Abstract
Objective
Rare disease research requires data sharing networks to power translational studies. We describe novel use of Research Electronic Data Capture (REDCap), a web application for managing clinical data, by the National Mesothelioma Virtual Bank, a federated biospecimen, and data sharing network.
Materials and Methods
National Mesothelioma Virtual Bank (NMVB) uses REDCap to integrate honest broker activities, enabling biospecimen and associated clinical data provisioning to investigators. A Web Portal Query tool was developed to source and visualize REDCap data in interactive, faceted search, enabling cohort discovery by public users. An AWS Lambda function behind an API calculates the counts visually presented, while protecting record level data. The user-friendly interface, quick responsiveness, automatic generation from REDCap, and flexibility to new data, was engineered to sustain the NMVB research community.
Results
NMVB implementations enabled a network of 8 research institutions with over 2000 mesothelioma cases, including clinical annotations and biospecimens, and public users’ cohort discovery and summary statistics. NMVB usage and impact is demonstrated by high website visits (>150 unique queries per month), resource use requests (>50 letter of interests), and citations (>900) to papers published using NMVB resources.
Discussion
NMVB’s REDCap implementation and query tool is a framework for implementing federated and integrated rare disease biobanks and registries. Advantages of this framework include being low-cost, modular, scalable, and efficient. Future advances to NVMB’s implementations will include incorporation of -omics data and development of downstream analysis tools to advance mesothelioma and rare disease research.
Conclusion
NVMB presents a framework for integrating biobanks and patient registries to enable translational research for rare diseases.
Keywords: National Mesothelioma Virtual Bank, REDCap, biorepository, rare disease, biobank, registry
INTRODUCTION
Malignant mesothelioma is an aggressive, rare cancer arising from serous outer linings of various organs including the lungs, heart, abdomen, and testes.1–3 Eighty percent of mesothelioma cases are associated with asbestos exposure, and related minerals and genetic variations are emerging as risk factors.4–7 Geographic locations and occupations also correlate with the risk of developing mesothelioma.8 Incidence is ∼3000 cases per year in United States and mesothelioma is therefore considered a rare disease.5 Mesothelioma can remain latent for 20–50 years, and despite reduced use of asbestos since the 1970s, the incidence is projected to rise.9 Standard treatment is trimodal therapy with chemotherapy, surgical resection, and radiation, but the prognosis is unfortunately grim with a median survival of 18 months and 5-year survival rate of 14%.10,12,13 Unfortunately, advancements in novel therapies for mesothelioma have historically been slow and 40% of patients reported not having any treatment.14,15 There may be several reasons for these challenges including regionality of the disease.16 Most patients with mesothelioma are treated at community hospitals that may not have access to experimental treatments. Conversely, large academic centers may not have the accrual of cases needed to conduct rigorous clinical trials and translational research using biospecimens. The inherent challenges posed by investigating diseases with low incidence such as mesothelioma are shared by rare disease communities.
To overcome these challenges, the National Mesothelioma Virtual Bank (NMVB) was established in 2006.17–19 NMVB is a consortium of 8 research institutions united with the mission of creating a national resource of mesothelioma data and associated biospecimens for basic and translational research. Consortium members consent patients, collect biospecimens, provide clinical annotations, and pool the data for public research. With institutions distributed geographically across the United States, including areas of high asbestos exposure, NMVB has coalesced over 2000 cases capable of powering rigorous biomedical research.20 Support from the National Institute of Occupational Health and Safety of the Centers for Disease Control and Prevention has enabled NMVB to flourish for more than 16 years and grow into the largest resource for mesothelioma biospecimens including contributions to The Cancer Genome Atlas (TCGA) program.21 Moreover, NMVB has implemented an open-access Tissue Microarray Data Exchange Specification, making it the largest publicly shareable biospecimen and richly annotated resource available to investigators at no cost in the United States.22–25
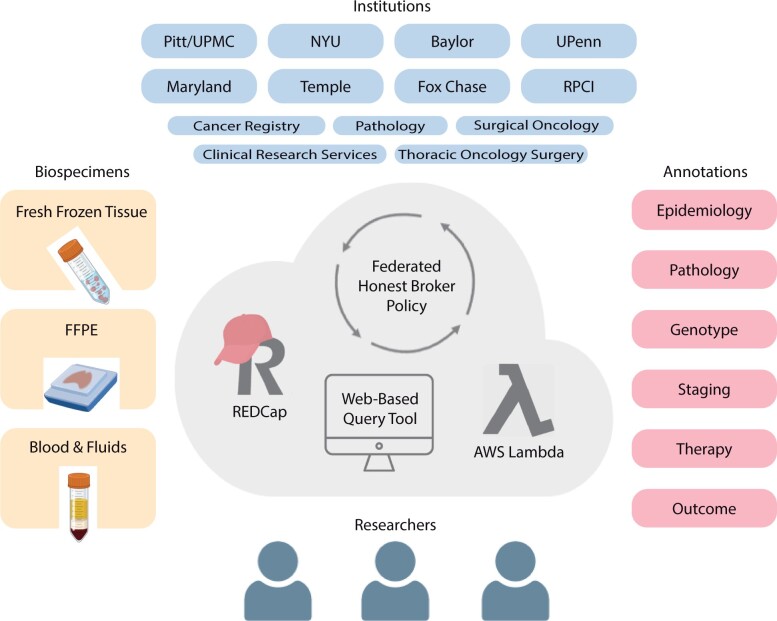
Creating complex biorepositories at scale requires sophisticated informatics solutions. The NVMB is a federated, virtual biorepository with physical specimens residing at individual institutions and a centralized database (Figure 1). To construct the combined biobank and registry, the NMVB previously developed the Clinical Annotation Engine (caTISSUE).18 While the caTISSUE system successfully integrated federated data, the platform required specialized training accumulating in an undesirably high cost burden and limitations for scaling. NMVB’s new Research Electronic Data Capture (REDCap) and AWS web-based architecture has solved the data collection and provisioning via 3 levels of access. The first is an honest broker system in REDCap that is used to deidentify and transmit data between distinct institutions, the national database, and users in compliance with Health Insurance Portability and Accountability Act (HIPAA). This includes identifying and annotating retrospective cases and communicating with the clinical team for consent. The second is data manager access to query and edit stored data. The third is the public query tool for population-level statistics.
Figure 1.
Schematic of the National Mesothelioma Virtual Bank (NMVB). NMVB is comprised of 8 research institutions that collect and contribute mesothelioma biospecimens and clinical annotations. The database is maintained virtually through honest brokers and is available to investigators.
