Abstract
Introduction:
Low vision rehabilitation improves quality-of-life for visually impaired patients, but referral rates fall short of national guidelines. Automatically identifying, from electronic health records (EHR), patients with poor visual prognosis could allow targeted referrals to low vision services. The purpose of this study was to build and evaluate deep learning models that integrate EHR data that is both structured and free-text to predict visual prognosis.
Methods:
We identified 5547 patients with low vision (defined as best documented visual acuity (VA) <20/40) on ≥ 1 encounter from EHR from 2009–2018, with ≥ 1 year of follow-up from the earliest date of low vision, who did not improve to > 20/40 over 1 year. Ophthalmology notes on or prior to the index date were extracted. Structured data available from the EHR included demographics, billing and procedure codes, medications, and exam findings including VA, intraocular pressure, corneal thickness, and refraction. To predict whether low vision patients would still have low vision a year later, we developed and compared deep learning models that used structured inputs and free-text progress notes. We compared three different representations of progress notes, including 1) using previously developed ophthalmology domain-specific word embeddings, and representing medical concepts from notes as 2) named entities represented by one-hot vectors and 3) named entities represented as embeddings. Standard performance metrics including area under the receiver operating curve (AUROC) and F1 score were evaluated on a held-out test set.
Results:
Among the 5547 low vision patients in our cohort, 40.5% (N=2246) never improved to better than 20/40 over one year of follow-up. Our single-modality deep learning model based on structured inputs was able to predict low vision prognosis with AUROC of 80% and F1 score of 70%. Deep learning models utilizing named entity recognition achieved an AUROC of 79% and F1 score of 63%. Deep learning models further augmented with free-text inputs using domain-specific word embeddings, were able to achieve AUROC of 82% and F1 score of 69%, outperforming all single- and multiple-modality models representing text with biomedical concepts extracted through named entity recognition pipelines.
Discussion:
Free text progress notes within the EHR provide valuable information relevant to predicting patients’ visual prognosis. We observed that representing free-text using domain-specific word embeddings led to better performance than representing free-text using extracted named entities. The incorporation of domain-specific embeddings improved the performance over structured models, suggesting that domain-specific text representations may be especially important to the performance of predictive models in highly subspecialized fields such as ophthalmology.
Keywords: ophthalmology; natural language processing; deep learning; vision, low; named entity recognition
1. Introduction
Almost 1.5 million (3.5% of individuals over the age of 65) are visually impaired and estimated to be candidates for low vision services [1]. These services help patients navigate their daily activities to maximize the function of their remaining. Without low vision services, many patients are unable to read standard print or maintain safety and independence in their daily activities [1], suffering enormous reductions in quality of life [2], with increased risk of falls and fractures [3], depression and anxiety [4], and mortality [5]. Maximizing functional vision is therefore a vital determinant of living and aging well [6].
Despite proven effectiveness in aiding patients in their activities of daily life [2,7], referral to low vision resources is greatly underutilized. Barriers to referral are multifactorial, including time constraints during appointments, and difficulty in predicting whether low vision patients might improve with treatment. Thus, there is a critical need to identify and educate patients who can benefit from low vision services, potentially in an automated manner, which may bypass the clinic-visit bottleneck and improve access to low vision services. If predictive algorithms could automatically identify patients from the electronic health record (EHR) with long-term poor vision, these algorithms could ultimately contribute to a clinical decision support system by offering targeted education and referrals to these critically important low vision services.
In order to build algorithms that capture and leverage the wealth of data available, researchers have explored utilizing unstructured data in the EHR systems. Clinical narratives reflect the main form of communication within healthcare, allowing providers to record richer and more personalized information. Because there is an immense wealth of data in clinical free text, there has been increasing interest in using natural language processing (NLP) to incorporate information from unstructured text data in predictive models [8]. However, there has been little prior work incorporating clinical free text into predictive models specifically for ophthalmic outcomes [9].
Previous work in our group explored using neural word embeddings as a method of representing clinical free text to predict low vision prognosis [10]. Using custom-trained ophthalmology domain-specific neural word embeddings, every word in the text is mapped to a vector which is then inputted into a deep learning model. These predictive models were able to achieve an AUROC of 81% for predicting low vision prognosis. An alternative representation of text involves named entity recognition (NER), where important concepts within the notes can be extracted and mapped to existing health ontologies, forming a feature set which can be used for prediction models [11–13]. The purpose of this study was to build and evaluate models to predict low vision prognosis by combining information from EHR in both structured and free-text formats, and comparing NER and neural word embeddings as potential approaches for representing ophthalmology clinical free text.
