Abstract
The proliferating cell nuclear antigen (PCNA) is now recognized as one of the key proteins in DNA metabolic events because of its direct interactions with many proteins involved in important cellular processes. We have determined the crystal structure of PCNA from a hyperthermophilic archaeon, Pyrococcus furiosus (PfuPCNA), at 2.1 Å resolution. PfuPCNA forms a toroidal, ring-shaped structure consisting of homotrimeric molecules, which is also observed in the PCNA crystals from human and yeast. The overall structure of PfuPCNA is highly conserved with other PCNA proteins, as well as with the bacterial β clamp and the bacteriophage gp45. This result shows that the three-dimensional structure of the sliding clamp is conserved in the three domains of life. PfuPCNA has two remarkable features compared with the human and yeast PCNA molecules: it has more ion pairs and fewer intermolecular main chain hydrogen bonds. The former may contribute to the thermal stability of PfuPCNA, and the latter may be the cause of the stimulatory effect of PfuPCNA on the DNA synthesizing activity of P. furiosus DNA polymerases in the absence of the clamp loader replication factor C in vitro.
Keywords: Archaea, hyperthermophiles, intermolecular contacts, PCNA, thermal stability, X-ray diffraction
DNA replication is a fundamental event for the maintenance of life. The molecular apparatus acting in this process has been well characterized in Bacteria and Eucarya. The proteins with common roles in the DNA replication mechanism have been identified from these two biological domains, even though the amino acid sequences between the proteins from Bacteria and Eucarya are quite different (Kornberg and Baker 1992; Stillman 1994; Kelman and O'Donnell 1995; Waga and Stillman 1998). The replicative DNA polymerases require an elongation factor, called the sliding clamp, for processive DNA synthesis. Proliferating cell nuclear antigen (PCNA) plays this role in the eukaryotic system. Bacterial DNA polymerase III β subunit (Stukenberg et al. 1991) and bacteriophage T4 gene 45 protein (gp45) (Huang et al. 1981) have long been known to be the sliding clamps for their own DNA polymerases. The amino acid sequences of these sliding clamps are very different from each other. The crystal structures of PCNA (Krishna et al. 1994; Gulbis et al. 1996), β subunit (Kong et al. 1992), gp45 (Moarefi et al. 2000), and bacteriophage RB69 gp43 (Shamoo and Steitz 1999) have been solved, and they all have in common a ring-shaped structure with a pseudo-sixfold symmetry. The homotrimer of PCNA, gp45, and gp43 or the homodimer of β subunit molecules encircles the DNA strand in its central cavity and can slide freely along double-stranded DNA. The sliding clamps directly interact with the replicative DNA polymerases (Polymerase [Pol] δ and Pol ɛ for PCNA, Pol III for β subunit, T4 Pol for gp45, and RB69 Pol for gp43) and enhance their processive DNA synthesis.
In addition to its role as the sliding clamp for DNA polymerases, PCNA communicates with a wide variety of proteins involved in the important cellular processes, including cell cycle control, DNA repairs, and an apoptotic pathway (Jonsson and Hübscher 1997; Kelman 1997; Kelman and Hurwitz 1998; Tsurimoto 1998). In an effort to understand the molecular recognition mechanisms between PCNA and these proteins, many studies on the structure–function relationships have been reported to date.
Recent genome analyses of Archaea, the third domain of life, showed that the proteins related to genetic information, including DNA replication, are structurally more similar to eukaryotic proteins than to those from Bacteria (Edgell and Doolittle 1997; Ishino and Cann 1998; Cann and Ishino 1999). We cloned a gene with a sequence homologous to eukaryotic PCNA from the hyperthermophilic euryarchaeote Pyrococcus furiosus, expressed it in Escherichia coli, and characterized the purified gene product (Cann et al. 1999). The protein interacted with both DNA polymerase I and II in this organism and enhanced their DNA synthesizing activities; therefore, we named it PfuPCNA. A chemical cross-linking experiment showed that PfuPCNA can also form a trimeric structure. In a characterization of the PCNA homologs in the other archaea, Sulfolobus solfataricus and Methanobacterium thermoautotrophicum, it has been reported that they also stimulate DNA polymerization (De Felice et al. 1999; Kelman and Hurwitz 2000). These results suggest that the basic mechanism for processive DNA synthesis is conserved in the three biological domains.