Data sharing platforms for rare diseases come with a unique set of requirements different from platforms for common diseases.26 Rare disease research and personalized medicine necessitate data management systems capable of linking biobanks and patient data.27,28 Rare diseases require multi-institutional collaboration to assemble sufficient cases for study. Managing multi-institutional data can pose additional challenges such as: controlling identifying data privately and confidentially via multiple honest brokers, precise case definitions, ongoing customization of fields, long-term maintenance under changing data requirements, and challenge of balancing maximal accessibility via public facing cohort discovery tools with statistical disclosure control. The importance of addressing these challenges is well recognized, with groups developing methods such as the open-source Registry for Rare Diseases (OSSE) to link multiple rare disease registries in an interoperable environment.29–32 As precision medicine grows, patient stratification will become more granular and more conditions may be considered rare. Additionally, data from multi-institutional collaborations spanning geographical regions will be required to inform robust research. Thus, the informatics community will face increased demand for data sharing platforms and software tailored for rare disease research.
A popular software tool to manage secure capture of clinical data for research is The REDCap system.26,33,34 Since its development at Vanderbilt University in 2004, REDCap has been widely adopted by the biomedical research community with over 1 million projects to date, users spanning thousands of institutions across 150 countries, and over 22 000 journal article citations.35,36 It is commonly used to create registries, which are valuable collections of observational patient data used to identify high-risk groups, examine disease patterns, and inform public health policy. REDCap-based registries span a range of domains including adult diseases,11,37–39 pediatrics populations,40,41 medical products,42 and veterinary medicine,43 among others. Registries are especially valuable for rare diseases because they help aggregate patients. Another important but less frequent application of REDCap is managing biobanks.34 Biobanks involve collection, processing, storage, and distribution of biological specimens. Biobanks may be disease-oriented banks that collect samples from patients with certain diseases to identify biomarkers, or population-based banks that store biomaterial and other phenotypic data to identify markers of susceptibility in a general population, or tissue banks which harvest tissues for transplantation or research. Because NMVB has dual-functionality as both a patient registry and a biobank, we explored using REDCap in capturing its data.
We present a novel implementation extending REDCap to create a federated virtual biobank with public query access. We describe the creation of the NMVB database using the REDCap platform, and present our workflow as a model for rare disease management for translational research. We discuss the advantages of a network system for creating a low-cost, scalable, and efficient data collection model, the development of a federated data sharing standard, and challenges of implementing a federated and integrated structure for rare diseases. NVMB presents a framework for future integration of a national biobank and patient registry for rare diseases.
OBJECTIVE
Our objective is to describe the implementation of REDCap in the NMVB, discuss the advantages and limitations of the system, and propose our framework as a model for data sharing in the rare disease community.
MATERIALS AND METHODS
Consortium institutions
NMVB is currently comprised of 8 institutions: University of Pittsburgh/University of Pittsburgh Medical Center (UPMC), University of Pennsylvania (UPenn), New York University (NYU), Mount Sinai School of Medicine (MSSM), University of Maryland, Temple University/Fox Chase Cancer Center, Roswell Park Cancer Institute (RPCI), and Baylor College of Medicine. The collaborative consortium allows collection of cases and biospecimens from medical settings across diverse urban and rural regions.
Identification of cases
Inclusion criteria for participation in NMVB are individuals who are: (1) seeking or receiving medical care for mesothelioma and (2) 18 years or older and able to provide consent. Exclusion criteria are individuals who are: (1) younger than 18 years of age or are (2) prisoner-patients not able to provide consent according to federal guidelines.
Prospective collection
Prospective cases are identified in clinical settings including out-patient clinical visits and in-patient hospital admissions. Anyone receiving medical care for mesothelioma at a participating institution is screened for participation. Due to the wide inclusion and narrow exclusion criteria, the demographics of the individuals approached for participation in NMVB is representative of the disease population.
Retrospective collection
Retrospective cases are identified using an honest broker system. The honest broker at each participating site reviews medical records to identify patients who are eligible but were not previously approached. The honest broker is also utilized by the tissue bank to retrieve excess samples from archived biospecimens that were collected for clinical use.
Common data elements
Common data elements (CDEs) were developed by the NMVB Coordinating Committee with consensus from participating institutions and domain experts for annotations at the patient, specimen, and block level as previously described.17 The CDEs are harmonious with accepted standards such as NAACCR Data Standards, CAP Cancer Protocol and Checklist, Association of Directors of Anatomic and Surgical Pathology, and American Joint Committee on Cancer Cancer Staging System. The use of CDEs permits annotations that are uniform, consistent, and interoperable.
Regulatory considerations
NMVB is designed to protect patient privacy in compliance with HIPAA requirements, and all NMVB activities are Institutional Review Board (IRB) approved.
REDCap
Multiple instances of REDCap software were used to build the NMVB database. See the “Introduction” and “Results” sections and Figure 2 for details.
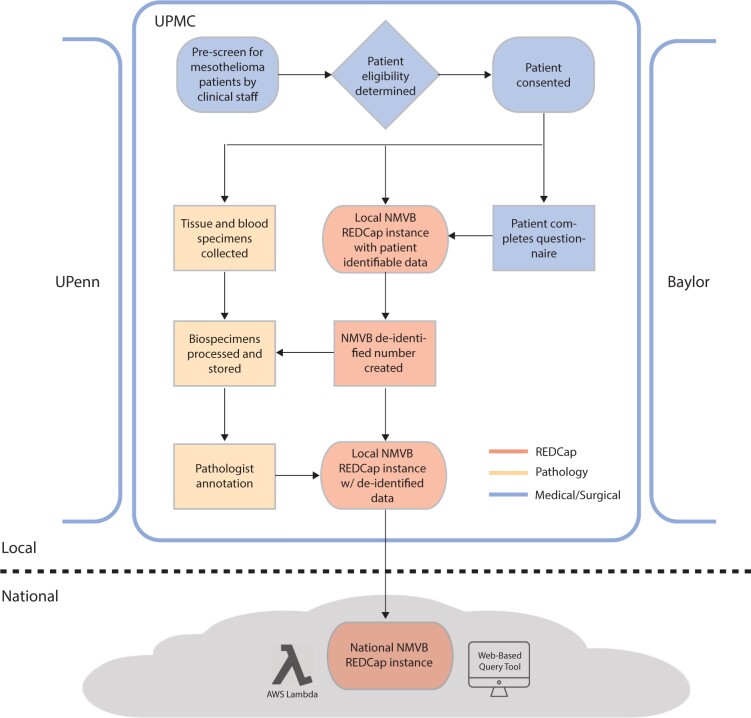
Figure 2.
