phz+, produces pyocyanin | Lab collection |
| Pseudomonas aureofaciens strains | ||
| 30–84 | Phz+, produces PCA, 2-OH-PCA, 2-OH-PHZ | 42 |
| ATCC 17415 | Phz+, produces PCA, 2-OH-PCA, 2-OH-PHZ | ATCC |
| ATCC 13985 | Phz+, produces PCA, 2-OH-PCA, 2-OH-PHZ | 49 |
| TX-1 | Phz+, produces PCA, 2-OH-PCA, 2-OH-PHZ | Eco-Soils |
| BS1391 | Phz+, produces PCA, 2-OH-PCA, 2-OH-PHZ | V. Kotchetkov |
| BS1393 | Phz+, produces PCA, 2-OH-PCA, 2-OH-PHZ | V. Kotchetkov |
| PGS12 | Phz+, produces PCA, 2-OH-PCA, 2-OH-PHZ | 23 |
| Pseudomonas chlororaphis strains | ||
| ATCC 17411 | Phz+, produces PCN | 57 |
| ATCC 17809 | Phz+, produces PCN | ATCC |
| ATCC 9446 | Phz+, produces PCN | 49 |
| Pseudomonas fluorescens strains | ||
| 2–79 | Phz+ Rifr, produces PCA | 61 |
| UN15 | Phz+, produces PCA | G. Botelho |
| UN4127 | Phz+, produces PCA | G. Botelho |
| UQ112 | Phz+, produces PCA | G. Botelho |
| M4-80R | Phz− Rifr | 24 |
| Escherichia coli strains | ||
| JM109 | F′ traD36 proA+proB+lacIqlacZΔM15/recA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) mcrA | Promega Corp. |
| S17-1 | thi pro hsdR hsdM recA rpsL RP4-2 (Tcr::Mu) (Kmr::Tn7) | Lab collection |
| Plasmids | ||
| pUC19 | ColE1 bla | 67 |
| pUCP26 | pUC18-derived broad-host-range vector, Tetr | 62 |
| pMOB3 | Kanrcat sacBR | H. P. Schweizer |
| pUCGM | Source of gentamicin cassette ColE1 aacC1 | H. P. Schweizer |
| pNOT19 | ColE1 bla accessory plasmid | H. P. Schweizer |
| p137, p143 | pUC19 containing 2.4-kb PstI-EcoRI fragment with phzCDE genes | This study |
| 1G5 | s-cos containing approximately 15-kb fragment of P. aeruginosa PAO1 genomic DNA | S. Lory |
| pUCP-1G5 | pUCP26 containing 10-kb BglII-NotI fragment with phzA1B1C1D1E1F1G1 genes from 1G5 | This study |
| pUCP-A1G1 | pUCP26 containing 6.9-kb NdeI-EcoRV fragment with phzA1B1C1D1E1F1G1 genes | This study |
| 1H4 | s-cos containing approximately 25-kb fragment of P. aeruginosa PAO1 genomic DNA | S. Lory |
| pUCP-A2G2 | pUCP26 containing 7.4-kb HindIII fragment with phzA2B2C2D2E2F2G2 genes | This study |
| M13-10116 | M13mp18 containing 4.5-kb fragment of P. aeruginosa PAO1 genomic DNA | M. Bangera |
| pUCP-phnAB | pUCP26 containing 2.8-kb SacI-KpnI fragment from M13-10116 with phnAB genes | This study |
| pUCP-M | pUCP26 containing PCR fragment with phzM | This study |
| pUCP-S | pUCP26 containing PCR fragment with phzS | This study |
| pUCP-H | pUCP26 containing PCR fragment with phzH | This study |
| pUCP-MS | pUCP26 containing tandem of phzM and phzS genes | This study |
| pNOT-M-Gm | pNOT19 containing phzM with Gmr cassette inserted into EcoRV site | This study |
| pNOT-S-Gm | pNOT19 containing phzS with Gmr cassette inserted into ScaI site | This study |
| pNOT-M-Gm-MOB | pUCP-M-Gm ligated with MOB cassette | This study |
| pNOT-S-Gm-MOB | pUCP-S-Gm ligated with MOB cassette | This study |
| Primers | ||
| PHZ-UP | TAAGGATCCGGTAGTTCCAAGCCCCAGAAAC | |
| PHZ-LOW | CACATTTGATCTAGATGGGTCACGGCTATTCAG | |
| METHYL1 | TTTTTGAATTCTTTCGGACGCAGGAAAAG | |
| METHYL2 | TTTTTGGATCCGTTGAAAGTTCCGATTCA | |
| OXY1 | TCGAACACTCTAGAAAAGGAAGCACC | |
| OXY2 | TTTTTTGCATGCTAGCGTGGCCGTTCC | |
| phzHup | CGCACGGATCCTTTCAGAATGTTC | |
| phzHlow | GCCACGCCAAGCTTCACGCTCA | |
| BLA1 | GGCCCCAGTGCTGCAATGATAC | |
| BLA2 | GAGTATTCAACATTTCCGTGTCGC | |
| SAC1 | GATGTTTTCTTGCCTTTGATGTTC | |
| SAC2 | GTCTTTGCATTAGCCGGAGATC |
Phz+, strain produces phenazines; Phz−, strain does not produce phenazines; Phz(?) strains was not tested for phenazine production; iodinin, 1,6-dihydroxyphenazine 5,10-di-N-oxide; 2-OH-PCA, 2-hydroxyphenazine-1-carboxylic acid; 2-OH-PHZ, 2-hydroxyphenazine; aeruginosin A, 5-methyl-7-amino-1-carboxyphenazinium betaine; aeruginosin B, 5-methyl-7-amino-1-carboxy-3-sulfophenazinium betaine; bla, β-lactamase; Rifr, rifampin resistance; Tetr, tetracycline resistance.
ATCC, American Type Culture Collection.
DNA manipulations.
Standard methods were used for plasmid DNA isolation, restriction enzyme digestion, agarose gel electrophoresis, ligation, and transformation (2). Pseudomonad cells were electroporated with a Gene Pulser system (Bio-Rad Laboratories, Hercules, Calif.) by using the procedure of Enderle and Farwell (17). Total DNA from P. aeruginosa was isolated and purified by using a cetyltrimethylammonium bromide miniprep procedure (2).
A 6.4-kb DNA probe containing the entire phz locus from P. fluorescens 2-79 (GenBank accession number L48616) was generated by PCR performed with oligonucleotide primers PHZ-UP and PHZ-LOW (Table 1). The amplification was carried out by using a 50-μl reaction mixture containing 1× eLONGase buffer (Life Technologies, Inc., Rockville, Md.), 2 mM MgSO4, 3.0% dimethyl sulfoxide (Sigma Chemical Co., St. Louis, Mo.), 200 μM (each) dGTP, dATP, dTTP, and dCTP (Perkin-Elmer, Norwalk, Conn.), 10 pmol of each primer, 0.7 μl of eLONGase enzyme mixture (Life Technologies, Inc.), and 20 ng of purified genomic DNA from strain 2-79. All amplifications were performed with a PTC-200 thermal cycler (MJ Research, Inc., Watertown, Mass.). The cycling program included a 30-s initial denaturation step at 94°C, followed by 30 cycles of 94°C for 30 s, 64°C for 30 s, and 72°C for 7 min. The PCR product was gel purified and labeled with a random primer biotin labeling kit (NEN Life Science Products Inc., Boston, Mass.). Other PCR amplifications were carried out by using 25-μl reaction mixtures that contained 1× Taq DNA polymerase buffer (Life Technologies, Inc.), 1.5 mM MgCl2, 200 μM (each) dGTP, dATP, dTTP, and dCTP, 20 pmol of each primer, 1.2 U of Platinum Taq DNA polymerase (Life Technologies, Inc.), and 20 ng of target DNA.
DNA samples used for Southern hybridization were digested with restriction endonucleases, separated by electrophoresis in 0.8% agarose gels, and transferred to BrightStar-Plus nylon membranes (Ambion, Inc., Austin, Tex.) in 0.4 M NaOH, and there was subsequent cross-linking of the DNA by exposure of the membranes to UV radiation (2). Hybridization analyses were carried out as described previously (13). DNA-DNA hybrids were detected with a BrightStar nonisotopic detection kit (Ambion, Inc.) by using the manufacturer's protocol.
Insertional inactivation of phzM and phzS genes.