In this study, we crystallized the PfuPCNA and solved its three-dimensional structure. The ring-shaped structure with the pseudo-sixfold symmetry of the sliding clamps was clearly conserved in the archaeal molecule.
Results
Mutation for Met73 to Leu
We made an overexpression system for the PfuPCNA gene in E. coli. The PfuPCNA protein was purified with ease by heating the crude cell extract to denature most of the E. coli proteins, followed by anion exchange chromatography (Cann et al. 1999). However, as we described previously, an extensive amount of another thermostable protein with a molecular mass of 20 kD was produced, as determined by SDS-PAGE analysis. The N-terminal amino acid sequence suggested that the protein was the product of an unexpected translational initiation at the ATG codon for Met73 in E. coli cells. This 20-kD product was not detected in the P. furiosus cells from the Western blot analysis using anti-PfuPCNA antiserum. It was predicted that the truncated protein would interfere with the formation of homogeneous crystals appropriate for structure analysis. To avoid this inconvenience, we disrupted the internal ATG codon of the structural gene for PfuPCNA. The sequence alignment of the P. furiosus, yeast, and human PCNAs showed that the yeast and human PCNAs have Leu at the position corresponding to Met73 of PfuPCNA (Fig. 1 ▶). Therefore, we changed the ATG to CTG at the corresponding site by site-directed mutagenesis, as described in Materials and Methods. The truncated product was not detected in the process of expression and purification of the mutant protein PfuPCNA(M73L). The purified PfuPCNA(M73L) stimulated the primer extension activity of P. furiosus Pol I, as in the case of the wild-type PfuPCNA.
Fig. 1.
Amino acid sequence alignment of PfuPCNA with yeast and human PCNA. The secondary structure elements determined from the crystal structure are labeled and indicated (black for α helices and gray for β strands).
Overall structure of Pfu PCNA(M73L)