Database structure of the NMVB using REDCap. The NMVB database is federated with activities of individual institutions at the “local” level and a unifying centralized database at the “national” level. The figure highlights Pitt/UPMC’s local workflow with steps occurring among medical/surgical teams colored in blue, pathology department in yellow, and REDCap in red.
User options
The NMVB national REDCap project contains hundreds of data elements on thousands of patients. The Web Portal Query (WPS) tool extracts data from the national REDCap project and transforms it into a public facing visualization that is available for cohort identification and summary statistics. Investigators can request biospecimens and access individual patient data for research studies by submitting a letter of interest (LOI) that is reviewed for approval by the NMVB coordinating committee.
Design requirements for Web Portal Query tool
The NMVB Web Portal Query tool, with the goal of enabling public faceted search of the NMVB resource, was constructed using the following design imperatives:
1) Per IRB authorization, it may allow querying and reporting of detailed counts of patients for non-identifying characteristics to the general public (statistical disclosure control is counts only, no record-level data provided, but counts allowed down to 1 patient).
2) The underpinning patient-level record data, even though deidentified and countable for key features, must not be distributed. This was achieved by an innovative use of the AWS Lambda function hidden behind an API that calculates, at high performance, the necessary counts for the faceted interface (rather than counting facets within the user’s browser which would expose record-level data to the user’s computer).
3) Interactive use requiring no prior training and few sentence explanations for a public user.
4) Speed of responsiveness in hundreds of milliseconds and intuitive graphical interface showing bar graphs of characteristics.
5) Display of 16 facets selected by investigators who frequently request NMVB resources.
RESULTS
NMVB project structure in REDCap and workflow
The NMVB is a federated, multi-level organization which currently has 8 coordinating sites with activities at the “local” level and centralized activities at the national level, as depicted in Figure 2. At Pitt/UPMC, we have implemented 3 REDCap projects, 2 at the local institution level (honest broker and deidentified) and one at the national level. At the local level, REDCap is used to collect, store, and deidentify data. At the national level, it is used to merge data from multiple institutions into a unified project. Note, each institution or honest broker service has the option to deploy their own workflow variation at the local level.
The workflow for prospective cases begins with clinical teams identifying potential mesothelioma patients by review of their clinic schedule. The coordinator and physician then screen the patient to determine eligibility for enrollment in NMVB. After eligibility screen, the patient is approached for participation and collection of tissue, blood, fluids, and genomic testing. When a patient consents, a questionnaire is administered to collect sociodemographic variables; environmental exposures; and health, occupational, and family medical history. A local honest broker creates a unique NMVB study ID number for the patient. Each site maintains a log of enrolled patients along with their NMVB study ID number that is maintained and only accessible by the local honest broker in accordance with best practices recommended by the REDCap consortium. Information from the questionnaire and EHR is then entered into a separate local REDCap project by NMVB ID in a patient deidentified manner.
Blood and tissue specimens collected by the surgical team are transferred to pathology for processing, annotation, and storage. Specimens are labeled with NMVB study ID and stored by tissue banking specialists. A pathologist reviews the biospecimens and enters predetermined pathomolecular CDEs (ie, Tumor-Node-Metastasis staging) into the local deidentified REDCap instance using the NMVB study ID. Finally, the local deidentified REDCap data are merged with the national REDCap data. Therefore, only deidentified, structured data are ever transmitted to the national NMVB project.
Note, for retrospective case collection, the workflow begins with a search of the medical records by local honest brokers for mesothelioma patients that have had a surgical resection or biopsy as part of standard care within the last year. The list of cases is cross checked with NMVB cases to identify unenrolled patients and sent to Pathology to obtain FFPE tissue blocks. If the tissue block is determined to be of sufficient quality to include in NMVB, an NMVB study ID is generated by the honest broker and the remaining steps are similar to prospective collection.
REDCap database
The national, deidentified REDCap project database contains 17 data collection instruments, covering the following domains: enrollment, demographics, patient’s cancer history, pre-existing conditions (comorbidities), recurrence data, therapy (chemo, radiation, surgery), physical characteristics and imaging history, occupation history, exposure history, tobacco and alcohol usage, family cancer history, vital status, specimen accession, block, specimen availability, and staging. The project uses a single data collection instrument for patient demographics with subsidiary instruments for each domain. The separate local, identified projects contain only the honest broker table, not all the forms.
Data transformation
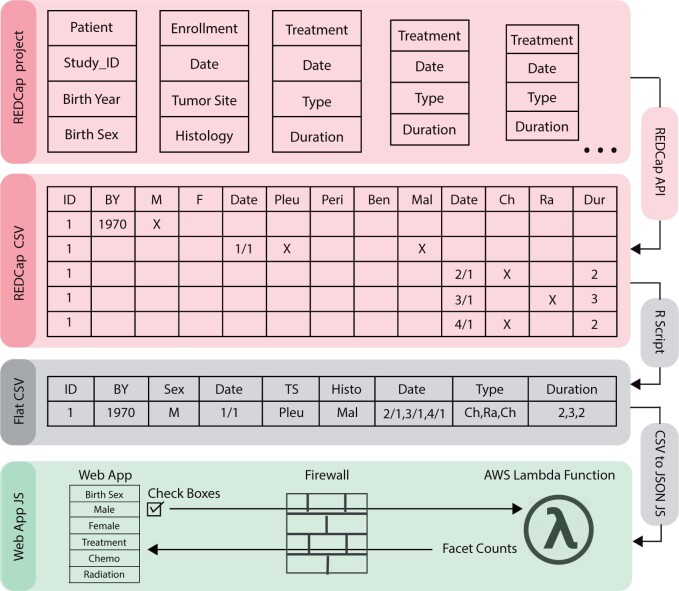
To produce data for the Web Portal Query tool, a REDCap data transformation tool was built in R that transforms any REDCap database standard API output into a matrix with 1 row per patient and 1 column per question. It generates a JSON file compatible with the WPQ tool and a CSV for honest brokers and LOI servicers. Figure 3 displays a theoretical REDCap Project with 5 forms for 1 patient. A generic call to the REDCap API generates a CSV with each row representing a form, and each column representing a nominal option for a field. The CSV is flattened to 1 row per patient and collapsed to 1 column per field by the transformation tool in R, generating a Flat CSV. This enables a 1-row-per-patient record-level dataset for detailed data investigations. A routine CSV-to-JSON conversion in JavaScript further transforms it as backend JSON data for the AWS Lambda Function. Parameters of the WPQ API call include Check Boxes with checked status in the Web App UI. The Lambda responds with the count values for each CDE, enabling the Web App to display data without user access to raw data.
Figure 3.