P. aeruginosa PAO1 phzM and phzS knockout mutants were generated by using a gene replacement strategy previously described by Schweizer (50). To mutagenize phzM, the gene was amplified by PCR with oligonucleotide primers METHYL1 and METHYL2 (Table 1) from cosmid 1G5 DNA of P. aeruginosa. The 1.26-kb PCR product was digested with EcoRI and BamHI, cloned into pNOT19, and inactivated by insertion of an 879-bp aacC1 cassette from pUCGM into a unique EcoRV site (Fig. 1A). The resulting plasmid, pNOT-ORF1-Gm, was digested with NotI and ligated with the 5.3-kb MOB3 sacB cassette (50).
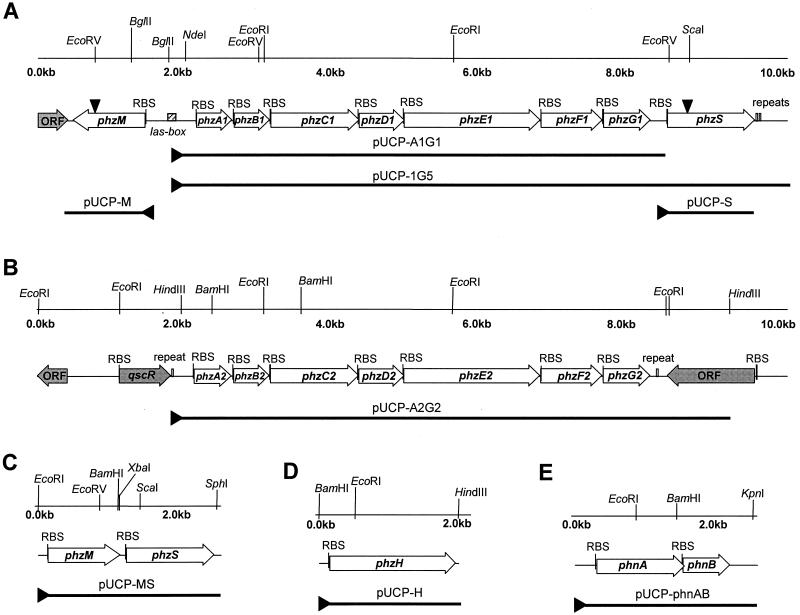
FIG. 1.
Restriction maps and locations of individual genes in regions of the P. aeruginosa PAO1 genome containing the phzA1B1C1D1E1F1G1(A) and phzA2B2C2D2E2F2G2 (B) operons, related DNA fragments contained in plasmids used in this study, and physical maps of plasmids pUCP-MS (C), pUCP-H (D), and pUCP-phnAB (E). DNA fragments contained in plasmids used in this study are indicated by thick lines. ▸, positions of the lac promoter from pUCP26; ▾, positions of insertions of the Gmr cassette. The shaded arrows indicate the positions of open reading frames (ORF) that were not relevant to the present study. RBS, ribosome binding site.
To inactivate phzS, the gene was amplified from cosmid 1G5 target DNA with oligonucleotide primers OXY1 and OXY2 (Table 1). The 1.24-kb PCR product was digested with XbaI and SphI, gel purified, and cloned into pNOT-19. The aacC1 cassette was then inserted into a unique ScaI site within phzS (Fig. 1A). The resulting plasmid, pNOT-ORF2-Gm, was ligated to the MOB3 sacB cassette and used for mutagenesis.
Plasmids containing the sacB cassette and the inactivated phzM or phzS gene were mobilized in P. aeruginosa PAO1 from E. coli S17-1. Following selection for double crossovers, isolates were screened by PCR for the presence of plasmid-borne bla and sacB genes in the genome. The β-lactamase gene was detected by PCR performed with primers BLA1 and BLA2 (Table 1), which amplified a 744-bp fragment of bla. The sacB cassette was detected by PCR performed with primers SAC1 and SAC2 (Table 1), which amplified a 1.05-kb DNA fragment. The cycling program included a 1-min initial denaturation step at 94°C, followed by 25 cycles of 94°C for 45 s, 53°C for 45 s, and 72°C for 1.25 min. The presence of mutated alleles of phzS and phzM was confirmed by PCR performed with oligonucleotide primers MET1 and MET2 and oligonucleotide primers ORF2UP and ORF2LOW (Table 1), respectively.
DNA sequencing and analysis.
DNA was sequenced by using an ABI PRISM dye terminator cycle sequencing kit (Perkin-Elmer, Norwalk, Conn.). All oligonucleotides were obtained from Operon Technologies, Inc. (Alameda, Calif.). Sequence data were compiled and analyzed with the Genetics Computer Group package (22) and the OMIGA 2.0 software package (Oxford Molecular Ltd., Oxford, United Kingdom). A database search for similar protein sequences was carried out with OMIGA's BLAST tool. A probable domain similarity search was performed by using the PROSITE (European Molecular Biology Laboratory, Heidelberg, Germany) (3) and ISREC ProfileScan (Swiss Institute for Experimental Cancer Research, Epalinges, Switzerland) (4) web servers. The significance of the similarity of a predicted protein to known proteins was determined by calculating the binary comparison score (Z score) as described elsewhere (13). Similarities with Z scores grater than 9 were considered significant.
Multiple sequence alignments were constructed with OMIGA's ClustalW. Phylogenetic analyses were carried out with PHYLIP 3.57c software (20). Distance matrices were generated with PROTDIST. Trees were inferred from the distances by using NEIGHBOR and FITCH with global rearrangement. Protein parsimony analyses were carried out with the PROTPARS program of the PHYLIP package. PHYLIP's SEQBOOT program was used for bootstrap analyses.
The present study was greatly facilitated by the use of the P. aeruginosa genome web site resources (http://www.pseudomonas.com).
Phenazine transformation assays.
Cultures of E. coli JM109 bearing pUCP26, pUCP-M, pUCP-S, pUCP-H, or pUCP-phnAB (Table 1) were grown in 2× YT (2) supplemented with tetracycline and were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 600 nm of 0.6. Immediately after induction, PCA was added to a final concentration of 0.3 mg/ml from a 25 mM stock solution in 5% (wt/vol) NaHCO3, and the cultures were grown for 6 h. Samples were extracted with chloroform at 3-h intervals and analyzed for phenazine composition by reverse-phase high-performance liquid chromatography (HPLC).
Cross-feeding experiments.
Overnight cultures of E. coli JM109 bearing pUCP26-based plasmids were diluted 1:100 in fresh 2× YT broth supplemented with tetracycline, grown with shaking to an optical density at 600 nm of 0.6, and induced with 0.5 mM IPTG. Immediately after induction, PCA was added to a concentration of 0.3 mg/ml, and the cultures were grown for 6 h to allow phenazine metabolites to accumulate. Finally, E. coli cells were removed by centrifugation and passage through a 0.22-μm-pore-size filter, and the filtrates were added to broth cultures of E. coli JM109 bearing pUCP26, pUCP-S, or pUCP-M. After 6 h of bacterial growth, phenazines were extracted with chloroform and analyzed by reverse-phase HPLC and mass spectrometry.
Analyses of phenazine compounds.