The crystal structure of PfuPCNA(M73L) was determined by the molecular replacement method, using the structure of yeast PCNA (Krishna et al. 1994) as a search model. As predicted from the sequence alignment (Cann et al. 1999), PfuPCNA(M73L) has two β–α–β–β–β–β–β–α–β–β–β structural domains linked to each other with an interdomain connecting loop (Fig. 2 ▶). However, the three-dimensional structure of the connecting loop could not be modeled in this study because of poor electron density in this region.
Fig. 2.
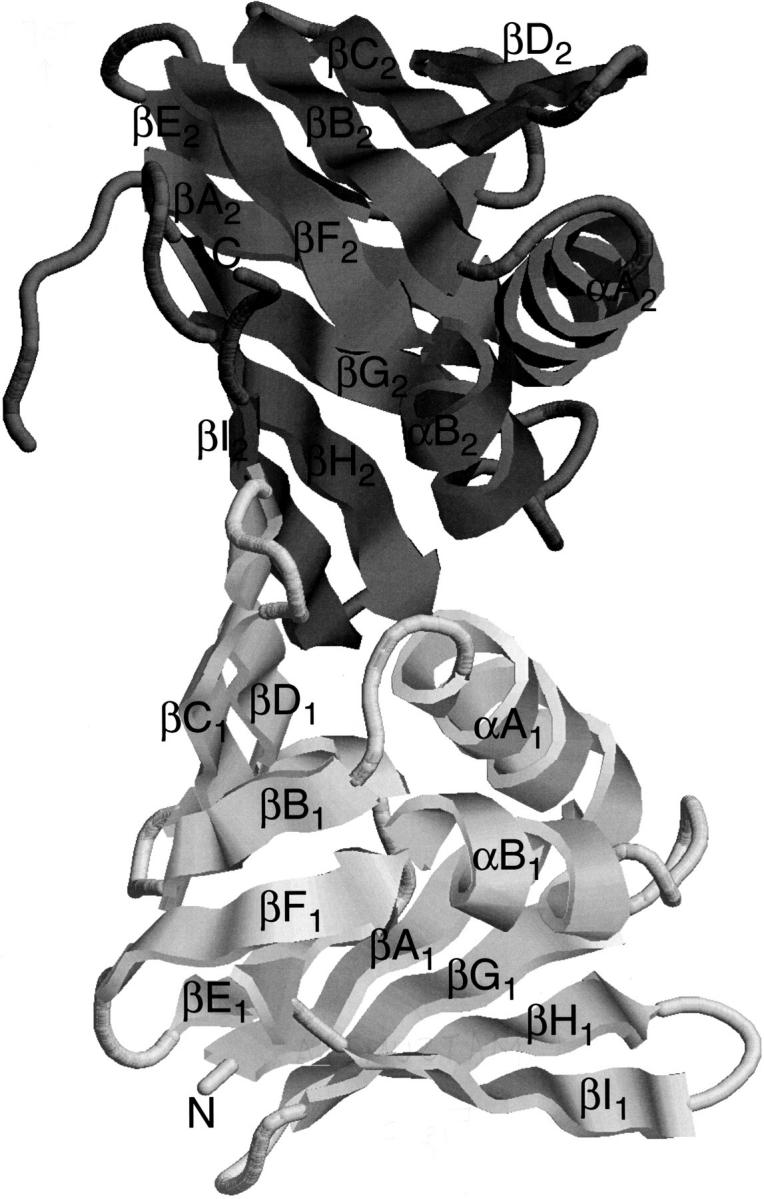
A ribbon diagram of PfuPCNA. The N-terminal domain is shown in light gray, and the C-terminal domain is shown in dark gray.
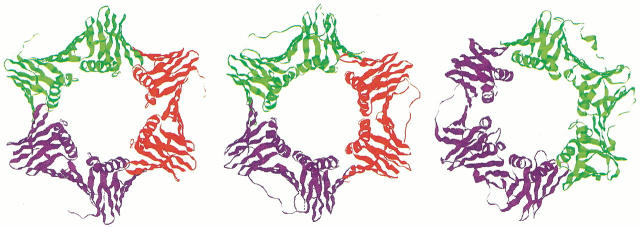
PfuPCNA(M73L) forms a trimer around the crystallographic threefold axis, which adopts a pseudo-sixfold, ring-shaped architecture similar to the eukaryotic PCNA and the bacterial β subunit (Fig. 3 ▶).
Fig. 3.
Comparison of sliding clamps from the three domains of life. (Left) PfuPCNA trimer, (center) yeast PCNA trimer (Krishna et al. 1994, PDB code 1PLQ), and (right) E. coli β subunit dimer (Kong et al. 1992, PDB code 2POL).
Structural basis for thermal stability
P. furiosus is a hyperthermophilic archaeon that grows optimally at 100°C (Fiala and Stetter 1986). There have been many explanations for the stability of proteins from thermophiles (Vogt and Argos 1997; Jaenicke and Böhm 1998), and PfuPCNA adopts some of the features found in those surveys.
The number of amino acids constituting PfuPCNA is 249, which is smaller than the number constituting the PCNAs of yeast (258) and human (261). PfuPCNA has shorter loops between βH1–βI1 and βD2–βE2, and at the C terminus (Fig. 1 ▶). A decrease in loop length is also observed in P. furiosus citrate synthase (Russell et al. 1997) and may contribute to the enhancement of the thermal stability.
PfuPCNA contains a 4%–6% higher percentage of charged residues and a 5%–10% lower percentage of polar/uncharged residues compared with the yeast and human PCNAs (Table 1). This fact agrees with the trend observed for thermophilic proteins (Deckert et al. 1998), although this statistical ``traffic rule'' is not necessarily applicable to individual proteins (Böhm and Jaenicke 1994).
Table 1.