Data transformation. Data from the REDCap project are transformed into a format compatible with the Web Portal Query tool. First, the REDCap API is used to export data from REDCap as a CSV file which is then transformed into a flat CSV using an R script. The flat CSV is converted to a JSON file using JavaScript, which can then be accessed by the WPQ API via AWS Lambda to calculate counts and statistics and display in real time.
Web Portal Query and data visualization tool
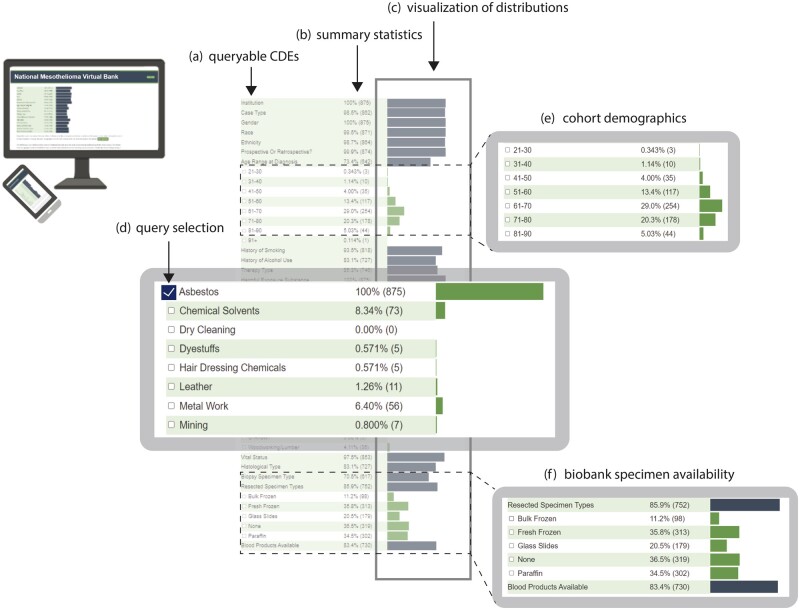
To enable fast, intuitive visualization of query results, we developed the NMVB Web Portal Query Tool shown in Figure 4. It is accessible to the general public on the NMVB website: https://data.mesotissue.org. The initial view displays 16 CDE categories which can be expanded to reveal more than 100 CDEs including demographics (ie, sex, age range), diagnosis (ie, histologic subtype), exposures (ie, asbestos), medical history (ie, smoking), treatment (ie, chemotherapy, immunotherapy), and specimen availability in the biobank (ie, FFPE tissue, blood products). Counts and percentages of the total NMVB cohort for which each data element is available are displayed with accompanying bar charts. Queries can be made using checkboxes for any number of CDEs. Upon selection or deselection of checkboxes, summary statistics and bar charts immediately update for all CDEs to reflect the new cohort. By utilizing this public view, investigators can efficiently assess the availability of annotations or biospecimens needed to power their studies and decide whether to submit an LOI for data and specimens.
Figure 4.
NMVB Web Portal Query and visualization tool. NMVB Web Portal Query interface contains a list of (a) CDE categories that can be expanded to reveal related CDEs. For each CDE, (b) summary statistics and (c) visualization of the distribution of counts is displayed as bar charts. Upon (d) CDE selections (query), summary statistics and bar charts update automatically. Example query results of a selected cohort are highlighted such as (e) age range and (f) availability of the specimens. Tool accessible at: data.mesotissue.org.
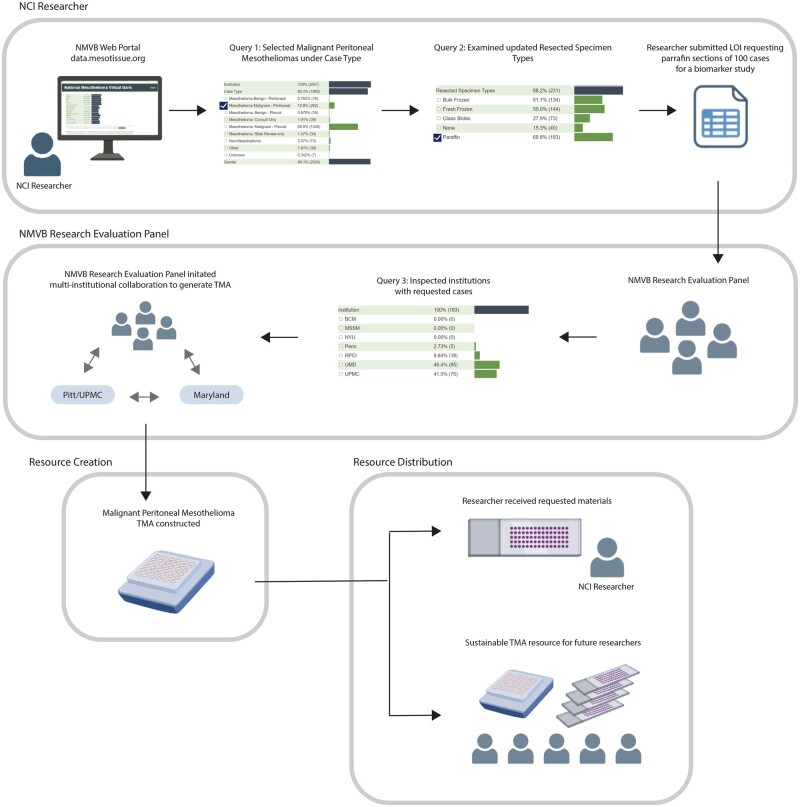
Case study
To demonstrate the impact of the NVMB Web Portal Query, we present a case study of a National Institutes of Cancer (NCI) researcher who used the tool to acquire biospecimens for a biomarker study on malignant peritoneal mesotheliomas (Figure 5). The researcher accessed the NMVB Web Portal, selected the checkbox for malignant peritoneal mesotheliomas, navigated to the Resected Specimen Types section, and identified 183 available paraffin specimens in the NMVB database. The researcher submitted an LOI requesting paraffin sections from 100 cases, which was evaluated and approved by the NMVB Research Panel. The panel used the Web Portal Query Tool to identify institutions from which to obtain the required specimens. However, as no single institution had the required number of cases, the panel approached UPMC and Maryland, which had 75 and 85 cases, respectively. To optimize rare disease resources, the panel also initiated a multi-institutional collaboration to create a TMA resource for malignant peritoneal mesotheliomas, which provided the researcher with ample samples while simultaneously creating a sustainable resource for future researchers.
Figure 5.
NMVB user case study. The case study illustrates how an NCI researcher utilized the tool to request paraffin sections from 100 cases of malignant peritoneal mesotheliomas for a biomarker study. The researcher submitted an LOI that was evaluated and approved by the NMVB Research Panel, which then used the Web Portal Query tool to identify institutions with the required specimens. As no single institution had the necessary samples, the panel initiated a multi-institutional collaboration to create a TMA resource. The resulting TMA provided ample samples for the researcher’s study and a sustainable resource for future studies.