2. Methods
2.1. Data Source/Study Cohort:
This study has been approved by the Stanford Institutional Review Board. Using retrospective data (structured and free-text) from 2009–2018 from the Stanford Clinical Data Warehouse [14], we previously identified all documented visual acuity measurements (N=553,184) belonging to 88,692 unique adult patients [15]. The patients’ gender is determined by their health records, which are based on self-report and not necessarily on examination of body characteristics or genetic testing. Visual acuity measurements are available from labeled fields in the EHR, including measurements for distance, near, with refraction, with or without habitual glasses or contacts for either eye [15]. Low vision on a particular encounter date was defined as visual acuity worse than 20/40 on all measurements documented for that encounter.
In total there were 13,847 patients with at least one documented encounter with low vision. The first date of low vision was determined for each patient (hereafter referred to as the index date). We included patients with follow-up for at least one year from the index date (N=5612). For these patients, we extracted all ophthalmology free-text clinical notes on or prior to the index date (N=5547 patients with available notes), as shown in Figure 1. 40.5% of these patients still had low vision one year later.
Figure 1. Cohort Selection Process.
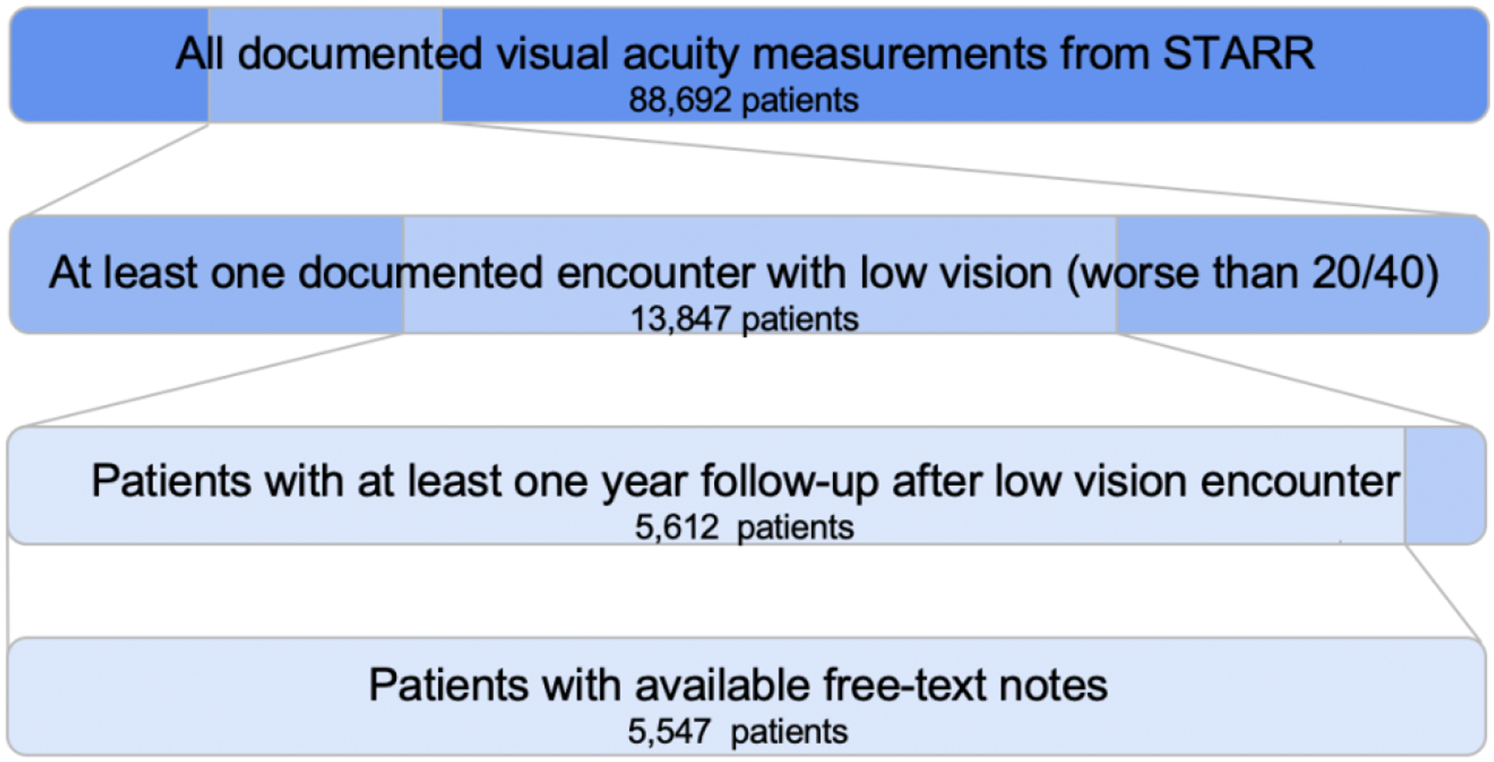
We started with 88,692 patients with documented visual acuity measurements, and only included patients who had at least one documented encounter with low vision worse than 20/40 and at least one-year follow-up (defined as greater than or equal to one visit with documented visual acuity measurement greater than or equal to 365 days from the index date) with free-text notes, ultimately resulting in 5,547 patients for our cohort.
