Abstract
Soil organisms are a crucial part of the terrestrial biosphere. Despite their importance for ecosystem functioning, few quantitative, spatially explicit models of the active belowground community currently exist. In particular, nematodes are the most abundant animals on Earth, filling all trophic levels in the soil food web. Here we use 6,759 georeferenced samples to generate a mechanistic understanding of the patterns of the global abundance of nematodes in the soil and the composition of their functional groups. The resulting maps show that 4.4 ± 0.64 × 1020 nematodes (with a total biomass of approximately 0.3 gigatonnes) inhabit surface soils across the world, with higher abundances in sub-Arctic regions (38% of total) than in temperate (24%) or tropical (21%) regions. Regional variations in these global trends also provide insights into local patterns of soil fertility and functioning. These high-resolution models provide the first steps towards representing soil ecological processes in global biogeochemical models and will enable the prediction of elemental cycling under current and future climate scenarios.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All raw data, sampled covariate layer data, models and maps are available at https://gitlab.ethz.ch/devinrouth/crowther_lab_nematodes. The total nematode abundance map is accessible online at https://www.crowtherlab.com.
Code availability
All source code and models are available at https://gitlab.ethz.ch/devinrouth/crowther_lab_nematodes.
References
Cameron, E. K. et al. Global gaps in soil biodiversity data. Nat. Ecol. Evol. 2, 1042–1043 (2018).
Bardgett, R. D. & van der Putten, W. H. Belowground biodiversity and ecosystem functioning. Nature 515, 505–511 (2014).
Paul, E. A. Soil Microbiology, Ecology, and Biochemistry 4th edn (Elsevier, 2015).
Wieder, W. R., Bonan, G. B. & Allison, S. D. Global soil carbon projections are improved by modelling microbial processes. Nat. Clim. Change 3, 909–912 (2013).
Bradford, M. A. et al. A test of the hierarchical model of litter decomposition. Nat. Ecol. Evol. 1, 1836–1845 (2017).
Crowther, T. W. et al. Quantifying global soil carbon losses in response to warming. Nature 540, 104–108 (2016).
Delgado-Baquerizo, M. et al. A global atlas of the dominant bacteria found in soil. Science 359, 320–325 (2018).
Thompson, L. R. et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017).
Lozupone, C. A. & Knight, R. Global patterns in bacterial diversity. Proc. Natl Acad. Sci. USA 104, 11436–11440 (2007).
Fierer, N. & Jackson, R. B. The diversity and biogeography of soil bacterial communities. Proc. Natl Acad. Sci. USA 103, 626–631 (2006).
Bahram, M. et al. Structure and function of the global topsoil microbiome. Nature 560, 233–237 (2018).
Tedersoo, L. et al. Global diversity and geography of soil fungi. Science 346, 1256688 (2014).
Davison, J. et al. Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Science 349, 970–973 (2015).
Nielsen, U. N. et al. Global-scale patterns of assemblage structure of soil nematodes in relation to climate and ecosystem properties. Glob. Ecol. Biogeogr. 23, 968–978 (2014).
Wu, T., Ayres, E., Bardgett, R. D., Wall, D. H. & Garey, J. R. Molecular study of worldwide distribution and diversity of soil animals. Proc. Natl Acad. Sci. USA 108, 17720–17725 (2011).
Boag, B. & Yeates, G. W. Soil nematode biodiversity in terrestrial ecosystems. Biodivers. Conserv. 7, 617–630 (1998).
Song, D. et al. Large-scale patterns of distribution and diversity of terrestrial nematodes. Appl. Soil Ecol. 114, 161–169 (2017).
Xu, X., Thornton, P. E. & Post, W. M. A global analysis of soil microbial biomass carbon, nitrogen and phosphorus in terrestrial ecosystems. Glob. Ecol. Biogeogr. 22, 737–749 (2013).