Comparisons of amino acid compositions (%)
| Amino acid | P. furiosus | Yeast | Human |
| Charged residues (DEKRH) | 32.5 | 28.7 | 26.1 |
| Polar/uncharged residues (GSTNQYC) | 22.1 | 27.5 | 32.2 |
| Hydrophobic residues (LMIVWPAF) | 45.4 | 43.8 | 41.7 |
The enhancement of electrostatic interactions through ion pairs is thought to be one of the main causes of the increased thermostability of proteins (Vogt and Argos 1997). PfuPCNA has 27 short ion pairs (distance ≤4.0 Å) per molecule between oppositely charged groups, whereas yeast PCNA (Krishna et al. 1994) has 9 per molecule, and human PCNA (Gulbis et al. 1996) has 39 per trimer. In the case of PfuPCNA, 10 out of 27 ion pairs are intermolecular, and they are localized at the trimer-forming interface. On the other hand, yeast PCNA has 2 and human PCNA has 5 (per trimer) intermolecular ion pairs at the interface.
Intermolecular interface
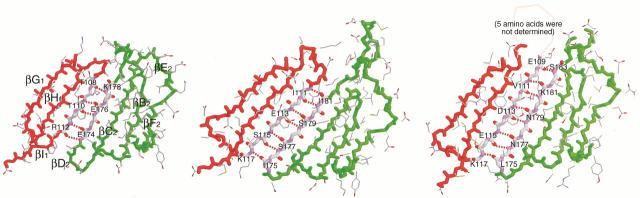
The PCNA trimer is formed through intermolecular main chain amide-to-carbonyl hydrogen bonds between the antiparallel β strands βI1 and βD2. PfuPCNA(M73L) possesses four hydrogen bonds (N···O ≤ 3.3 Å), whereas seven hydrogen bonds were observed in yeast PCNA and eight in human PCNA (Table 2; Fig. 4 ▶). The reduction of the amino acids in PfuPCNA occurs at the loops adjacent to βI1 and βD2, and these differences in the loop lengths may be the cause of the distinct intermolecular hydrogen bond patterns.
Table 2.
N···O distances at the intermolecular interface ( d < 4 Å)
| Hydrogen bonded atoms | d/Å |
| P. furiosus | |
| Thr108 O···Lys178 N | 3.87 |
| Thr110 N···Glu176 O | 2.90 |
| Thr110 O···Glu176 N | 2.90 |
| Arg112 N···Glu174 O | 2.83 |
| Arg112 O···Glu174 N | 3.19 |
| Yeast | |
| Ile111 N···Ile181 O | 3.11 |
| Ile111 O···Ile181 N | 3.14 |
| Glu113 N···Ser179 O | 2.88 |
| Glu113 O···Ser179 N | 2.94 |
| Ser115 N···Ser177 O | 2.79 |
| Ser115 O···Ser177 N | 3.06 |
| Lys117 N···Ile175 O | 3.01 |
| Human (average value of three non-crystallographic symmetry-related sites) | |
| Glu109 O···Ser183 N | 2.98 |
| Val111 N···Lys181 O | 3.15 |
| Val111 O···Lys181 N | 2.89 |
| Asp113 N···Asn179 O | 2.81 |
| Asp113 O···Asn179 N | 2.88 |
| Glu115 N···Asn177 O | 3.03 |
| Glu 115 O···Asn177 N | 3.30 |
| Lys117 N···Leu175 O | 2.84 |
Fig. 4.
Comparison of main-chain hydrogen bonds at the intermolecular interface. (Left) PfuPCNA, (center) yeast PCNA, and (right) human PCNA (Gulbis et al. 1996, PDB code 1AXC).
The buried surface area per molecule at intermolecular interface is 1.44 × 103 Å2, which is 7% smaller than the 1.55 × 103 Å2 of the yeast PCNA trimer. This value additionally supports a weaker intermolecular interaction within PfuPCNA.