Usage results
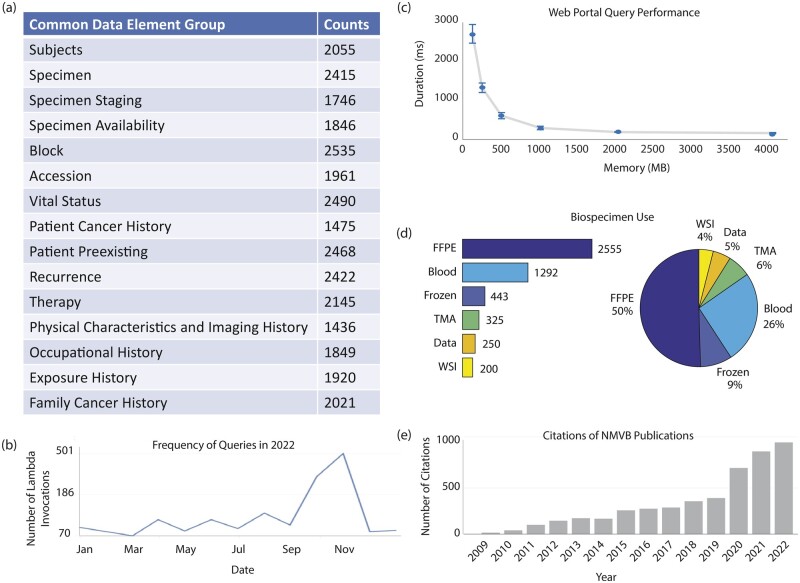
The successful implementation of REDCap has facilitated the expansion of NMVB into a comprehensive and widely used database, comprising over 2000 cases sourced from 8 institutions. The high completeness across all CDE groups is reflected in Figure 6a. Additionally, the usage of the web query tool is demonstrated in Figure 6b, which exhibits an average of ∼170 lambda invocations, or unique queries, per month in 2022 with a peak of ∼500 in November.
Figure 6.
Usage results of REDCap implementation in NMVB. Size and comprehensiveness of the database is reflected in the high (a) form completeness. Usage and performance of the database is quantified by (b) number of queries over time, (c) query response time, and (d) LOI and biospecimen utilization. Impact on mesothelioma research is reflected by the growing number of (e) citations of research papers using resources from NMVB. All data as of December 31, 2023.
To ensure optimal performance of the query tool, we conducted tests by adjusting the memory allocation in the AWS environment and measuring query response times, as depicted in Figure 6c. The results indicated that at a memory allocation of 2048 megabytes (MB), each query was completed within ∼157 ms (SD = 7). This response time falls well below the standard human response time of 200 ms44 for a typical response to stimuli and is also significantly faster than that of comparable clinical databases.45 Furthermore, response times did not show any significant improvement beyond the 2048 MB allocation, thus making it the optimal setting for the query tool.
Since inception, NMVB has enabled 56 researchers from 45 different institutions to conduct research projects. To date, greater than 50 LOIs have been requested by researchers, and greater than 4000 biospecimens from the biobank have been utilized (Figure 6d). Research publications fueled by the NMVB have been widely cited, as shown in Figure 6e. Biomedical advancements propelled by NMVB include: identification of BAP1 mutations as a risk factor,7,21,46 discovery of CD30 as a new potential therapeutic target,47 update of diagnostic guidelines for malignant mesothelioma,1,48,49 development of a novel gene expression prognostic test,50 development of deep learning methods,51 and contributions to TCGA.
DISCUSSION
Initially used for diagnostics purposes in pathology settings, biobanks have evolved to complex organizations that enable large-scale omics studies, personalized medicine, and translational research. REDCap software is a key tool for managing these data.52–57 In the rare disease domain, REDCap has enabled multicenter biobanks to aggregate enough samples to conduct evidence-based studies on diseases of low incidence. Moreover, it has enabled a novel “virtual” biobank framework where new data elements can be easily added over time. NMVB implemented REDCap in data collection, storage, and management and utilized it to perform data transformation and visualization for efficient data sharing.
Using REDCap to create a federated data sharing network has several advantages:
Modular. The federated design of NMVB with single institutions serving as a module to the overall architecture allows for maximum flexibility. An institution can have varying underlying clinical workflows for specimen collection and storage while data capture for the NMVB database can be standardized through CDEs in REDCap.
Scalable. The modular design allows for efficient addition of new institutions. As a case study, we recently added Baylor College of Medicine to NMVB, and the onboarding process took only 3 months, with 295 cases comprising over 300 data elements in the initial upload.
Low costs. REDCap is an open-source software, and usage is free. The WPQ tool uses JavaScript in a single html file deployed on a simple web server as the interface, backed by an AWS API Gateway and an AWS Lambda Function (also in JavaScript). WPQ is extremely efficient both for cost (pennies on AWS) and performance (under 200 ms latency).
Standardized data elements. Standardized data elements are easily implemented using REDCap’s project structure and are beneficial for gathering uniform, consistent, and interoperable data from various institutions.58
Evolving data types. REDCap’s flexibility59 allows NMVB to add new data elements over time supporting long-term sustainability and provides a collaborative framework to add partners from the PCORI CDRN and NCATS networks where REDCap is ubiquitous.
Interactive, real-time data exploration. WPQ and associated data transformations enable direct use of the REDCap API to solve one of REDCap’s missing features: interactive faceted data exploration.
Data privacy. REDCap has built-in features for patient confidentiality and HIPAA compliance requirements.60 The honest broker approach ensures identifiable data from 1 institution is not shared with honest brokers from another institution. Each site manager can only see the deidentified data for their site, and WPQ only includes deidentified data, adherent to Safe Harbor.
Federated data sharing. By simplifying data management for the federated NMVB network, using REDCap, we successfully addressed the following issues via methods generalizable to any similar rare disease network61: controlling identifying data privately and confidentially via multiple honest brokers; precise case definitions; ongoing customization of fields in data collection; long-term maintenance under changing data requirements; and in particular, the challenge of balancing public facing cohort discovery with maximal accessibility and minimal statistical disclosure control.