Serna-Chavez, H. M., Fierer, N. & van Bodegom, P. M. Global drivers and patterns of microbial abundance in soil. Glob. Ecol. Biogeogr. 22, 1162–1172 (2013).
Geisen, S. et al. Integrating quantitative morphological and qualitative molecular methods to analyse soil nematode community responses to plant range expansion. Methods Ecol. Evol. 9, 1366–1378 (2018).
Darby, B. J., Todd, T. C. & Herman, M. A. High-throughput amplicon sequencing of rRNA genes requires a copy number correction to accurately reflect the effects of management practices on soil nematode community structure. Mol. Ecol. 22, 5456–5471 (2013).
Carini, P. et al. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2, 16242 (2016).
Blazewicz, S. J., Barnard, R. L., Daly, R. A. & Firestone, M. K. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 7, 2061–2068 (2013).
Platt, H. M. in The Phylogenetic Systematics of Freeliving Nematodes (ed. Lorenzen, S.) i–ii (The Ray Society, 1994).
Ingham, R. E., Trofymow, J. A., Ingham, E. R. & Coleman, D. C. Interactions of bacteria, fungi, and their nematode grazers: effects on nutrient cycling and plant growth. Ecol. Monogr. 55, 119–140 (1985).
Ferris, H. Contribution of nematodes to the structure and function of the soil food web. J. Nematol. 42, 63–67 (2010).
Crowther, T. W., Boddy, L. & Jones, T. H. Species-specific effects of soil fauna on fungal foraging and decomposition. Oecologia 167, 535–545 (2011).
Neher, D. A. Role of nematodes in soil health and their use as indicators. J. Nematol. 33, 161–168 (2001).
Procter, D. L. Global overview of the functional roles of soil-living nematodes in terrestrial communities and ecosystems. J. Nematol. 22, 1–7 (1990).
Fierer, N., Strickland, M. S., Liptzin, D., Bradford, M. A. & Cleveland, C. C. Global patterns in belowground communities. Ecol. Lett. 12, 1238–1249 (2009).
Sohlenius, B. Abundance, biomass and contribution to energy flow by soil nematodes in terrestrial ecosystems. Oikos 34, 186–194 (1980).
Jobbágy, E. G. & Jackson, R. B. The vertical distribution of soil organic carbon and its relation to climate and vegetation. Ecol. Appl. 10, 423–436 (2000).
Ettema, C. H. Soil nematode diversity: species coexistence and ecosystem function. J. Nematol. 30, 159–169 (1998).
Ferris, H. Nematode Ecophysiological Parameters. http://nemaplex.ucdavis.edu/Ecology/EcophysiologyParms/EcoParameterMenu.html (2018).
Mulder, C. & Vonk, J. A. Nematode traits and environmental constraints in 200 soil systems: scaling within the 60–6000 μm body size range. Ecology 92, 2004 (2011).
Bar-On, Y. M., Phillips, R. & Milo, R. The biomass distribution on Earth. Proc. Natl Acad. Sci. USA 115, 6506–6511 (2018).
Schlesinger, W. H. & Bernhardt, E. S. in Biogeochemistry 419–444 (2013).
Powers, T. O. et al. Tropical nematode diversity: vertical stratification of nematode communities in a Costa Rican humid lowland rainforest. Mol. Ecol. 18, 985–996 (2009).
Yeates, G. W., Bongers, T., De Goede, R. G., Freckman, D. W. & Georgieva, S. S. Feeding habits in soil nematode families and genera—an outline for soil ecologists. J. Nematol. 25, 315–331 (1993).
Chavent, M., Kuentz-Simonet, V., Liquet, B. & Saracco, J. ClustOfVar: an R package for the clustering of variables. J. Stat. Softw. 50, 1–16 (2012).
Hengl, T., Nussbaum, M., Wright, M. N., Heuvelink, G. B. M. & Gräler, B. Random forest as a generic framework for predictive modeling of spatial and spatio-temporal variables. PeerJ 6, e5518 (2018).