Discussion
We have presented the crystal structure of P. furiosus PCNA as the first sliding clamp molecule from Archaea. This molecule's striking structural similarity to sliding clamp molecules from other biological domains shows that the basic mechanism for processive DNA synthesis in DNA replication is conserved in the three domains of life. Loading the sliding clamp onto the DNA strand requires the protein factor called the clamp loader. Eukaryotic replication factor C (RFC) (Waga and Stillman 1998), bacterial γ complex (Kelman and O'Donnell 1995), and bacteriophage gp44/62 (Young et al. 1992) are known to have this role. We characterized the RFC homolog of P. furiosus, which enhanced the PCNA-dependent DNA synthesis of Pol I and Pol II (I.K.O. Cann, S. Ishino, M. Yuasa, M. Daiyasu, H. Toh, and Y. Ishino, unpubl.). A topic of common interest in the replication field is understanding how the clamp loaders recognize sliding clamps. Extensive biochemical analyses to elucidate the mechanism of clamp loading have recently been reported (Yuzhakov et al. 1999; Turner et al. 1999). Structural analyses are now necessary for progress in this subject. The archaeal RFC possibly consists of a hetero-tetramer or pentamer of two proteins, small subunit (RFCS) and large subunit (RFCL). This simpler complex, compared with those from Eucarya and Bacteria, may yield some advantage for analysis of the interactions between the sliding clamp and the clamp loader.
Although the overall structure of PfuPCNA is very similar to that of eukaryotic PCNAs, remarkable differences in amino acid composition and three-dimensional structure are profound. The increment of charged residues, the shortening of loop lengths, and the gain of ion pairs found in PfuPCNA structure generally agree with the trends observed in plenty of proteins from thermophiles.
In regard to subunit interaction, the shortening of loops and the increase in ion pairs in PfuPCNA conversely affect the intermolecular interface. The shortening of loops occurs in the vicinity of the β strands involved in the assembly of the subunits (Fig. 4 ▶). This structural difference in the interface of PfuPCNA results in the deformation of the intermolecular β-sheet, as reflected in the decrease in numbers of hydrogen bonds. On the other hand, the increase in intermolecular ion pairs should stabilize the interface.
PfuPCNA can stimulate the DNA polymerization reaction of P. furiosus DNA polymerases with a circular DNA template in vitro (Cann et al. 1999). This means that the PfuPCNA trimer ring can open and load onto circular DNA by itself. The instability by the loop shortenings is presumed not to be fully compensated by the increased ion pairs. This weakened intermolecular interaction could account for the self-loading phenomenon in PfuPCNA. As an extreme case, the crystal structure of the gp45 from bacteriophage T4 showed that there are four intermolecular main chain hydrogen bonds and that the buried surface area at the intermolecular interface is only 0.87 × 103 Å2 (Moarefi et al. 2000). This protein exists as an open-form trimer in solution (Alley et al. 1999).
In this study, even the final 2.1 Å resolution map could not reveal defined electron density for the residues in the interdomain connecting loop and the C-terminal loop. However, these disordered regions are of great interest, because RFC, DNA polymerases, and other proteins that utilize PCNA as a carrier are known to bind to the interdomain connecting loop and/or the C-terminal loop (Tsurimoto 1998). The three-dimensional structures of the higher order complexes, such as PCNA/DNA polymerase and PCNA/RFC, will provide a wealth of knowledge about the basic molecular recognition mechanisms in DNA metabolism.
Materials and methods
Site-directed mutagenesis for Met73 to Leu
Polymerase chain reaction (PCR)–mediated mutagenesis was performed to change the ATG codon for Met73 to CTG. The plasmid pTPCNA (the structural gene for PfuPCNA is inserted into the pET21a vector [Novagen, Madison, WI]), which was made previously (Cann et al. 1999), was directly amplified by PCR using the two primers M73LF (5′-CAATTGGAGTTAACCTGGACCACC TAAAG-3′) and M73LR (5′-CTTTAGGTGGTCCAGGTTAACT CCAATTG-3′), which bind to the region containing the target codon (underlined). The PCR conditions involved 30 cycles of denaturation for 30 sec at 95°C, annealing for 1 min at 55°C, and extension for 10 min at 68°C with Pyrobest DNA polymerase (Takara Shuzo, Kyoto, Japan). The amplified plasmid was purified by agarose gel electrophoresis and used for transformation. The plasmids isolated from several independent transformants were sequenced. The plasmid that contained the gene with only one mutation at the target codon was selected and designated as pTPCNAL73. The production and purification of PfuPCNA(M73L) from E. coli BL21(DE3)/pTPCNAL73 were performed as described for PfuPCNA (Cann et al. 1999). The purified PfuPCNA(M73L) solution was concentrated to 20 mg/mL by ultrafiltration, using Microcon YM-30 (Millipore, Bedford, MA), and was stored at 4°C.