Limitations
REDCap has some limitations when considering its use as a data management system. Most notably, it does not provide a comprehensive set of features for biobanks, such as sample management, tracking, and storage. For managing physical biospecimen inventory, most biobanks currently use commercial LIMS systems. Moreover, REDCap does not support deep annotation of pathology specimens, imaging such as MRI or H&E, or genomics. Federated systems more equipped to handle such omics data are used and developed within national wide consortia such as MIRACUM62–64 and DIFUTURE65 leveraging multiple sophisticated open-source tools such as cBioportal.66,67
Comparison to current literature
Several groups have attempted to implement REDCap for virtual biorepositories, but none have comprehensively met the needs of NMVB as a virtual, federated, and combined biobank and registry. Fisher et al26 described a federated REDCap approach serving an international consortium studying primary tumors of the spine containing nearly 1500 patients from 8 consortia sites. However, no public query tool was described. Felmeister et al68 described an open-source biorepository portal toolkit with an electronic honest broker partially constructed in REDCap that scaled to 8 institutions, managing multiple protocols and over 60 000 biospecimens. The system facilitates automated deidentification of clinical and genomic data and allows for electronic health records (EHR) integration. The advantage is that biospecimens are deidentified and cohort identification is available through a Harvest framework69 to consortium members. This modular, scalable system illustrates the utility of REDCap in biorepository management serving Children’s Hospital of Pennsylvania biorepository portal and brain cancer researchers via the Kids First Data Resource Portal70 and the Children’s Brain Tumor Network (CBTN).71 Gluski et al72 described a federated hydrocephalus biobank using REDCap containing 50 data elements of 293 samples from 228 patients. However, no honest broker or advanced analytics were utilized. Willers et al73 described the A3BC which developed a REDCap-based registry and biobank model focusing on optimizing its biobank collection for downstream analytics by linking a broad array of datasets to biospecimens. They compared several open-source and commercial biobanking solutions and determined that REDCap was the most sustainable and low cost.
Suggestions for future work
The use of REDCap in NMVB has been successful in creating a virtual, federated biobank and registry. However, further development is required to meet the evolving needs of NMVB. Future directions include adding new data elements to NMVB to support emerging technologies and data types (particularly -omics and real-world evidence). Expanding NMVB’s capabilities to enable automated inventory of specimens stored at multiple sites will also optimize biospecimen querying. Finally, a key area of focus will be to optimize the system for downstream data analytics using REDCap’s External Module framework. By doing this, NMVB could leverage machine learning and artificial intelligence to achieve a greater impact on mesothelioma and human health.
CONCLUSION
Rare disease data sharing networks require participation from multiple institutions. The adoption of REDCap to create a federated database structure has multiple advantages including being flexible, scalable, low cost, and sustainable to changing data types. The framework implemented by NMVB serves as a model for other data sharing networks for rare diseases.
ACKNOWLEDGMENTS
We thank Paul Harris for helpful discussions.
Contributor Information
Rumana Rashid, Department of Biomedical Informatics, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA; Medical Scientist Training Program, University of Pittsburgh-Carnegie Mellon University, Pittsburgh, Pennsylvania, USA.
Susan Copelli, Department of Biomedical Informatics, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Jonathan C Silverstein, Department of Biomedical Informatics, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
Michael J Becich, Department of Biomedical Informatics, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA.
FUNDING
This work was supported by the Centers for Disease Control and Prevention (CDC) in association with the National Institute for Occupational Safety and Health (NIOSH) grant number 5U24OH009077-14 to MJB, the National Institutes of Health grant number UL1 2UL1TR001857 to MJB and JCS, and by the National Institute of General Medical Sciences of the National Institutes of Health grant number 5 T32 GM 8208-33 and the National Library of Medicine grant number 2T15LM007059-36 to RR.
AUTHOR CONTRIBUTION
RR and MJB contributed to conception, design, and all drafts of the manuscript; and all other authors provided critical input and approved the final version of the manuscript to be published. JCS contributed to the technical description, design, coding, and deployment of the system.
CONFLICT OF INTEREST STATEMENT
MJB is a founder, patent owner, investor, and stock owner of PredxBio (formerly SpIntellx). All other authors have no competing interests to declare.
DATA AVAILABILITY
All NMVB clinical data as well as available biospecimens are publicly accessible as counts through the NMVB query tool at http://www.mesotissue.org/data. The code repository for the data transformation and statistical disclosure control for NMVB can be provided by contacting j.c.s@pitt.edu.
REFERENCES
- 1. Dacic S. Pleural mesothelioma classification—update and challenges. Mod Pathol 2022; 35 (Suppl 1): 51–6. [DOI] [PubMed] [Google Scholar]
- 2. Sinn K, Mosleh B, Hoda MA.. Malignant pleural mesothelioma: recent developments. Curr Opin Oncol 2021; 33 (1): 80–6. [DOI] [PubMed] [Google Scholar]
- 3. Churg A, Galateau-Salle F, Roden AC, et al. Malignant mesothelioma in situ: morphologic features and clinical outcome. Mod Pathol 2020; 33 (2): 297–302. [DOI] [PubMed] [Google Scholar]
- 4. Attanoos RL, Churg A, Galateau-Salle F, Gibbs AR, Roggli VL.. Malignant mesothelioma and its non-asbestos causes. Arch Pathol Lab Med 2018; 142 (6): 753–60. [DOI] [PubMed] [Google Scholar]
- 5. Bianchi C, Bianchi T.. Malignant mesothelioma: global incidence and relationship with asbestos. Ind Health 2007; 45 (3): 379–87. [DOI] [PubMed] [Google Scholar]
- 6. Selikoff IJ, Churg J, Hammond EC.. RELATION BETWEEN EXPOSURE TO ASBESTOS AND MESOTHELIOMA. N Engl J Med1965; 272: 560–5. [DOI] [PubMed] [Google Scholar]