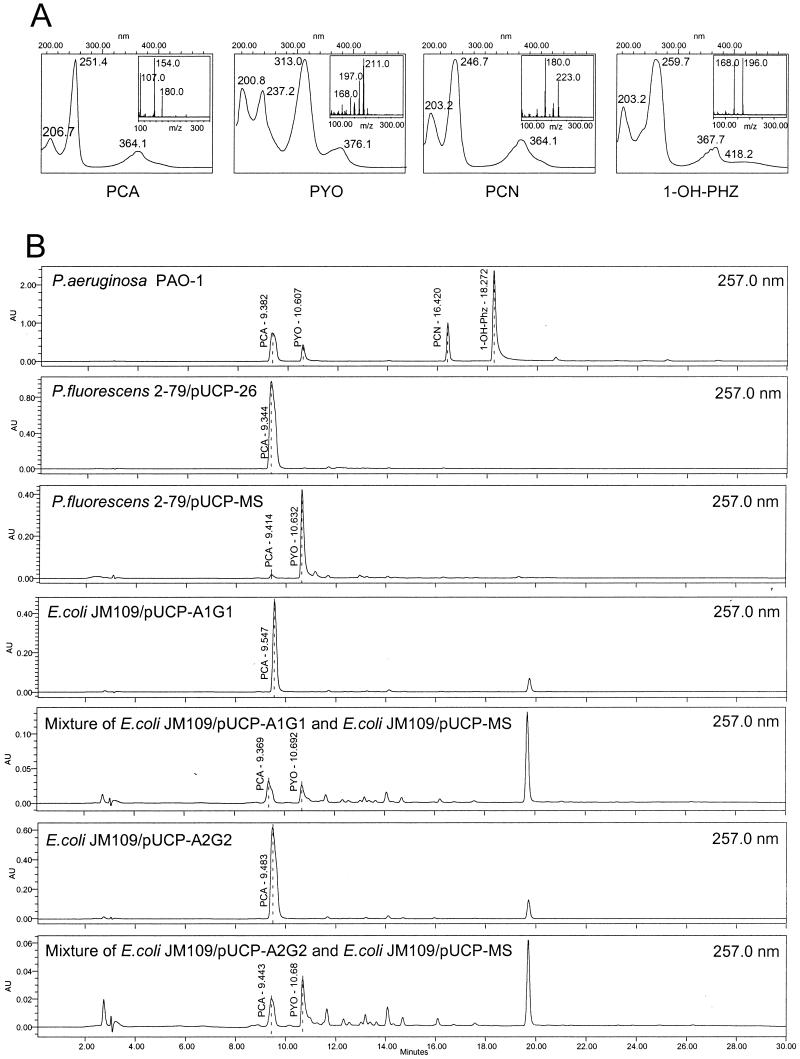
Phenazine compounds were extracted from bacterial cultures with chloroform, filtered (pore size, 0.2 μm), and subjected to C18 reverse-phase HPLC. Analyses were performed by using a Waters HPLC Integrity system consisting of an Alliance 2690 separation module, a 996 photodiode array detector, and an electron ionization ThermaBeam mass detector. Phenazine compounds were separated on a Symmetry C18 column (5 μm; 3.0 by 150 mm). The solvent flow rate was 350 μl min−1, and the flow consisted of 2 min of 8% acetonitrile–25 mM ammonium acetate, followed by a 25-min linear gradient to 80% acetonitrile–25 mM ammonium acetate. A filtered 1 M ammonium acetate solution was used to prepare all solvents. HPLC gradient profiles were monitored at spectral peak maxima of 257.0 and 313.0 nm, which are characteristic of PCA and pyocyanin in the solvent system used. Mass spectrometry total-intensity chromatogram analyses were performed from m/z 100 to 355 at a rate of 1 scan s−1. The nebulizer, ion source, and expansion region temperatures were 84, 220, and 80°C, respectively, with a helium flow rate of 15 lb/in2. The UV spectra and mass spectral characteristics of the major phenazine compounds detected are summarized in Fig. 2A.
FIG. 2.
UV and mass spectra of PCA, pyocyanin (PYO), PCN, and 1-OH-PHZ produced by P. aeruginosa PAO1 (A). (B) HPLC analyses of phenazine compounds produced by wild-type P. aeruginosa PAO1; P. fluorescens 2-79 harboring pUCP26; P. fluorescens 2-79 harboring pUCP-MS; E. coli JM109 harboring pUCP-A1G1, containing phzA1B1C1D1E1F1G1; a mixture of E. coli JM109 harboring pUCP-A1G1 and E. coli JM109 harboring pUCP-MS; E. coli JM109 harboring pUCP-A2G2, containing phzA2B2C2D2E2F2G2; and a mixture of E. coli JM109 harboring pUCP-A2G2 and E. coli JM109 harboring pUCP-MS. The identities of phenazine compound peaks were confirmed by spectral analysis and mass spectrometry. The retention times for PCA, pyocyanin, PCN, and 1-OH-PHZ with the solvent system used in this study were 9.4, 10.6, 16.4, and 18.2 min, respectively. The absorption maxima for PCA were at 251 and 364 nm; the absorption maxima for pyocyanin were at 237, 313, and 376 nm; the absorption maxima for PCN were at 246 and 364 nm; and the absorption maxima for 1-OH-PHZ were at 259 and 367 nm. AU, absorbance units.
Reference samples of pyocyanin and PCA were purchased from Colour Your Enzyme (Bath, Ontario, Canada). Mass spectrometry analyses of protonated (molecular weight, 211) and unprotonated (molecular weight, 210) forms of synthetic pyocyanin revealed the presence of an unknown m/z 224 ion peak [m/z (relative intensity) 224(13) 211(58) 210(100, M+) 197(11) 196(14) 182(60, M+-CO) 181(44, M+-·CHO) 168(18) 167(29, M+-NCHO) 149(14) 102(7) 91(16, C7H7+) 79(13) 78(8) 77(16, C6H5+) 76(10) 75(5)] that was not detected (Fig. 2) when the same reference samples were analyzed with a Voyager matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometer (PerSeptive Biosystems, Framingham, Mass.), in which no heating of the sample occurred. The predominant peak detected was at m/z 211, which corresponded to the protonated form of pyocyanin. The MALDI-TOF analyses were carried out in dihydrobenzoic and α-cyano-4-hydroxycinnamic acids with an acceleration voltage of 2,500 V (positive ion), a grid voltage of 60%, a guide wire voltage of 0.03%, and a delay of 150. We concluded that the m/z 224 peak was the product of a thermally promoted intermolecular reaction in the ThermoBeam mass spectral detector, which is consistent with the observations of Watson et al. (60).
Phenazine compounds extracted from P. chlororaphis ATCC 17809 and ATCC 17411 were used as a source of reference material for PCN because the synthetic compound was not available. Both of these strains are well characterized and are known to produce large amounts of PCN (52). The retention time, UV spectra, and mass spectral characteristics of the major phenazine compounds produced by each of these strains were identical to those of the other and to those of the putative PCN peak observed in extracts from P. aeruginosa or E. coli expressing phzH from PAO1 in the presence of PCA (data not shown).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the nucleotide sequence data for the phzA1B1C1D1E1F1G1 and phzA2B2C2D2E2F2G2 operons from P. aeruginosa PAO1 are AF005404 and AE004616, respectively.
RESULTS
Detection, cloning, and sequence analysis of phzA1B1C1D1E1F1G1 and phzA2B2C2D2E2F2G2.
The phzA1B1C1D1E1F1G1 genes were initially detected in P. aeruginosa PAO1 by Southern hybridization of total DNA restriction digests with a probe containing the phzCD genes from P. fluorescens 2-79. The analysis identified a 2.4-kb PstI-EcoRI DNA fragment that hybridized strongly to the 2-79 phzCD probe (data not shown). The fragment was cloned into pUC19, and two positive clones, plasmids p137 and p143, were identified by colony hybridization with the phenazine-specific probe (Fig. 1). Sequence analysis indicated that the cloned DNA fragment contained a complete sequence similar to phzD and partial flanking sequences similar to phzC and phzE of 2-79. The DNA from plasmid p143 was then used as a probe to screen a P. aeruginosa PAO1 genomic library (a membrane with arrayed cosmid DNA was a kind gift from Stephen Lory). Four positive cosmid clones were identified, and clone 1G5 was chosen for further study. After restriction mapping, a 10-kb BglII-NotI DNA fragment containing the phenazine biosynthetic genes was subcloned into pUCP26, and the resulting plasmid, pUCP-1G5, was used to determine the complete nucleotide sequence of the phenazine biosynthetic locus phzA1B1C1D1E1F1G1 from P. aeruginosa PAO1 (Fig. 1A).
The complete DNA sequence of the operon (GenBank accession number AF005404) is identical to the corresponding region of the P. aeruginosa PAO1 genome that was described by Stover et al. (53). Computer analyses of the DNA sequence of clone 1G5 revealed seven open reading frames, designated phzA1 through phzG1, located in the P. aeruginosa PAO1 genome at positions 4,713,795 to 4,720,062 (Fig. 1A). A well-conserved ribosome binding site precedes each gene. The stop and start codons of open reading frames phzC1, phzD1, and phzE1 overlap, possibly reflecting translational coupling. A 16-bp region of imperfect dyad symmetry (5′-CTACCAGATCTTGTAG-3′) located 372 bp upstream of phzA1 is similar to putative lux boxes found upstream of the phzA gene in P. fluorescens 2-79 and P. aureofaciens 30-84 (36, 43), suggesting that expression of the gene cluster is regulated in a cell density-dependent manner. Sequence comparisons indicated that phzA1B1C1D1E1F1G1 from P. aeruginosa is structurally and functionally homologous to the phenazine biosynthetic loci of P. fluorescens 2-79 (36), P. aureofaciens 30-84 (43), and P. chlororaphis PCL1391 (10) (data not shown).