2.2. Data Pre-Processing and Feature Engineering
2.2.1. Structured Inputs
Structured features available from the research warehouse were processed either as boolean variables or as continuous numeric variables. Boolean variables included demographic data, billing codes (ICD and CPT) indicating prior diagnoses and procedures, and current active medication usage. Continuous numeric variables included eye exam information for both eyes, summarized with high, low, most recent and mean values. To capture visual acuity, we calculated the logarithm of minimum angle of resolution from the best corrected visual acuity (BCVA logMAR). logMAR is recognized as the most reliable and discriminative visual acuity measurement [16]. All features with less than 1% variance were removed, and missing value indicator variables were created to indicate whether an individual clinical measurement was missing. In total there were 556 structured input features.
2.2.2. Free-text Clinical Progress Note Inputs
We identified and extracted all notes on or before the first date of low vision and combined all notes into one text file per patient.
Pubmed Word Embeddings:
All notes were lower-cased, tokenized, and had stopwords removed. See Supplementary Table A for more details. Words were mapped to 300-dimensional neural word embeddings customized for ophthalmology that were pre-trained on PubMed ophthalmology abstracts [10].
CLAMP Output Post-Processing:
In parallel with the PubMed word embeddings, we evaluated the ability of the Clinical Language Annotation, Modeling and Processing (CLAMP) tool [18] to identify ophthalmology-specific terminology by using a subset of ophthalmology terms and notes from the cohort. CLAMP is a clinical NLP toolkit that is trained on a dataset of generic clinical notes, namely, the i2b2 2010 challenge corpus. More validation details can be found in the supplementary materials. We then ran all notes through our customized NER pipeline (CLAMP) to identify the specific named entities as well as section headers, and conducted post-processing of these CLAMP outputs.
We first processed the section headers to categorize the named entities. Next, we removed entities under the section headers of: allergy, attestation, instruction, consent, and family/social history. These entities are not relevant to the patient themselves, and present as inconsistent data points. In addition to removing irrelevant entities, we also removed entities that were not mapped to a CUI, which were often unnecessary attributions of the named entities (drug dosage, body location, etc.). Thus, our final output from CLAMP per patient is a list of CUI’s with a negation marker.
CUI One-Hot Encoding:
As an input to our model, we created a one-hot encoding of the CUI’s, incorporating the negation by creating separate “terms” per CUI for the positive and negative occurrences of the entity. Similar to the structured inputs, features with less than 1% variance were removed, decreasing the vocabulary size of the CUI one-hot encoding from 22,164 to 1078.
Cui2vec Embeddings:
In addition to the baseline CUI one-hot encoding, we utilized the pretrained cui2vec word embeddings on the CLAMP output, which mapped CUI’s to 500-dimensional neural word embeddings [18]. Cui2vec is a comprehensive set of 108,477 clinical embeddings extracted from insurance claims, clinical notes, and biomedical journal articles [18]. Because cui2vec was not trained with negated terms, we removed all the negated entities from the output. Similar to before, features with less than 1% variance were removed, decreasing the input size from 4392 to 239.
2.3. Modeling Approach
Overview:
Eight models were constructed for comparison on predicting whether a low vision patient would see an improvement in their vision within a one-year follow-up. These include 1) a structured model which relied upon only structured input features, 2) free-text models that utilized only free-text clinical notes as input features, and 3) combination models which utilized both sets of features. As seen in Figure 2, we compared 2 different methods of extracting free text data- word embeddings and NER. Once we extracted the named entities, we explored onehot encoding and a pre-trained embedding on CUI’s. We also explored a combination of the four single-modality models, as described in Table 1. More information can be found in the supplementary materials, including detailed descriptions of each model architecture and depictions of combination model architectures in Supplementary Figure A. All code for this project is publicly available [19].
Figure 2. Overview of different model inputs.