Gorelick, N. et al. Google Earth Engine: planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 202, 18–27 (2017).
Hengl, T. et al. SoilGrids250m: global gridded soil information based on machine learning. PLoS ONE 12, e0169748 (2017).
Andrassy, I. Die Rauminhalts und Gewichtsbestimmung der Fadenwürmer (Nematoden). Acta Zool. Hung. 2, 1–15 (1956).
Mulder, C., Cohen, J. E., Setälä, H., Bloem, J. & Breure, A. M. Bacterial traits, organism mass, and numerical abundance in the detrital soil food web of Dutch agricultural grasslands. Ecol. Lett. 8, 80–90 (2005).
Persson, T. Influence of soil organisms on nitrogen mineralization in a northern Scots pine forest. In Proc. VIII Int. Colloq. Soil Zool. (eds Lebrun, P. et al.) 117–126 (1983).
Bongers, T. The maturity index: an ecological measure of environmental disturbance based on nematode species composition. Oecologia 83, 14–19 (1990).
Bongers, T. The Maturity Index, the evolution of nematode life history traits, adaptive radiation and cp-scaling. Plant Soil 212, 13–22 (1999).
Kleiber, M. Body size and metabolism. Hilgardia 6, 315–353 (1932).
West, G. B., Brown, J. H. & Enquist, B. J. A general model for the origin of allometric scaling laws in biology. Science 276, 122–126 (1997).
Atkinson, H. J. in Nematodes as Biological Models vol. 2 (ed Zuckerman, B. M.) 122–126 (Academic, 1980).
Klekowski, R. Z., Wasilewska, L. & Paplinska, E. Oxygen consumption in the developmental stages of Panagrolaimus rigidus. Nematologica 20, 61–68 (1974).
Klekowski, R. Z., Wasilewska, L. & Paplinska, E. Oxygen consumption by soil-inhabiting nematodes. Nematologica 18, 391–403 (1972).
Acknowledgements
This research was supported by a grant from DOB Ecology to T.W.C., a grant from the Netherlands Organization for Scientific Research (grant 016.Veni.181.078) to S.G., grants from NSF (OPP 1115245, 1341736, 0840979) to B.J.A., by a Ramon y Cajal fellow award (RYC-2016-19939) to R.C.H., a grant from UNEP & Global Environment Facility to J.E.C., a grant from NERC (NE/M017036/1) to T.C., a grant from FAPEMIG/FAPESP/VALE S.A.(CRA-RDP-00136-10) to L.B.C., through the strategic programme UID/BIA/04050/2013 (POCI-01-0145-FEDER-007569) awarded to S.R.C., a grant from CNPq PROTAX (562346/2010-4) to J.M.d.C.C., a grant from DFG (CRC990) to V.K. and S.S., a grant from the MSHE of Russia (AAAA-A17-117112850234-5) to A.A.K., grants from the Chinese Academy of Sciences (XDB15010402) and the National Natural Science Foundation of China (41877047) to Q.L., grants from the National Natural Science Foundation of China (31330011, 31170484) to W.L., grants from NERC (NE/M017036/1) to M.M., grants from the Spanish Ministry of Innovation (CGL2009-14686-C02-01/ 02, CGL2013-43675-P) to J.A.R.M., grants from NSF (DEB-0450537, DEB-1145440) to P.M., T.O.P. and K. Powers, grants from the German Academic Exchange Service (PKZ 91540366) and NAFOSTED (106.05 – 2017.330) to T.A.D.N., by an ARC Discovery project (DP150104199) to U.N.N., by the National Key Research and Development Program of China (2016YFC0502101) and the National Natural Science Foundation of China (31370632) to K. Pan, a grant from the Natural Environment Research Council (NERC) to D.G.W., a grant from BAPHIQ (106AS-9.5.1-BQ-B3) J.-i.Y. The James Hutton Institute receives financial support from the Scottish Government Rural and Environment Science and Analytical Services (RESAS) division. Investigations in northwest Russia were carried out under state order for IB KarRC RAS and are partially supported by the Russian Foundation for Basic Research (18-34-00849). We thank E. Clark and A. Orgiazzi for review of the manuscript; and R. Bouharroud, Z. Ferji, L. Jackson and E. Mzough for providing data.