Similarity searches performed with the PAO1 genome revealed a second copy of the phenazine cluster containing seven genes, designated phzA2B2C2D2E2F2G2 (Fig. 1B). As in the first copy, each gene is preceded by a well-conserved ribosome binding site, and the stop and start codons of phzC2, phzD2, and phzE2 overlap. A putative transcriptional terminator consisting of an 18-bp region of dyad symmetry (5′-ATAACCGCAAGCGGTTAT-3′) was identified 30 bp downstream of the phzG2 stop codon. The phzA2B2C2D2E2F2G2 gene cluster is located approximately 2.6 Mb from phzA1B1C1D1E1F1G1 at positions 2,070,685 to 2,076,985 of the P. aeruginosa PAO1 genome. The two phz operons are 98.3% identical at the DNA level. Essentially all of the sequence divergence occurs in the first pair of genes, phzA1B1 and phzA2B2; the regions containing phzC through phzG are nearly identical. Notably, the lux box in the putative promoter region upstream of phzA1 was not found upstream of phzA2. A cosmid clone bearing the full-length phzA2B2C2D2E2F2G2 gene cluster, clone 1H4, was identified by comparative Southern hybridization analyses performed with a 3.1-kb ScaI fragment containing phzE1, phzF1, and phzG1 (data not shown).
Identification of phzM, phzS, and phzH genes in the P. aeruginosa PAO1 genome database.
We identified two potential phenazine-modifying genes in P. aeruginosa PAO1 by analysis of the genomic region containing phzA1B1C1D1E1F1G1. This operon is flanked by genes designated PA4209 and PA4217 in the PAO1 database (53). PA4209, referred to in this paper as phzM, is preceded by a putative ribosome binding site, GAGAGA, and spans positions 4,713,098 to 4,712,094 in the PAO1 genome. The phzA1B1C1D1E1F1G1 operon and phzM are transcribed divergently and are separated by 695 bp (Fig. 1A). phzM encodes a 334-residue protein with a calculated molecular mass of 36.4 kDa that most closely resembles O-demethylpuromycin-O-methyltransferase (DmpM) from Streptomyces anulatus (NCBI accession number P42712; 30.1% identity; Z score, 72.4), a putative O-methyltransferase (SC5C11.09c) from Streptomyces coelicolor (accession number CAB76315; 30.9% identity; Z score, 64.4), O-methyltransferase (MmcR) from Streptomyces lavendulae (accession number AAD32742; 29.6% identity; Z score, 58.2), and carminomycin 4-O-methyltransferase (DauK) from Streptomyces sp. (accession number AAB16938; 28.9% identity; Z score, 44.9). A ProfileScan database search revealed a generic methyltransferase motif (residues 234 to 274) and a conserved S-adenosyl-l-methionine (SAM) binding domain (residues 164 to 273) within PhzM.
PA4217, referred to in this paper as phzS, is located 236 bp downstream from phzG1 and spans positions 4,720,300 to 4,721,508 of the P. aeruginosa PAO1 genome. phzS is preceded by a well-conserved ribosome binding site, AAGGAA, and encodes a 402-amino-acid protein with a molecular mass of 43.6 kDa. PhzS is similar to bacterial monooxygenases, including salicylate hydroxylase (NahW) from Pseudomonas stutzeri (NCBI accession number AAD02157; 27.7% identity; Z score, 38.9), a putative salicylate hydroxylase (SCE29.14c) from S. coelicolor (accession number T36193; 37.5% identity; Z score, 50.6), and putative salicylate hydroxylases from Bordetella pertussis (accession number AAC46266; 32.1% identity; Z score, 48.2) and Acinetobacter sp. (accession number AAC27110; 28.7% identity; Z score, 43.4). A Pfam database search revealed a monooxygenase motif (residues 157 to 366) and a putative N-terminal NAD binding site (residues 7 to 36) within PhzS.
A third potential phenazine-modifying gene was identified in the P. aeruginosa genome by using the sequence of the recently described phzH gene from P. chlororaphis PCL1391 (10) as a query sequence in a BLAST search (10). This search identified the PA0051 gene, which exhibits 80.3% identity to phzH and spans positions 66,303 to 68,135 in the P. aeruginosa genome (53). This gene, referred to in this paper as phzH, is preceded by a well-conserved ribosome binding site, GAGG, and encodes a 610-amino-acid protein with a calculated molecular mass of 69.5 kDa. The deduced amino acid sequence encoded by phzH most closely resembles the sequence of phenazine-modifying enzyme PhzH from P. chlororaphis (NCBI accession number AAF17502; 80.3% identity; Z score, 556.5) and is similar to the sequences of bacterial asparagine synthetases, including TcsG from Streptomyces aureofaciens (accession number BAB12569; 46.1% identity; Z score, 267.9), PA2084 from P. aeruginosa (accession number AAG05472; 45.4% identity; Z score, 291.6), and AsnO from Bacillus subtilis (accession number O05272; 40.2% identity; Z score, 273.3). Two well-conserved Pfam protein motifs commonly associated with asparagine synthetases were found in PhzH: the glutamine amidotransferase class II signature (residues 43 to 153) and an asparagine synthetase signature (residues 199 to 608).
Functional analyses of the core phenazine loci and the phnAB genes from P. aeruginosa PAO1.
To determine the function of the phenazine genes from P. aeruginosa, each of the seven-gene operons was cloned in pUCP26 under control of the lac promoter. The resulting plasmids, pUCP-A1G1 and pUCP-A2G2 (Fig. 1), and pUCP-1G5 were expressed in P. fluorescens M4-80, which does not produce phenazines, and in P. fluorescens 2-79, which produces a single phenazine compound, PCA. Extracts from each plasmid-bearing strain were analyzed by HPLC and mass spectrometry, and the data were compared to elution profiles and spectra generated with synthetic pyocyanin or extracts from wild-type P. fluorescens 2-79 and P. aeruginosa PAO1. Extracts from P. aeruginosa PAO1 contained a mixture of pyocyanin, 1-OH-PHZ, and PCA. Only PCA was detected in extracts from strain M4-80 containing either pUCP-A1G1 or pUCP-A2G2 (Table 2). Similar results were obtained for derivatives of strain 2-79, in which the presence of either plasmid led to increased levels of PCA accumulation (data not shown) but did not change the range of compounds synthesized (Table 2). In contrast, strain 2-79 bearing pUCP-1G5 produced large amounts of both PCA and the derivative 1-OH-PHZ (Table 2). Plasmid pUCP-1G5 contained the complete phzA1B1C1D1E1F1G1 operon and 3.3 kb of additional downstream DNA.
TABLE 2.
Detection by HPLC and HPLC-mass spectrometry of phenazine compounds produced by Pseudomonas strains
| Strain/plasmid | Phenazine compound(s) detected |
|---|---|
| P. fluorescens 2-79/pUCP26 | PCA |
| P. fluorescens 2-79/pUCP-1G5 | PCA, 1-OH-PHZ |
| P. fluorescens 2-79/pUCP-A1G1 | PCA |
| P. fluorescens 2-79/pUCP-A2G2 | PCA |
| P. fluorescens 2-79/pUCP-phnAB | PCA |
| P. fluorescens 2-79/pUCP-MS | PCA, pyocyanin |
| P. fluorescens M4-80/pUCP26 | None |
| P. fluorescens M4-80/pUCP-A1G1 | PCA |
| P. fluorescens M4-80/pUCP-A2G2 | PCA |
| P. fluorescens M4-80/pUCP-phnAB | None |
| P. aeruginosa PAO1 | PCA, pyocyanin, 1-OH-PHZ, PCN |
| P. aeruginosa PAOmxM | PCA, 1-OH-PHZ, PCN |
| P. aeruginosa PAOmxM/pUCP-M | PCA, pyocyanin, 1-OH-PHZ, PCN |
| P. aeruginosa PAOmxS | PCA, PCN, red pigment |
| P. aeruginosa PAOmxS/pUCP-S | PCA, pyocyanin, 1-OH-PHZ, PCN |
Previous studies of tryptophan biosynthesis in P. aeruginosa identified the phnAB genes(18), which encode an anthranilate synthase homologue that complements trpE and trpE(G) mutants of E. coli. PhnAB also contributes to pyocyanin synthesis in P. aeruginosa, and pyocyanin production by phnA mutants is greatly reduced (18). To further evaluate the role of phnAB in pyocyanin synthesis, the genes were excised from M13-10116 phage (the phage DNA was a kind gift from M. G. Bangera) by digestion with SacI and KpnI, treated with T4 DNA polymerase, and inserted into the SmaI site of pUCP26. The resulting plasmid, pUCP-phnAB, was introduced into P. fluorescens M4-80R and 2-79 by electroporation, and transformants were assayed for phenazine production by HPLC. In no case was pyocyanin produced, and strain 2-79 produced only PCA (Table 2) in quantities comparable to those detected in 2-79 containing pUCP26.
We also conducted phenazine transformation assays to determine whether phnAB conferred the ability to transform PCA, a precursor of pyocyanin and 1-OH-PHZ, into other phenazine compounds. This approach has proven to be useful in characterizing other genes encoding phenazine-modifying enzymes (13). Cultures of E. coli JM109 containing pUCP-phnAB or the control plasmid pUCP26 were induced with IPTG and incubated for 9 h in the presence of 0.3 mg of PCA ml−1. HPLC analyses revealed that the extracts from E. coli JM109(pUCP-phnAB) were identical to those from the control strain, and no conversion of PCA to pyocyanin or any other phenazine derivative was detected (data not shown).