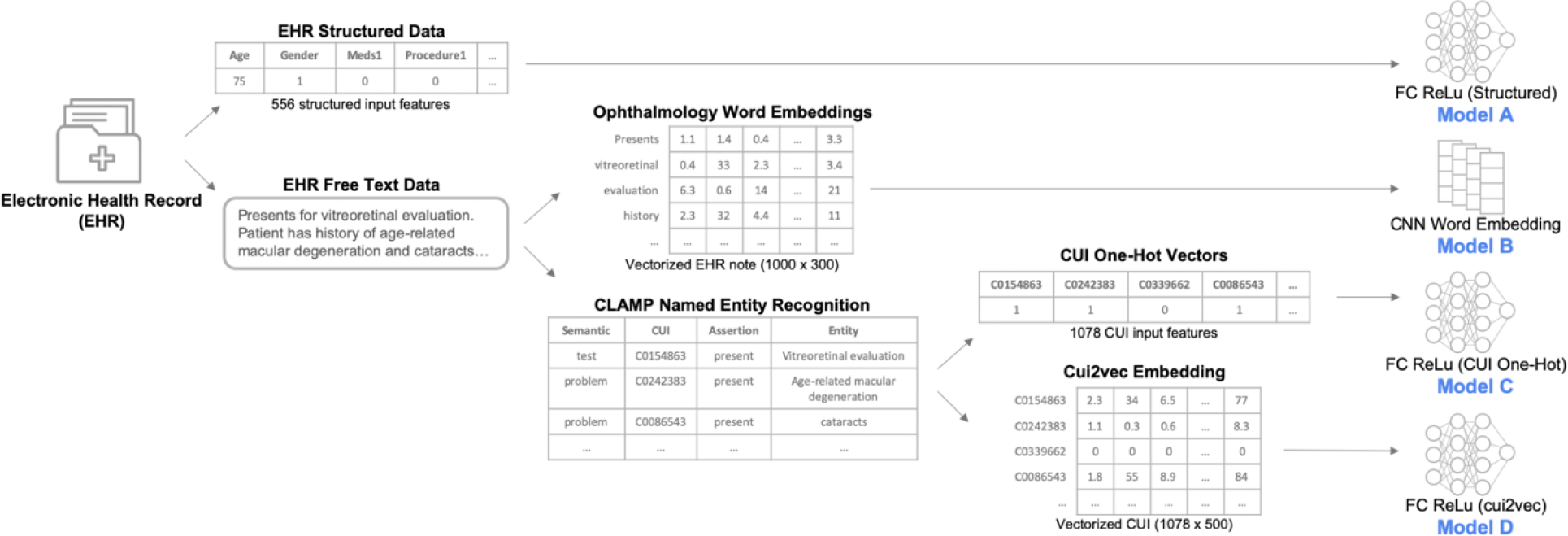
This figure depicts the different representations of the electronic health record data and their corresponding single-modality model architectures. Electronic health records (EHR) provided structured data as well as clinical free-text notes. The structured data were inputs to a fully connected (FC) deep neural network (Model A). Free-text notes were represented either with previously trained domain-specific word embeddings and modeled with a convolutional neural network (Model B), or processed through a biomedical named entity recognition pipeline (CLAMP) which mapped biomedical concepts to concept unique identifiers (CUIs ) in the Unified Medical Language System. The CUIs were then represented either as one-hot vectors (Model C) or as cui2vec vectors in an embedding space (Model D), both modeled using fully connected deep neural networks.
Table 1. Description of Models Combining Different Representations of Clinical Free-Text with Structured Inputs.
FC = Fully Connected neural network architecture; CNN = Convolutional Neural Network; CUI = Concept Unique Identifier; Cui2vec = representations of CUIs in an embedding space
| Model | Description | Concatenation Method |
|---|---|---|
| E | FC Structured (Model A) + CNN Word Embedding (Model B) | Late-Fusion |
| F | FC Structured (Model A) + FC CUI One-Hot (Model C) | Early-Fusion |
| G | FC Structured (Model A) + FC CUI Cui2vec (Model D) | Late-Fusion |
| H | FC Structured (Model A) + FC CUI Cui2vec (Model D) + FC Word Embedding | Late-Fusion |
All models were trained with hyperparameters and classification probability threshold tuned on a validation set to achieve optimal F1 score. To extract parameters with the best performance, we conducted hyperparameter tuning on the number of units for the dense layer, the dropout rate for the dropout layers, and the optimal learning rate.