Crystallization and data collection
Single crystals of PfuPCNA were prepared by the hanging drop vapor diffusion method at 20°C. The initial condition search was performed with Grid Screen ammonium sulfate (Hampton Research, Laguna Niguel, CA) and was optimized with a finer grid of pH, precipitant concentration, and additives. The optimal composition of the precipitation solution was 100 mM sodium citrate (pH 5.5), 2.4 M ammonium sulfate, 5% (v/v) 1,4-dioxane. Then, 1.0 μL of 20 mg/mL protein solution and 1.0 μL of precipitation solution were mixed and placed over 500 μL of precipitation solution. Colorless hexagonal needles of ∼1.0 × 0.2 × 0.2 mm were obtained in a week. The PfuPCNA crystal was mounted in a glass capillary for X-ray diffraction data collection.
Diffraction data were collected at 296 K on the BL-6A beamline at Photon Factory, using 1.0000 Å radiation with an ADSC Quantum 4R CCD detector (Area Detector Systems, Poway, CA). The data were processed using DPS/MOSFLM (Rossmann and van Beek 1999), scaled, and converted for structure determination using SCALA and TRUNCATE in the CCP4 suite (Collaborative Computational Project, Number 4 1994).
Structure determination
The initial crystal structure of PfuPCNA was determined by the molecular replacement method using AMoRe (Navaza 1994) with yeast PCNA (Krishna et al. 1994, PDB code 1PLQ) as the search model. The solution that gave the lowest R value of 0.559 after rotation and translation search using 25.0–3.5 Å data was used for the structure refinement. The molecular model was built using O (Jones et al. 1991) and was refined using CNS software (Brünger et al. 1998). Met1, Val118-Leu125, and Glu248-Glu249 were not modeled because of poor electron density at these regions. The results of the structure analysis are summarized in Table 3. The coordinates and structural factors have been deposited to the Protein Data Bank under the accession code 1GE8.
Table 3.
Summary of crystal structure analysis
| Data collection | |
| Space group | P63 |
| Unit cell (Å) | a = 89.682 |
| c = 63.269 | |
| Total reflections (25.00–2.00 Å) | 121,358 |
| Unique reflections (25.00–2.00 Å) | 19,680 |
| Completeness | |
| 25.00–2.00 Å (%) | 99.9 |
| 2.24–2.11 Å (%) | 99.9 |
| Average redundancy | |
| 25.00–2.00 Å | 6.2 |
| 2.24–2.11 Å | 6.2 |
| Average I/σ (I) | |
| 25.00–2.00 Å | 6.7 |
| 2.24–2.11 Å | 2.3 |
| Pmeas | |
| 25.00–2.00 Å | 0.091 |
| 2.24–2.11 Å | 0.357 |
| Structure refinement (25.00–2.10 Å) | |
| Reflections used for refinement | 15,291 |
| Reflections used for cross-validation | 1,717 |
| R | 0.2250 |
| Rfree | 0.2529 |
| Root mean square deviation (Rmsd) of bond lengths (Å) | 0.00627 |
| Rmsd of bond angles (°) | 1.120 |
| Rmsd of dihedral angles (°) | 25.8 |
| Rmsd of improper angles (°) | 0.681 |
| Average b factor (Å2) | 30.7 |
Comparison of three-dimensional structures
The multiple sequence alignment of the P. furiosus, yeast, and human PCNAs was performed using CLUSTAL X V1.8 (Thompson et al. 1994) with a slight modification by hand. Secondary structures were assigned using PROCHECK (Laskowski et al. 1993) based on the three-dimensional structures of PfuPCNA, yeast PCNA (Krishna et al. 1994, PDB code 1PLQ), and human PCNA (Gulbis et al. 1996, PDB code 1AXC). Hydrogen bond and ion pair distances were calculated using ACT in CCP4. Buried surface areas were calculated using SURFACE in CCP4, with a probe radius of 1.40 Å. The figures of the molecules were prepared using RasMol (Sayle and Milner-White 1995).