Functional analyses of phzM, phzS, and phzH
To determine whether phzM, phzS, and phzH encode phenazine-modifying enzymes, gene function was analyzed in E. coli transformed with pUCP-M, pUCP-S, pUCP-H, and pUCP-MS (Table 1). These plasmids were constructed by amplifying phzM, phzS, and phzH from PAO1 genomic DNA with oligonucleotide primers METHYL1 and METHYL2, oligonucleotide primers OXY1 and OXY2, and oligonucleotide primers phzHup and phzHlow, respectively (Table 1). The PCR products were digested with restriction endonucleases, gel purified, and cloned into pUCP26 under control of the lac promoter (Fig. 1). To construct pUCP-MS, pUCP-M was digested with XbaI and SphI, and the PCR fragment containing phzS was cloned immediately downstream of phzM. All plasmids were single-pass sequenced to confirm the integrity of cloned gene(s).
Induced cultures of E. coli bearing pUCP-MS converted PCA to pyocyanin within 6 h, whereas no such conversion occurred in control cultures harboring only pUCP26 (data not shown). We did not detect derivatives of PCA in supernatants of E. coli JM109(pUCP-M) cultures incubated under similar conditions, although these cultures were an intense dark red. Induced cultures of E. coli JM109 bearing pUCP-S and E. coli bearing pUCP-H converted PCA to 1-OH-PHZ and PCN, respectively (data not shown). We were able to restore the complete pyocyanin biosynthetic pathway in E. coli by mixing induced cultures containing either pUCP-A1G1 or pUCP-A2G2 with a culture containing pUCP-MS. HPLC analysis revealed that in both cases the mixed cultures produced small but detectable amounts of pyocyanin (Fig. 2B).
Plasmid pUCP-MS also was introduced into P. fluorescens 2-79. Overnight broth cultures of 2-79(pUCP-MS) were intensely blue, which was consistent with synthesis of pyocyanin, and the results of HPLC and mass spectrometry analyses (Fig. 2B) confirmed that large amounts of this compound were present.
Effect of phzM and phzS mutations on pyocyanin production.
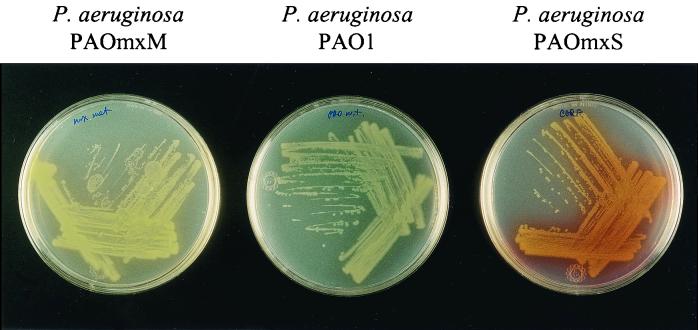
The gene replacement approach was used to evaluate the role of the phenazine-modifying genes phzM and phzS in pyocyanin biosynthesis in P. aeruginosa PAO1. Mutant derivatives with mutations in phzM (strain PAOmxM) or phzS (strain PAOmxS) exhibited unusual pigment phenotypes when they were cultivated on PIA. Whereas cultures of wild-type PAO1 were blue due to production of pyocyanin, cultures of P. aeruginosa PAOmxM were yellow, and P. aeruginosa PAOmxS produced large amounts of a dark red water-soluble pigment (Fig. 3). Chloroform extracts prepared from cultures of these mutants grown in PB medium were analyzed by HPLC and mass spectrometry (Table 2). Both mutants were able to synthesize PCA and PCN but did not produce pyocyanin. In fact, the phzM mutant produced and secreted more PCN than the parent strain (data not shown). PAOmxM produced 1-OH-PHZ, although it produced markedly less than the parental strain, while no 1-OH-PHZ was detected in extracts from PAOmxS. Finally, complementation of PAOmxM and PAOmxS with pUCP-M and pUCP-S, respectively, restored production of pyocyanin (Table 2).
FIG. 3.
Production of phenazine compounds by wild-type P. aeruginosa PAO1 (center plate) and marker exchange mutants P. aeruginosa PAOmxM and PAOmxS (left and right plates, respectively) on PIA plates.
DISCUSSION
The present study demonstrated clearly that the opportunistic human pathogen P. aeruginosa PAO1 contains two complete copies of a seven-gene operon shown previously (13, 36, 43) to be responsible for the synthesis of PCA. We also screened a collection of 30 bacterial strains, including isolates of P. aeruginosa, P. fluorescens, P. chlororaphis, P. aureofaciens, Pseudomonas putida, B. cepacia, B. phenazinium, and B. iodinum (Table 1), for the presence of these core phenazine biosynthetic genes by performing Southern hybridization with probes spanning phzA through phzG. Similar sequences were not detected in phenazine-producing isolates of B. cepacia, B. phenazinium, and B. iodinum under moderately stringent conditions (6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 55°C) despite the fact that the same mechanism of phenazine synthesis is thought to function in all of these genera (34, 52, 57). Only total DNA from 26 known phenazine-producing strains of P. aeruginosa, P. aureofaciens, P. chlororaphis, and P. fluorescens hybridized strongly to the phenazine gene probe (data not shown), indicating not only that the operon is conserved in phenazine-producing fluorescent pseudomonads but also that it may be unique to them.
Of the pseudomonads, only P. aeruginosa has been found to contain two copies of the phenazine operon. Both copies are homologous to phenazine biosynthetic loci from P. aureofaciens, P. fluorescens, and P. chlororaphis (13, 36, 43), and both are functional, as each enabled the non-phenazine-producing strain P. fluorescens M4-80R to synthesize PCA, which was implicated previously as a pyocyanin precursor (8). Although the two phenazine operons of P. aeruginosa PAO1 are 98% identical overall, they differ markedly in their promoter regions. In fluorescent Pseudomonas species, phenazine production occurs mainly during the stationary phase and is regulated by quorum sensing and the highly conserved, global, two-component signal transduction system, GacA-GacS (44, 47). The N-acyl-homoserine lactone-mediated regulation of phenazine synthesis in P. aeruginosa is complex and involves at least two different quorum-sensing circuits, las and rhl, organized in a hierarchical cascade (41). Whiteley et al. (63) recently identified the phzA1B1C1D1E1F1G1 operon among the genes controlled by quorum sensing in P. aeruginosa. This operon is preceded by a well-conserved putative promoter element required for quorum control, the las box (Fig. 1). Interestingly, no such element is present upstream of phzA2B2C2D2E2F2G2. On the other hand, analyses of the PAO1 genome have revealed a gene, qscR, that is similar to lasR and rhlR and is located upstream of phzA2B2C2D2E2F2G2 (Fig. 1). According to Chugani et al. (12), qscR encodes a protein that negatively regulates expression of a number of quorum-sensing-controlled genes via repression of lasI and lasI-regulated target genes, including phzA1B1C1D1E1F1G1. It is tempting to speculate that the presence of two differentially regulated phenazine biosynthetic operons may give the bacterium greater flexibility in modulating the amount or range of phenazine compounds produced depending on the phase of growth or in response to environmental signals.
Recently, Mahajan-Miklos et al. (35) demonstrated that phenazines are involved in the killing of C. elegans by the clinical isolate P. aeruginosa PA14. The pathogenicities of two pyocyanin-deficient mutants of P. aeruginosa PA14, both containing a TnphoA insertion in a gene similar to phzB1, were reduced in the Caenorhabditis killing model, in Arabidopsis leaf infiltration assays, and in the burned mouse model. Interestingly, inactivation of phzB reduced pyocyanin production in strain PA14 but did not eliminate it entirely (35). These results are consistent with expression of a second, differently regulated copy of the phz genes in the PA14 genome. We compared the proteins encoded by phzA and phzB from P. aeruginosa PA14 with the corresponding products of the P. aeruginosa PAO1 genes. The results of cluster analyses (data not shown) indicated that the products of the phzA and phzB genes of strain PA14 most closely resemble PhzA2 and PhzB2 from P. aeruginosa PAO1 (all bootstrap values were ≥81%). These data suggest that PAO1 is not unique in containing two homologous phz operons, and the trait may be common among strains of P. aeruginosa. It remains to be determined whether this unusual feature is typical of both environmental and clinical isolates and if it is relevant to the role in pathogenesis of pyocyanin and other phenazines produced by P. aeruginosa.