Author information
Authors and Affiliations
Contributions
J.v.d.H., S.G., D.R. and T.W.C. designed and performed the data analyses. J.v.d.H., D.R. and T.W.C. designed and performed geospatial analyses. J.v.d.H., S.G., H.F., R.G.M.d.G. and C.M. designed and performed biomass calculations. S.G., H.F., W.T., D.A.W., R.G.M.d.G., B.J.A., W.A., W.S.A., R.D.B., M.B., R.C.-H., J.E.C., T.C., X.C., S.R.C., R.C., J.M.d.C.C., M.D., L.d.B.C., D.D., M.E., B.S.G., C.G., K.H., D.K., P.K., A.K., G.K., V.K., A.A.K., Q.L., W.L., M. Magilton, M. Marais, J.A.R.M., E.M., E.H.M., C.M., P.M., R.N., T.A.D.N., U.N.N., H.O., J.E.P.R., K. Pan, V.P., L.P., J.C.P.d.S., C.P., T.O.P., K. Powers, C.W.Q., S.R., S.S.M., S.S., H.S., A.S., A.V.T., J.T., W.H.v.d.P., M.V., C.V., L.W., D.H.W., R.W., D.G.W. and J.-i.Y. contributed data. J.v.d.H., S.G. and T.W.C. wrote the first draft of the manuscript with input from D.A.W. All authors contributed to editing of the paper.
Corresponding authors
Ethics declarations
Competing interests
W.S.A. is an employee of Nature Communications, a sister journal from the same publisher; he did not have any access to or involvement with the editorial process at Springer Nature. All other authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Nico Eisenhauer, Deborah Neher and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Fig. 1 Model accuracy assessment and extent of interpolation and extrapolation across all terrestrial pixels in 73 global covariate layers.
a, Coefficient of variation (standard deviation as a fraction of the mean predicted value) as a measure of the prediction accuracy of our model. b, Proportional extent of interpolation (purple) versus extrapolation (red) in the univariate space. c, Percentage of pixels that fall within the convex hulls of the first 11 principal component spaces (collectively covering >80% of the sample space variation). d, Percentage of pixels interpolated as a function of the percentage of global environmental conditions covered by the sample set. On the global scale, 86% of the Earth’s pixels have at least 90% of the covariate bands falling within the sampled range of environmental conditions. e, Percentage of pixels falling within the 55 convex hull spaces of the first 11 principal components (collectively explaining >80% of the variation). On the global scale, 62% of the Earth’s pixels fell within 100% of 55 convex hull spaces. f, Percentage of terrestrial pixels falling within the sampled range, per covariate band.
Extended Data Fig. 2 Linear regression models of the most important variables from the final random forest model and annual mean temperature.
SOC and cation-exchange capacity have a positive correlation with total nematode abundance, whereas pH is negatively correlated. These linear regression models (n = 1,809) were not used to create global perspectives of nematode distribution patterns. The grey area represents the 95% confidence interval of the mean.
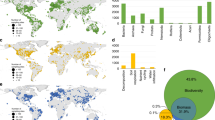
Extended Data Fig. 3 Global maps of nematode trophic group abundance.
a, Bacterivores. b, Fungivores. c, Herbivores. d, Omnivores. e, Predators. Scales differ per map. Most trophic groups show similar patterns, but predators (e) are predicted to be present in particularly high abundances in some arid soils—for example, in the Sahara and Arabian Desert. Pixel values were binned into seven quantiles to create the colour palette.