Acknowledgments
We thank S. Ishino and K. Komori for assistance with biochemical experiments. We also thank I.K.O. Cann for discussions. We are grateful to T. Oyama for assistance with crystallographic experiments.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ``advertisement'' in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.36401.
References
- Alley, S.C., Shier, V.K., Abel-Santos, E., Sexton, D.J., Soumillion, P., and Benkovic, S.J. 1999. Sliding clamp of the bacteriophage T4 polymerase has open and closed subunit interfaces in solution. Biochemistry 38 7696–7709. [DOI] [PubMed] [Google Scholar]
- Böhm, G. and Jaenicke, R. 1994. On the relevance of sequence statistics for the properties of extremophilic proteins. Int. J. Pept. Protein Res. 43 97–106. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54 905–921. [DOI] [PubMed] [Google Scholar]
- Cann, I.K.O. and Ishino, Y. 1999. Archaeal DNA replication: Identifying the pieces to solve a puzzle. Genetics 152 1249–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann, I.K.O., Ishino, S., Hayashi, I., Komori, K., Toh, H., Morikawa, K., and Ishino, Y. 1999. Functional interactions of a homolog of proliferating cell nuclear antigen with DNA polymerases in Archaea. J. Bacteriol. 181 6591–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. 1994. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D50 760–763. [DOI] [PubMed] [Google Scholar]
- Deckert, G., Warren, P.V., Gaasterland, T., Young, W.G., Lenox, A.L., Graham, D.E., Overbeek, R., Snead, M.A., Keller, M., Aujay, M., et al. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392 353–358. [DOI] [PubMed] [Google Scholar]
- De Felice, M., Sensen, C.W., Charlebois, R.L., Rossi, M., and Pisani, F.M. 1999. Two DNA polymerase sliding clamps from the thermophilic archaeon Sulfolobus solfataricus. J. Mol. Biol. 291 47–57. [DOI] [PubMed] [Google Scholar]
- Edgell, D.R. and Doolittle, W.F. 1997. Archaea and the origin(s) of DNA replication proteins. Cell 89 995–998. [DOI] [PubMed] [Google Scholar]
- Fiala, G. and Stetter, K.O. 1986. Pyrococcus furiosus, sp. nov., represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145 56–61. [Google Scholar]
- Gulbis, J.M., Kelman, Z., Hurwitz, J., O'Donnell, M., and Kuriyan, J. 1996. Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell 87 297–306. [DOI] [PubMed] [Google Scholar]
- Huang, C.-C., Hearst, J.E., and Alberts, B.M. 1981. Two types of replication proteins increase the rate at which T4 DNA polymerase traverses the helical regions in a single-stranded DNA template. J. Biol. Chem. 256 4087–4094. [PubMed] [Google Scholar]
- Ishino, Y. and Cann, I.K.O. 1998. The euryarchaeotes, a subdomain of Archaea, survive on a single DNA polymerase: Fact or farce? Genes Genet. Syst. 73 323–336. [DOI] [PubMed] [Google Scholar]
- Jaenicke, R. and Böhm, G. 1998. The stability of proteins in extreme environments. Curr. Opin. Struct. Biol. 8 738–748. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.-Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47 110–119. [DOI] [PubMed] [Google Scholar]
- Jonsson, Z.O. and Hübscher, U. 1997. Proliferating cell nuclear antigen: More than a clamp for DNA polymerase. Bioessays 19 967–975. [DOI] [PubMed] [Google Scholar]
- Kelman, Z. 1997. PCNA: Structure, function, and interactions. Oncogene 14 629–640. [DOI] [PubMed] [Google Scholar]