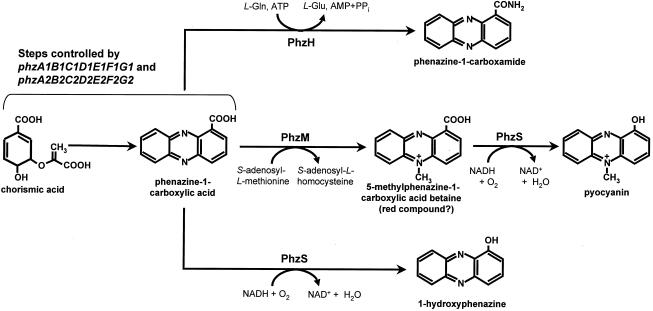
Our results indicate that specific genes in P. aeruginosa are required for conversion of PCA to pyocyanin and PCN. At least two modifications would be expected in the conversion of PCA to pyocyanin: addition of the N-methyl group, converting PCA to 5-methylphenazine-1-carboxylate betaine; and hydroxylative decarboxylation of the betaine to form pyocyanin (8, 52, 57). Using DNA sequence information from the Pseudomonas Genome Project (www.pseudomonas.com), we identified two potential genes in the vicinity of the phzA1B1C1D1E1F1G1 operon (Fig. 1) that could encode the required PCA-modifying enzymes. Several lines of evidence support the idea that phzM and phzS play essential roles in the pyocyanin biosynthetic pathway. Transformation of the PCA-producing strain P. fluorescens 2-79 with phzM and phzS triggered synthesis of large amounts of pyocyanin (Table 2). We also were able to reconstitute the full pathway for biosynthesis of pyocyanin in E. coli by mixing induced cultures expressing phzMS with cultures expressing phzA1B1C1D1E1F1G1 or phzA2B2C2D2E2F2G2 (Fig. 2B). Finally, E. coli expressing phzM and phzS efficiently converted exogenously supplied PCA to pyocyanin. These data are consistent with results of mutagenesis studies in which P. aeruginosa marker exchange mutants with mutations in phzM and phzS failed to produce detectable amounts of pyocyanin (Table 2).
Our results support the results of previous studies (8, 21, 51) that predicted the existence of a phenazine methyltransferase and a phenazine-specific hydroxylase in the pyocyanin biosynthetic pathway. Byng et al. (8) attempted to determine the exact relationship among the different phenazine compounds synthesized by P. aeruginosa. Their study yielded three different groups of mutant strains: one group that produced no phenazines; a second group that synthesized PCA and PCN; and a third group that produced PCA, PCN, and the dark red compounds aeruginosins A and B (8, 9). The members of the second group of mutants identified by Byng et al. appear to be phenotypically identical to the phzM knockout strain, while the members of the third group resemble the phzS knockout strain (Table 2). Our data raise questions concerning the identity of the dark red compound characterized previously as a complex of aeruginosin A (5-methyl-7-amino-1-carboxymethylphenazinium betaine) and aeruginosin B (5-methyl-7-amino-1-carboxy-3-sulfophenazinium betaine) and thought to be derived from PCA via a specific biosynthetic pathway (8). We were surprised to find that E. coli expressing phzM alone converted PCA to a dark red compound(s) similar in appearance to the product(s) synthesized by phzS mutants of P. aeruginosa (Fig. 3). Unfortunately, we were unable to identify this material by C18 reverse-phase HPLC because the pigment is extremely hydrophilic and could not be extracted with the solvents commonly used to recover phenazines. These properties are similar to those of the aeruginosins described previously by Herbert and Holliman (27) and Holliman (28). We hypothesize that the dark red pigment synthesized by P. aeruginosa PAOmxS mutants and by E. coli expressing phzM is not a complex of aeruginosins A and B but rather is a colored intermediate (probably 5-methylphenazine-1-carboxylic acid betaine) formed through methylation of PCA by PhzM.
PhzM is a predicted 36.4-kDa protein similar to enzymes involved in the methylation of polyketide antibiotics by Streptomyces spp. Streptomycetes produce a variety of bioactive polyketides, many of which contain methyl groups introduced via the action of N- and O-methyltransferases (16, 66). PhzM most closely resembles O-methyltransferases from Streptomyces spp. and shares with them a putative catalytic domain (residues 137 to 177) that might be universal among SAM-dependent methyltransferases (4). The presence of a potential SAM binding site in the N-terminal part of PhzM (residues 67 to 176) further supports the idea that PhzM functions as a methyltransferase.
The second putative phenazine-modifying gene, phzS, is located immediately downstream of phzA1B1C1D1E1F1G1 (Fig. 1) and encodes a predicted 43.6-kDa protein similar to putative salicylate hydroxylases from S. coelicolor (GenBank accession numbers T36193 and CAB95795), Bordetella pertussis (AAC46266), Acinetobacter sp. strain ADP1 (AAC27110), and P. aeruginosa (AAG06716) and NahW from P. stutzeri AN10 (AAD02157). NahW is of particular interest as it is the best-characterized enzyme related to PhzS. According to Bosch et al. (5), nahW encodes a flavoprotein monooxygenase with broad substrate specificity that catalyzes conversion of salicylic acid and its derivatives to catechol. In a detailed sequence analysis of NahW, they identified several conserved motifs present in bacterial salicylate hydroxylases. A sequence comparison of PhzS with NahW and other salicylate hydroxylases revealed that PhzS contains the consensus NADH binding domain (159-DVLVGADGIHSAVR-172), putative N-terminal flavin adenine dinucleotide binding site (11-GAGIGG-16), and substrate active site (303-GRITLLGDAAHLMYPMGANGA-323). Not only is PhzS clearly involved in the biosynthesis of pyocyanin, but it is also probably responsible for the production of 1-OH-PHZ by P. aeruginosa. E. coli expressing phzS efficiently converted exogenously supplied PCA to 1-OH-PHZ, and we also detected 1-OH-PHZ in extracts from the P. aeruginosa phzM mutant (Table 2). Thus, our results suggest that 1-OH-PHZ is formed from PCA via enzymatic synthesis and not through light-mediated decomposition of pyocyanin, as proposed previously(45). In fact, no traces of decomposition were found in solutions of synthetic pyocyanin after 5 days of incubation at room temperature under direct light (data not shown).
We suggest that two steps are involved in the synthesis of pyocyanin from PCA (Fig. 4). In the first step, catalyzed by the SAM-dependent methyltransferase PhzM, PCA is converted to 5-methylphenazine-1-carboxylic acid betaine. The second step, catalyzed by the NADH (or NADPH)-dependent flavoprotein monooxygenase PhzS, involves hydroxylative decarboxylation of 5-methylphenazine-1-carboxylic acid betaine to pyocyanin. In cross-feeding studies to determine the order of the two reactions, we detected pyocyanin only when cultures of E. coli expressing phzS were incubated with filtered extracts from cultures expressing phzM (data not shown). When cultures expressing phzM were incubated for 6 h with filtered extracts from cultures expressing phzS, only PCA and 1-OH-PHZ were detected. We speculate that the ability of PhzS to convert PCA to 1-OH-PHZ can be explained by the broad substrate specificity of the enzyme.
FIG. 4.
Proposed mechanism for the synthesis of pyocyanin, 1-OH-PHZ, and PCN in P. aeruginosa PAO1.
The gene for biosynthesis of PCN was identified in the P. aeruginosa database by using the recently discovered phzH gene from P. chlororaphis (10) as the query sequence. E. coli expressing phzH converted exogenously supplied PCA to PCN (data not shown), confirming that phzH from P. aeruginosa is functionally homologous to its counterpart from P. chlororaphis. The deduced product of these genes is similar to class II glutamine amidotransferases, namely, to asparagine synthases. The closest functionally characterized enzyme with similarity to PhzH is the asparagine synthetase AsnO from B. subtilis. Bacterial asparagine synthetases are grouped into two families (68); the first comprises enzymes similar to E. coli AsnA that accept only ammonia as the amino donor, whereas the members of the second family resemble E. coli AsnB and can use both ammonia and glutamine as amino donors (48). According to Yoshida et al. (68), asnO encodes an AsnB type of asparagine synthase crucial for sporulation in B. subtilis. Based on the sequence similarity and function data, we propose that phzH of P. aeruginosa encodes a glutamine-dependent, phenazine-specific amidotransferase that catalyzes the amidation of PCA to PCN (Fig. 4).