- 7. Testa JR, Cheung M, Pei J,. et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet 2011; 43 (10): 1022–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Robinson BM. Malignant pleural mesothelioma: an epidemiological perspective. Ann Cardiothorac Surg 2012; 1 (4): 491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhai Z, Ruan J, Zheng Y, et al. Assessment of global trends in the diagnosis of mesothelioma from 1990 to 2017. JAMA Netw Open 2021; 4 (8): e2120360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robinson BWS, Lake RA.. Advances in malignant mesothelioma. N Engl J Med 2005; 353 (15): 1591–603. [DOI] [PubMed] [Google Scholar]
- 11. Cummings KJ, Becich MJ, Blackley DJ, et al. Workshop summary: potential usefulness and feasibility of a US National Mesothelioma Registry. Am J Ind Med 2020; 63 (2): 105–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mutti L, Peikert T, Robinson BWS, et al. Scientific advances and new frontiers in mesothelioma therapeutics. J Thorac Oncol 2018; 13 (9): 1269–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Janes SM, Alrifai D, Fennell DA.. Perspectives on the treatment of malignant pleural mesothelioma. N Engl J Med 2021; 385 (13): 1207–18. [DOI] [PubMed] [Google Scholar]
- 14. Espinoza-Mercado F, Borgella JD, Berz D, et al. Disparities in compliance with national guidelines for the treatment of malignant pleural mesothelioma. Ann Thorac Surg 2019; 108 (3): 889–96. [DOI] [PubMed] [Google Scholar]
- 15. Saddoughi SA, Abdelsattar ZM, Blackmon SH.. National trends in the epidemiology of malignant pleural mesothelioma: A National Cancer Data Base Study. Ann Thorac Surg 2018; 105 (2): 432–7. [DOI] [PubMed] [Google Scholar]
- 16. Amin W, Linkov F, Landsittel DP, et al. Factors influencing malignant mesothelioma survival: a retrospective review of the National Mesothelioma Virtual Bank cohort. F1000Res 2018; 7: 1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mohanty SK, Mistry AT, Amin W, et al. The development and deployment of common data elements for tissue banks for translational research in cancer—an emerging standard based approach for the Mesothelioma Virtual Tissue Bank. BMC Cancer 2008; 8: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amin W, Parwani AV, Schmandt L, et al. National Mesothelioma Virtual Bank: a standard based biospecimen and clinical data resource to enhance translational research. BMC Cancer 2008; 8: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Amin W, Parwani AV, Melamed J, et al. National Mesothelioma Virtual Bank: a platform for collaborative research and mesothelioma biobanking resource to support translational research. Lung Cancer Int 2013; 2013 (2013): 765748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Amin W, Singh H, Pople AK, et al. A decade of experience in the development and implementation of tissue banking informatics tools for intra and inter-institutional translational research. J Pathol Inform 2010; 1 (1): 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hmeljak J, Sanchez-Vega F, Hoadley KA, et al. ; TCGA Research Network. Integrative molecular characterization of malignant pleural mesothelioma. Cancer Discov 2018; 8 (12): 1548–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berman JJ, Datta M, Kajdacsy-Balla A, et al. The tissue microarray data exchange specification: implementation by the cooperative prostate cancer tissue resource. BMC Bioinformatics 2004; 5: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kang HP, Borromeo CD, Berman JJ, Becich MJ.. The tissue microarray OWL schema: an open-source tool for sharing tissue microarray data. J Pathol Inform 2010; 1 (1): 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amin W, Srinivasan M, Song SY, Parwani AV, Becich MJ.. Use of automated image analysis in evaluation of mesothelioma tissue microarray (TMA) from National Mesothelioma Virtual Bank. Pathol Res Pract 2014; 210 (2): 79–82. [DOI] [PubMed] [Google Scholar]
- 25. Kajdacsy-Balla A, Geynisman JM, Macias V, et al. ; Cooperative Prostate Cancer Tissue Resource. Practical aspects of planning, building, and interpreting tissue microarrays: the cooperative prostate cancer tissue resource experience. J Mol Histol 2007; 38 (2): 113–21. [DOI] [PubMed] [Google Scholar]
- 26. Fisher CG, Goldschlager T, Boriani S, et al. An evidence-based medicine model for rare and often neglected neoplastic conditions: clinical article. J Neurosurg Spine 2014; 21 (5): 704–10. [DOI] [PubMed] [Google Scholar]
- 27. Semler SC, Wissing F, Heyder R.. German medical informatics initiative. Methods Inf Med 2018; 57 (S 01): e50–e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beyan O, Choudhury A, van Soest J, et al. Distributed analytics on sensitive medical data: the personal health train. Data Intell 2020; 2 (1–2): 96–107. [Google Scholar]
- 29. Storf H, Schaaf J, Kadioglu D, et al. Registries for rare diseases: OSSE—an open-source framework for technical implementation. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 2017; 60 (5): 523–31. [DOI] [PubMed] [Google Scholar]
- 30. Scheible R, Kadioglu D, Ehl S, et al. Enabling external inquiries to an existing patient registry by using the open source registry system for rare diseases: demonstration of the system using the European Society for Immunodeficiencies Registry. JMIR Med Inform 2020; 8 (10): e17420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vasseur J, Zieschank A, Göbel J,. et al. Development of an Interactive Dashboard for OSSE Rare Disease Registries. Stud Health Technol Inform 2022; 293: 187–8. [DOI] [PubMed] [Google Scholar]
- 32. Schueler K, Zieschank A, Göbel J,. et al. A Medical report feature for OSSE Rare Disease Registries. Stud Health Technol Inform 2021; 281: 1085–6. [DOI] [PubMed] [Google Scholar]
- 33. Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42 (2): 377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barnes R, Votova K, Rahimzadeh V,. et al. Biobanking for genomic and personalized health research: participant perceptions and preferences. Biopreserv Biobank 2020; 18 (3): 204–12. [DOI] [PubMed] [Google Scholar]
- 35. Harris PA, Taylor R, Minor BL, et al. ; REDCap Consortium. The REDCap consortium: building an international community of software platform partners. J Biomed Inform 2019; 95: 103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.REDCap. https://www.project-redcap.org/. Accessed April 17, 2021.