- Kelman, Z. and Hurwitz, J. 1998. Protein–PCNA interactions: A DNA-scanning mechanism? Trends Biochem. Sci. 23 236–238. [DOI] [PubMed] [Google Scholar]
- Kelman, Z, and Hurwitz, J. 2000. A unique organization of the protein subunits of the DNA polymerase clamp loader in the archaeon Metanobacterium thermoautotrophicum ΔH. J. Biol. Chem. 2757327–7336. [DOI] [PubMed] [Google Scholar]
- Kelman, Z. and O'Donnell, M. 1995. DNA polymerase III holoenzyme: Structure and function of a chromosomal replicating machine. Annu. Rev. Biochem. 64 171–200. [DOI] [PubMed] [Google Scholar]
- Kong, X.P., Onrust, R., O'Donnell, M., and Kuriyan, J. 1992. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell 69 425–437. [DOI] [PubMed] [Google Scholar]
- Kornberg, A. and Baker, T.A. 1992. DNA replication, 2nd ed. W.H. Freeman, New York.
- Krishna, T.S.R., Kong, X.P., Gary, S., Burgers, P.M., and Kuriyan, J. 1994. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79 1233–1243. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., and Thornton, J.M. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26 283–291. [Google Scholar]
- Moarefi, I., Jeruzalmi, D., Turner, J., O'Donnell, M., and Kuriyan, J. 2000. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J. Mol. Biol. 296 1215–1223. [DOI] [PubMed] [Google Scholar]
- Rossmann, M.G. and van Beek, C.G. 1999. Data processing. Acta Crystallogr. D55 1631–1640. [DOI] [PubMed] [Google Scholar]
- Russell, R.J.M., Ferguson, J.M.C., Hough, D.W., Danson, M.J., and Taylor, G.L. 1997. The crystal structure of citrate synthase from the hyperthermophilic archaeon Pyrococcus furiosus at 1.9 Å resolution. Biochemistry 36 9983–9994. [DOI] [PubMed] [Google Scholar]
- Sayle, R.A. and Milner-White, E.J. 1995. RasMol: Biomolecular graphics for all. Trends Biochem. Sci. 20 374–376. [DOI] [PubMed] [Google Scholar]
- Shamoo, Y. and Steitz, T.A. 1999. Building a replisome from interacting pieces: Sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell 99 155–166. [DOI] [PubMed] [Google Scholar]
- Stillman, B. 1994. Smart machines at the DNA replication fork. Cell 78 725–728. [DOI] [PubMed] [Google Scholar]
- Stukenberg, P.T., Studwell-Vaughan, P.S., and O'Donnell, M. 1991. Mechanism of the sliding b-clamp of DNA polymerase III holoenzyme. J. Biol. Chem. 266 11328–11334. [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto, T. 1998. PCNA, a multifunctional ring on DNA. Biochim. Biophys. Acta 1443 23–39. [DOI] [PubMed] [Google Scholar]
- Turner, J., Hingorani, M.M., Kelman, Z., and O'Donnell, M. 1999. The internal workings of a DNA polymerase clamp-loading machine. EMBO J. 18 771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, G. and Argos, P. 1997. Protein thermal stability: Hydrogen bonds or internal packing? Fold. Des. 2 S40–S46. [DOI] [PubMed] [Google Scholar]
- Waga, S. and Stillman, B. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67 721–751. [DOI] [PubMed] [Google Scholar]
- Young, M.C., Reddy, M.K., and von Hippel, P.H. 1992. Structure and function of the bacteriophage T4 DNA polymerase holoenzyme. Biochemistry 31 8675–8690. [DOI] [PubMed] [Google Scholar]
- Yuzhakov, A., Kelman, Z., and O'Donnell, M. 1999. Trading places on DNA—A three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell 96 153–163. [DOI] [PubMed] [Google Scholar]