A question then arises concerning the role of the phnAB genes, which have been implicated previously in pyocyanin production. These genes initially were characterized by Essar et al. (18) and were found to encode products resembling the anthranilate synthase subunits TrpE and TrpG. However, unlike trpE and trpG, which encode a typical tryptophan-specific anthranilate synthase in P. aeruginosa, phnA and phnB are cotranscribed and expressed during the stationary phase (18). Essar et al. also demonstrated that phnAB could complement trpE and trpE(G) mutants of E. coli and that PhnAB was not feedback inhibited by tryptophan. Insertional inactivation of phnA significantly reduced pyocyanin synthesis, an observation confirmed later by other workers (1, 35, 46); however, production was not eliminated completely, and mutants synthesized pyocyanin at levels that were 22 to 34% of the wild-type levels, which was attributed to the activity of TrpEG (18). Essar et al. suggested that PhnAB functions as a phenazine-specific anthranilate synthase, providing anthranilic acid as a precursor for pyocyanin production.
To further evaluate the role of these genes, we expressed phnAB in P. fluorescens 2-79, which produces only PCA, and in M4-80R, which does not produce phenazine. The introduced genes neither enhanced nor enabled PCA production in these strains (Table 2), nor did E. coli JM109 expressing phnAB convert PCA to other phenazines. Recent studies of the mechanism of phenazine nucleus assembly indicate that PCA is synthesized via the conversion, catalyzed by PhzE and PhzD, of 2-amino-2-deoxyisochorismic acid to 2,3-dihydroxy-3-hydroxyanthranilate (M. G. McDonald, D. V. Mavrodi, L. S. Thomashow, and H. G. Floss, unpublished data). Thus, anthranilic acid is not a precursor in the formation of the phenazine nucleus. Therefore, it seems likely that PhzE1 and PhzE2 in P. aeruginosa are largely responsible for the conversion of chorismic acid to PCA via the same mechanism proposed (36) for PhzE in P. fluorescens 2-79. There is ample evidence that PhnA and PhnB influence pyocyanin production in P. aeruginosa (1, 18, 35, 46), but it seems unlikely that they participate directly in assembly of the phenazine nucleus. The precise role of these enzymes in P. aeruginosa metabolism, the identities of intermediates, and the products involved remain to be discovered. These uncertainties reinforce the fact that despite numerous biochemical studies of pyocyanin biosynthesis, the actual enzymology and genetic control are still not well understood and require further investigation.
ACKNOWLEDGMENTS
We are grateful to Stephen Lory and M. G. Bangera for providing the cosmids used in this study and to William Siems of the Laboratory for Biotechnology and Bioanalysis at Washington State University for the MALDI-TOF analysis of pyocyanin samples. We thank David M. Weller for his help, valuable discussions, and critical review of the manuscript and Karen Hansen for technical assistance.
REFERENCES
- 1.Anjaiah V, Koedam N, Nowak-Thompson B, Loper J E, Hofte M, Tambong J T, Cornelis P. Involvement of phenazines and anthranilate in the antagonism of Pseudomonas aeruginosa PNA1 and Tn5 derivatives toward Fusarium spp. and Pythiumspp. Mol Plant-Microbe Interact. 1998;11:847–854. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. J. New York, N. Y: Wiley and Sons; 1995. [Google Scholar]
- 3.Bairoch A, Bucher P, Hofmann K. The PROSITE database, its status in 1995. Nucleic Acids Res. 1995;24:189–196. doi: 10.1093/nar/24.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L. The Pfam protein families database. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch R, Moore E R B, Garcia-Valdes E, Pieper D H. NahW, a novel, inducible salicylate hydroxylase involved in mineralization of naphthalene by Pseudomonas stutzeriAN10. J Bacteriol. 1999;181:2315–2322. doi: 10.1128/jb.181.8.2315-2322.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britigan B E, Rasmussen G T, Cox C D. Augmentation of oxidant injury to human pulmonary epithelial cells by the Pseudomonas aeruginosasiderophore pyochelin. Infect Immun. 1997;65:1071–1076. doi: 10.1128/iai.65.3.1071-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budzikiewicz H. Secondary metabolites from fluorescent pseudomonads. FEMS Microbiol Rev. 1993;104:209–228. doi: 10.1111/j.1574-6968.1993.tb05868.x. [DOI] [PubMed] [Google Scholar]
- 8.Byng G S, Eustice D C, Jensen R. Biosynthesis of phenazine pigments in mutant and wild-type cultures of Pseudomonas aeruginosa. J Bacteriol. 1979;138:846–852. doi: 10.1128/jb.138.3.846-852.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carson M, Jensen R A. Phenotypic recognition of pyocyanin mutants in Pseudomonas aeruginosa. J Bacteriol. 1974;117:312–314. doi: 10.1128/jb.117.1.312-314.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin-A-Woeng T. Molecular basis of biocontrol of tomato foot and root rot by Pseudomonas chlororaphis strain PCL1391. Ph.D. thesis. Leiden, The Netherlands: University of Leiden; 2000. [DOI] [PubMed] [Google Scholar]
- 11.Chin-A-Woeng T F C, Bloemberg G V A, van der Bij J, van der Drift K M G M, Schripsema J, Kroon B, Scheffer R J, Keel C, Bakker P A H M, Tichy H-V, de Bruijn F J, Thomas-Oates J E, Lugtenberg B J J. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant-Microbe Interact. 1998;11:1069–1077. [Google Scholar]
- 12.Chugani S A, Whiteley M, Lee K M, D'Argenio D, Manoil C, Greenberg E P. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delaney S M, Mavrodi D V, Bonsall R F, Thomashow L S. phzO, a gene for biosynthesis of hydroxylated phenazine compounds in Pseudomonas aureofaciens30-84. J Bacteriol. 2001;183:318–327. doi: 10.1128/JB.183.1.318-327.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denning G M, Wolleweber L A, Railsback M A, Cox C D, Stoll L L, Britigan B E. Pseudomonaspyocyanin increases interleukin-8 expression by human airway epithelial cells. Infect Immun. 1998;66:5777–5784. doi: 10.1128/iai.66.12.5777-5784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denning G M, Railsback M A, Rasmussen G T, Cox C D, Britigan B E. Pseudomonaspyocyanine alters calcium signaling in human airway epithelial cells. Am J Physiol. 1998;274:L893–L900. doi: 10.1152/ajplung.1998.274.6.L893. [DOI] [PubMed] [Google Scholar]
- 16.Dickens M L, Ye J, Strohl W R. Analysis of clustered genes encoding both early and late steps in daunomycin biosynthesis by Streptomycessp. strain C5. J Bacteriol. 1995;177:536–543. doi: 10.1128/jb.177.3.536-543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enderle P J, Farwell M A. Electroporation of freshly plated Escherichia coli and Pseudomonas aeruginosacells. BioTechniques. 1998;25:954–958. doi: 10.2144/98256bm05. [DOI] [PubMed] [Google Scholar]
- 18.Essar D W, Eberly L, Hadero A, Crawford I P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans K, Passador L, Srikumar R, Tsang E, Nezezon J, Poole K. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1998;180:5443–5447. doi: 10.1128/jb.180.20.5443-5447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 21.Flood M E, Herbert R B, Holliman F G. Pigments of Pseudomonas species. V. Biosynthesis of pyocyanin and the pigments of Ps. aureofaciens. J Chem Soc Perkin Trans I. 1972;1972:622–626. doi: 10.1039/p19720000622. [DOI] [PubMed] [Google Scholar]
- 22.Genetics Computer Group. Program manual for the Wisconsin package, version 8. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 23.Georgakopoulos D G, Hendson M, Panopoulos N J, Schroth M N. Cloning of a phenazine biosynthetic locus of Pseudomonas aureofaciensPGS12 and analysis of its expression in vitro with the ice nucleation reporter gene. Appl Environ Microbiol. 1994;60:2931–2938. doi: 10.1128/aem.60.8.2931-2938.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamdan H, Weller D M, Thomashow L S. Relative importance of fluorescent siderophores and other factors in biological control of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens2-79 and M4-80R. Appl Environ Microbiol. 1991;57:3270–3277. doi: 10.1128/aem.57.11.3270-3277.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassan H M, Fridovich I. Mechanism of the antibiotic action of pyocyanine. J Bacteriol. 1980;141:156–163. doi: 10.1128/jb.141.1.156-163.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassett D J, Schweizer H P, Ohman D E. Pseudomonas aeruginosa sodA and sodBmutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177:6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herbert R B, Holliman F G. Pigments of Pseudomonasspecies. II. Structure of aeruginosin B. J Chem Soc. 1969;C:2517–2520. doi: 10.1039/j39690002517. [DOI] [PubMed] [Google Scholar]