- 37. Huang H, Ng MY, Wu JT,. et al. Automating the Renal Cell Carcinoma Registry in Singapore: a case study on the integration of the research electronic data capture system with the enterprise data warehouse. J Registry Manag 2018; 45 (4): 156–60. [PubMed] [Google Scholar]
- 38. Pang X, Kozlowski N, Wu S,. et al. Construction and management of ARDS/sepsis registry with REDCap. J Thorac Dis 2014; 6 (9): 1293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thomas E, Grace SL, Boyle D, et al. Utilising a data capture tool to populate a cardiac rehabilitation registry: a feasibility study. Heart Lung Circ 2020; 29 (2): 224–32. [DOI] [PubMed] [Google Scholar]
- 40. Bahr TM, Christensen RD, Agarwal AM, George TI, Bhutani VK.. The Neonatal Acute Bilirubin Encephalopathy Registry (NABER): background, aims, and protocol. Neonatology 2019; 115 (3): 242–6. [DOI] [PubMed] [Google Scholar]
- 41. Mouttalib S, Rice HE, Snyder D,. et al. Evaluation of partial and total splenectomy in children with sickle cell disease using an Internet-based registry. Pediatr Blood Cancer 2012; 59 (1): 100–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. da Silva KR, Costa R, Crevelari ES,. et al. Glocal clinical registries: pacemaker registry design and implementation for global and local integration-methodology and case study. PLoS One 2013; 8 (7): e71090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hall KE, Boller M, Hoffberg J, et al. ; ACVECC’s Veterinary Committee on Trauma (VetCOT) Registry Subcommittee. ACVECC-Veterinary Committee on Trauma Registry Report 2013–2017. J Vet Emerg Crit Care (San Antonio) 2018; 28 (6): 497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stone NJ, Chaparro A, Keebler JR, Chaparro BS, McConnell DS.. Introduction to Human Factors: Applying Psychology to Design. Boca Raton: CRC Press; 2017. [Google Scholar]
- 45. Paris N, Mendis M, Daniel C, Murphy S, Tannier X, Zweigenbaum P.. i2b2 implemented over SMART-on-FHIR. AMIA Jt Summits Transl Sci Proc 2018; 2017: 369–78. [PMC free article] [PubMed] [Google Scholar]
- 46. Nasu M, Emi M, Pastorino S, et al. High incidence of somatic BAP1 alterations in sporadic malignant mesothelioma. J Thorac Oncol 2015; 10 (4): 565–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dabir S, Kresak A, Yang M, et al. CD30 is a potential therapeutic target in malignant mesothelioma. Mol Cancer Ther 2015; 14 (3): 740–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Husain AN, Colby TV, Ordóñez NG,. et al. Guidelines for pathologic diagnosis of malignant mesothelioma 2017 update of the consensus statement from the international mesothelioma interest group. Arch Pathol Lab Med 2018; 142 (1): 89–108. [DOI] [PubMed] [Google Scholar]
- 49. Beasley MB, Galateau-Salle F, Dacic S.. Pleural mesothelioma classification update. Virchows Arch 2021; 478 (1): 59–72. [DOI] [PubMed] [Google Scholar]
- 50. De Rienzo A, Cook RW, Wilkinson J, et al. Validation of a gene expression test for mesothelioma prognosis in formalin-fixed paraffin-embedded tissues. J Mol Diagn 2017; 19 (1): 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hartman D, Le Douget J-E, Ye Y, et al. Application of deep learning models on whole slide images uncover new histological markers related to high-risk malignant pleural mesothelioma. J Clin Oncol 2022; 40 (16_suppl): e13580. [Google Scholar]
- 52. Becich MJ. The role of the pathologist as tissue refiner and data miner: the impact of functional genomics on the modern pathology laboratory and the critical roles of pathology informatics and bioinformatics. Mol Diagn 2000; 5 (4): 287–99. [DOI] [PubMed] [Google Scholar]
- 53. Drake TA, Braun J, Marchevsky A, et al. ; Shared Pathology Informatics Network. A system for sharing routine surgical pathology specimens across institutions: the shared pathology informatics network. Hum Pathol 2007; 38 (8): 1212–25. [DOI] [PubMed] [Google Scholar]
- 54. Gilbertson JR, Gupta R, Nie Y, Patel AA, Becich MJ.. Automated clinical annotation of tissue bank specimens. Stud Health Technol Inform 2004; 107 (Pt 1): 607–10. [PubMed] [Google Scholar]
- 55. Patel AA, Gilbertson JR, Parwani AV,. et al. An informatics model for tissue banks-lessons learned from the Cooperative Prostate Cancer Tissue Resource. BMC Cancer 2006; 6: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Patel AA, Gupta D, Seligson D, et al. ; The Shared Pathology Informatics Network. Availability and quality of paraffin blocks identified in pathology archives: a multi-institutional study by the Shared Pathology Informatics Network (SPIN). BMC Cancer 2007; 7 (1): 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Eriksson J, Andersson S, Appelqvist R, et al. Merging clinical chemistry biomarker data with a COPD database—building a clinical infrastructure for proteomic studies. Proteome Sci 2016; 15: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Obeid JS, McGraw CA, Minor BL, et al. Procurement of shared data instruments for research electronic data capture (REDCap). J Biomed Inform 2013; 46 (2): 259–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nicolas-Boluda A, Oppenheimer A, Bouaziz J, Fauconnier A.. Patient-reported outcome measures in endometriosis. J Clin Med 2021; 10 (21): 5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lawrence CE, Dunkel L, McEver M, et al. A REDCap-based model for electronic consent (eConsent): moving toward a more personalized consent. J Clin Transl Sci 2020; 4 (4): 345–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jacobson RS, Becich MJ, Bollag RJ, et al. A federated network for translational cancer research using clinical data and biospecimens. Cancer Res 2015; 75 (24): 5194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Prokosch H-U, Acker T, Bernarding J, et al. MIRACUM: medical informatics in research and care in university medicine. Methods Inf Med 2018; 57 (S 01): e82–e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Walther T, Farin E, Boeker M,. et al. RECUR – Aufbau eines automatisierten digitalen Registers für Patient*innen mit rezidivierenden Steinen des oberen Harntraktes [RECUR- Establishment of An Automated Digital Registry for Patients with Recurrent Stones in the Upper Urinary Tract]. Gesundheitswesen 2021; 83 (S 01): S27–32. [DOI] [PubMed] [Google Scholar]
- 64. RECUR: A Nationwide Registry for Recurrent Urolithiasis builds on MIRACUM infrastructure. Medizininformatik-Initiative. https://www.medizininformatik-initiative.de/de/recur-nationwide-registry.
- 65. Prasser F, Kohlbacher O, Mansmann U, Bauer B, Kuhn KA.. Data integration for future medicine (DIFUTURE). Methods Inf Med 2018; 57 (S 01): e57–e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2 (5): 401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6 (269): l1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Felmeister AS, Masino AJ, Rivera TJ, Resnick AC, Pennington JW.. The biorepository portal toolkit: an honest brokered, modular service oriented software tool set for biospecimen-driven translational research. BMC Genomics 2016; 17 (S4): 434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pennington JW, Ruth B, Italia MJ, et al. Harvest: an open platform for developing web-based biomedical data discovery and reporting applications. J Am Med Inform Assoc 2014; 21 (2): 379–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kids First Data Portal. https://portal.kidsfirstdrc.org/login. Accessed July 16, 2023.
- 71. Children’s Brain Tumor Network (CBTN). Children’s Hospital of Philadelphia® Center for Data-Driven Discovery in Biomedicine. https://d3b.center/our-research/cbtn/.
- 72. Gluski J, Zajciw P, Hariharan P, et al. Characterization of a multicenter pediatric-hydrocephalus shunt biobank. Fluids Barriers CNS 2020; 17 (1): 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Willers C, Lynch T, Chand V, et al. A versatile, secure, and sustainable all-in-one biobank-registry data solution: the A3BC REDCap model. Biopreserv Biobank 2022; 20 (3): 244–59. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All NMVB clinical data as well as available biospecimens are publicly accessible as counts through the NMVB query tool at http://www.mesotissue.org/data. The code repository for the data transformation and statistical disclosure control for NMVB can be provided by contacting j.c.s@pitt.edu.