Extended Data Fig. 4 Global map of total nematode abundance per unit area (m2).
Correcting for the lower bulk density in soils that are high in organic matter, this map shows the same global patterns of nematode abundance as in Fig. 3. Hence, it is not the low bulk density of soils in boreal regions that result in the observed patterns, but rather the high nematode abundances. Pixel values were binned into seven quantiles to create the colour palette.
Extended Data Fig. 5 Global maps of nematode trophic group abundance per unit area (m2).
a, Bacterivores. b, Fungivores. c, Herbivores. d, Omnivores. e, Predators. Scales differ per map. Correcting for the lower bulk density in soils that are high in organic matter, these maps show the same global patterns of nematode trophic group abundance as in Extended Data Fig. 3a–e. Pixel values were binned into seven quantiles to create the colour palette.
Extended Data Fig. 6 Community types and driving variables of community type composition.
a, Correlations between trophic groups. Overall, correlations of predators with other trophic groups are the least positive. b, On the basis of the relative abundance of each trophic group, soil nematode communities can be classified in four distinct types. We find that these soil nematode communities are dominated by herbivores (type 1), herbivores and bacterivores (type 2), bacterivores (type 3) or have a mixed composition (type 4). c, Non-metric multidimensional scaling to highlight environmental conditions that drive the composition of each of the four main community types. Vegetation-type indices, such as the normalized difference vegetation index (NDVI) and enhanced vegetation index (EVI), drive the dominance of herbivores in nematode communities (type 1), whereas edaphic characteristics are correlated with communities dominated by microbivores (types 3 and 4). The names of the environmental variables are listed in Supplementary Table 3.
Supplementary information
41586_2019_1418_MOESM2_ESM.csv
Supplementary Table Supplementary Table 1 | Nematode abundance data and corresponding metadata values. Abundance data for each trophic group and associated metadata from 1,876 1-km2 pixels that were used for geospatial modelling and abundance data from 39 1-km2 pixels from Antarctica. (.csv file).
41586_2019_1418_MOESM3_ESM.csv
Supplementary Table Supplementary Table 2 | Summary of mean, median and sample size values per biome. The number of sites corresponds to the number of 1-km2 pixels into which the samples were aggregated. (.csv file).
41586_2019_1418_MOESM4_ESM.xlsx
Supplementary Table Supplementary Table 3 | Global covariate layers used for geospatial modelling. A total of 73 global covariate layers was used in our modelling approach. The 7 Nadir Reflectance Band layers (i.e., MCD43A4.005 BRDF-Adjusted Reflectance 16-Day Global 500m) are summarised as one entry in the table. (.xlsx file).
41586_2019_1418_MOESM5_ESM.xlsx
Supplementary Table Supplementary Table 4 | Variable importance metrics. Edaphic characteristics emerged as the most important variables. As the full dataset includes collinear variables leading to a false representation of the variable importance metrics, analysis was performed on a selection of main variables. (.xlsx file).
41586_2019_1418_MOESM6_ESM.csv
Supplementary Table Supplementary Table 5 | Number of soil nematodes per trophic group, per biome. Summing the predicted number of nematodes per 1 km2 pixel across biomes we estimate a total of 4.4 × 1020 nematodes are present in the top 15 cm of soil across the globe. (.csv file).
41586_2019_1418_MOESM7_ESM.csv
Supplementary Table Supplementary Table 6 | Relative abundance of soil nematodes per trophic group, per biome. (.csv file).
41586_2019_1418_MOESM8_ESM.csv
Supplementary Table Supplementary Table 7 | Nematode biomass per trophic group, per biome. Note that values are presented in megatons (106 tons) carbon. (.csv file).
Rights and permissions
About this article
Cite this article
van den Hoogen, J., Geisen, S., Routh, D. et al. Soil nematode abundance and functional group composition at a global scale. Nature 572, 194–198 (2019). https://doi.org/10.1038/s41586-019-1418-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1418-6