- 28.Holliman F G. Pigments of Pseudomonasspecies. I. Structure and synthesis of aeruginosin A. J Chem Soc. 1969;C:2514–2516. doi: 10.1039/j39690002514. [DOI] [PubMed] [Google Scholar]
- 29.Hussain A S, Bozinovski J, Maurice D H, McLaughlin B E, Marks G S, Brien J F, Nakatsu K. Inhibition of the action of nitric oxide prodrugs by pyocyanin: mechanistic studies. Can J Pharmacol. 1996;75:398–406. [PubMed] [Google Scholar]
- 30.Jander G, Rahme L G, Ausubel F M. Positive correlation between virulence of Pseudomonas aeruginosamutants in mice and insects. J Bacteriol. 2000;182:3843–3845. doi: 10.1128/jb.182.13.3843-3845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korth H, Römer A, Budzikiewicz H, Pulverer G. 4,9-Dihydroxy-phenazine-1,6-dicarboxylic acid dimethylester and the “missing link” in phenazine biosynthesis. J Gen Microbiol. 1978;104:299–303. doi: 10.1099/00221287-104-2-299. [DOI] [PubMed] [Google Scholar]
- 32.Latifi A, Wilson M K, Foglino M, Bycroft B W, Stewart G S A B, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosaPAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 33.Lauredo I T, Sabater J R, Ahmed A, Botvinnikova Y, Abraham W N. Mechanism of pyocyanin- and 1-OH-PHZ-induced lung neutrophilia in sheep airways. J Appl Physiol. 1998;85:2298–2304. doi: 10.1152/jappl.1998.85.6.2298. [DOI] [PubMed] [Google Scholar]
- 34.Leisinger T, Margraff R. Secondary metabolites of the fluorescent pseudomonads. Microbiol Rev. 1979;43:422–442. doi: 10.1128/mr.43.3.422-442.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahajan-Miklos S, Tan M-W, Rahme L G, Ausubel F M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis eleganspathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 36.Mavrodi D V, Ksenzenko V N, Bonsall R F, Cook R J, Boronin A M, Thomashow L S. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens. J Bacteriol. 1998;180:2541–2548. doi: 10.1128/jb.180.9.2541-2548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazzola M, Cook R J, Thomashow L S, Weller D M, Pierson L S., III Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl Environ Microbiol. 1992;58:2616–2624. doi: 10.1128/aem.58.8.2616-2624.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller R, Rasmussen G T, Cox C D, Britigan B E. Protease cleavage of iron-transferrin augments pyocyanin-mediated endothelial cell injury via promotion of hydroxyl radical formation. Infect Immun. 1996;64:182–188. doi: 10.1128/iai.64.1.182-188.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parke J L. Population dynamics of Pseudomonas cepacia in the pea spermosphere in relation to biocontrol of Pythium. Phytopathology. 1990;80:1307–1311. [Google Scholar]
- 40.Parke J L, Rand R E, Joy A E, King E B. Biological control of Pythium damping-off and Aphanomyces root rot of peas by application of Pseudomonas cepacia or Pseudomonas fluorescensto seed. Plant Dis. 1991;75:987–992. [Google Scholar]
- 41.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierson L S, III, Thomashow L S. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens30-84. Mol Plant-Microbe Interact. 1992;5:330–339. doi: 10.1094/mpmi-5-330. [DOI] [PubMed] [Google Scholar]
- 43.Pierson L S, III, Gaffney T, Lam S, Gong F. Molecular analysis of genes encoding phenazine biosynthesis in the biological control bacterium Pseudomonas aureofaciens30-84. FEMS Microbiol Lett. 1995;134:299–307. doi: 10.1111/j.1574-6968.1995.tb07954.x. [DOI] [PubMed] [Google Scholar]
- 44.Pierson L S, Wood D W, Pierson E A. Homoserine lactone-mediated gene regulation in plant-associated bacteria. Annu Rev Phytopathol. 1998;36:207–225. doi: 10.1146/annurev.phyto.36.1.207. [DOI] [PubMed] [Google Scholar]
- 45.Propst C, Lubin L. Light-mediated changes in pigmentation of Pseudomonas aeruginosacultures. J Gen Microbiol. 1979;113:261–266. doi: 10.1099/00221287-113-2-261. [DOI] [PubMed] [Google Scholar]
- 46.Rahme L G, Tan M-W, Le L, Wong S M, Tompkins R G, Calderwood S B, Ausubel F M. Use of model plant hosts to identify Pseudomonas aeruginosavirulence factors. Proc Natl Acad Sci USA. 1997;94:13245–13250. doi: 10.1073/pnas.94.24.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 48.Reitzer L J, Magasanik B. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Neidhardt F, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: ASM Press; 1987. pp. 302–320. [Google Scholar]
- 49.Römer A, Herbert R B. Further observations on the source of nitrogen in phenazine biosynthesis. Z Naturforsch C. 1982;37:1070–1074. [Google Scholar]
- 50.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing portable oriT and the counter-selectable Bacillus subtilis sacBmarker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 51.Sheikh N M, MacDonald J C. Biogenesis of the N-methyl group of pyocyanine. Can J Microbiol. 1964;10:861–866. doi: 10.1139/m64-112. [DOI] [PubMed] [Google Scholar]
- 52.Smirnov V V, Kiprianova E A. Bacteria of Pseudomonas genus. Kiev, USSR: Naukova Dumka; 1990. pp. 100–111. [Google Scholar]
- 53.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkmann F S L, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K-S, Wu Z, Paulsen I T, Reizer J, Saier M H, Hancock R E W, Lory S, Olson M V. Complete genome sequence of Pseudomonas aeruginosaPAO1: an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 54.Suh S-J, Silo-Suh L, Woods D E, Hassett D J, West S E H, Ohman D E. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomashow L S, Weller D M. Current concepts in the use of introduced bacteria for biological disease control: mechanisms and antifungal metabolites. In: Stacey G, Keen N T, editors. Plant-microbe interactions. New York, N.Y: Chapman and Hall; 1996. pp. 187–235. [Google Scholar]
- 56.Thomashow L S, Essar D W, Fujimoto D K, Pierson III L S, Thrane C, Weller D M. Genetic and biochemical determinants of phenazine antibiotic production in fluorescent pseudomonads that suppress take-all disease of wheat. In: Nester E W, Verma D P S, editors. Advances in molecular genetics of plant-microbe interactions—1993. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1993. pp. 535–541. [Google Scholar]
- 57.Turner J M, Messenger A J. Occurrence, biochemistry and physiology of phenazine pigment production. Adv Microb Physiol. 1986;27:211–275. doi: 10.1016/s0065-2911(08)60306-9. [DOI] [PubMed] [Google Scholar]
- 58.Vukomanovic D V, Zoutman D E, Stone J A, Marks G S, Brien J F, Nakatsu K. Electrospray mass-spectrometric, spectrophotometric and electrochemical methods do not provide evidence for the binding of nitric oxide by pyocyanine at pH 7. Biochem J. 1997;322:25–29. doi: 10.1042/bj3220025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warren J B, Loi R, Rendell N B, Taylor G W. Nitric oxide is inactivated by the bacterial pigment pyocyanin. Biochem J. 1990;266:921–923. [PMC free article] [PubMed] [Google Scholar]
- 60.Watson D, MacDermot J, Wilson R, Cole P J, Taylor G W. Purification and structural analysis of pyocyanin and 1-OH-PHZ. Eur J Biochem. 1986;159:309–313. doi: 10.1111/j.1432-1033.1986.tb09869.x. [DOI] [PubMed] [Google Scholar]
- 61.Weller D M. Colonization of wheat roots by a fluorescent pseudomonad suppressive to take-all. Phytopathology. 1983;73:1548–1553. [Google Scholar]
- 62.West S E H, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 63.Whiteley M, Lee K M, Greenberg E P. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson R, Sykes D A, Watson D, Rutman A, Taylor G W, Cole P J. Measurement of Pseudomonas aeruginosaphenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect Immun. 1988;56:2515–2517. doi: 10.1128/iai.56.9.2515-2517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yabuuchi E, Ohyama A. Characterization of “pyomelanin”-producing strains of Pseudomonas aeruginosa. Int J Syst Bacteriol. 1972;22:53–64. [Google Scholar]
- 66.Yang S-W, Lin L-J, Cordell G A, Wang P, Corley D G. O- and N-methylation in the biosynthesis of staurosporine. J Nat Prod. 1999;62:1551–1553. doi: 10.1021/np990261q. [DOI] [PubMed] [Google Scholar]
- 67.Yanisch-Perron C, Viera J, Messing J. Improved M13 phage vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 68.Yoshida K-I, Fujita Y, Ehrich S D. Three asparagine synthetase genes of Bacillus subtilis. J Bacteriol. 1999;181:6081–6091. doi: 10.1128/jb.181.19.6081-6091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]