2.4. Evaluation
We used sensitivity (recall), specificity, positive predictive value (precision), negative predictive value, F1-score (the harmonic mean of recall and precision), area under the receiver operating characteristic curve (AUROC), and area under the precision recall curve (AUPRC), as our evaluation metrics. These are all evaluated on an independent held-out test set of 300 patients. The F1 score was determined by iterating through 0.05 increments of thresholds to arrive at the highest F1 scores for the model. We obtained the 95% confidence interval for these performance metrics by bootstrapping with 10,000 replicates. In addition to calculating standard AUROC and AUPRC metrics, we also created binormal smoothened ROC and PRC curves using the R package pROC [20]. This method produces a smooth curve, free from parametric assumptions, that can fit arbitrarily complex distributions [21].
3. Results
Population characteristics are summarized in Table 2.
Table 2. Population Characteristics.
BCVA LogMAR = Logarithm of minimum angle of resolution from the best corrected visual acuity
| Total N=5547 | Vision Improved N = 3301 (59.5%) | Vision Not Improved N = 2246 (40.5%) | |||||
|---|---|---|---|---|---|---|---|
| N | Percent | N | Percent | N | Percent | ||
| Gender | Female | 3224 | 57.4% | 1924 | 57.8% | 1300 | 57.1% |
| Race | Asian | 1270 | 22.6% | 823 | 24.7% | 447 | 19.6% |
| Black | 222 | 4.0% | 113 | 3.4% | 109 | 4.8% | |
| Hispanic | 988 | 17.6% | 544 | 16.3% | 444 | 19.5% | |
| White | 2335 | 41.6% | 1416 | 42.5% | 919 | 40.3% | |
| Other | 797 | 14.2% | 438 | 13.1% | 359 | 15.8% | |
| Ethnicity | Non-Hispanic | 4442 | 79.2% | 2707 | 81.2% | 1735 | 76.2% |
| Hispanic/Latinx | 988 | 17.6% | 544 | 16.3% | 444 | 19.5% | |
| Unknown | 182 | 3.2% | 83 | 2.5% | 99 | 4.3% | |
| Mean | Std | Mean | Std | Mean | Std | ||
| Age (years) | 67.2 | 20.4 | 67.5 | 18.6 | 66.8 | 22.8 | |
| BCVA LogMAR Right | Best | 0.23 | 1.11 | −0.05 | 0.91 | 0.64 | 1.24 |
| Worst | 0.53 | 1.10 | 0.37 | 1.01 | 0.77 | 1.19 | |
| Median | 0.34 | 1.10 | 0.10 | 0.94 | 0.68 | 1.22 | |
| BCVA LogMAR Left | Best | 0.25 | 1.14 | −0.01 | 0.99 | 0.62 | 1.23 |
| Worst | 0.56 | 1.13 | 0.43 | 1.06 | 0.75 | 1.19 | |
| Median | 0.35 | 1.12 | 0.14 | 1.00 | 0.67 | 1.21 | |
3.1. CLAMP Validation
Upon validation, CLAMP was able to sufficiently extract relevant ophthalmology named entities from our clinical notes, resulting in high precision, recall, and F1 score greater than 80% as shown in Table 3. Based on these results, we proceeded with using CLAMP to extract named entities from our ophthalmology clinical notes. In addition, our CLAMP customization to detect ophthalmology note sections was able to identify the different sections in the clinical notes, resulting in high F1 scores, as seen in Table 4.
Table 3. CLAMP Validation Results for Ophthalmology-specific Terminology.
TP = True Positive, FP = False Positive, FN = False Negative
| Terms | TP | FP | FN | Total | Precision | Recall | F1 |
|---|---|---|---|---|---|---|---|
| Cataract/Nuclear Sclerosis/Cortical Clouding | 124 | 5 | 45 | 174 | 0.961 | 0.734 | 0.832 |
| Pseudophakia (posterior chamber intraocular lens) | 22 | 0 | 35 | 57 | 1 | 0.386 | 0.557 |
| Corneal edema | 7 | 0 | 3 | 10 | 1 | 0.700 | 0.824 |
| Diabetic retinopathy | 19 | 0 | 0 | 19 | 1 | 1 | 1 |
| Glaucoma | 74 | 2 | 10 | 86 | 0.974 | 0.881 | 0.925 |
| Macular degeneration | 62 | 0 | 5 | 67 | 1 | 0.925 | 0.961 |
| Macular edema | 33 | 0 | 4 | 37 | 1 | 0.892 | 0.943 |
| Retinal detachment | 52 | 1 | 12 | 65 | 0.981 | 0.813 | 0.889 |
| Poor/low/blurred/decreased/impaired vision; blindness | 128 | 1 | 9 | 138 | 0.992 | 0.934 | 0.962 |
| Eye pain | 53 | 0 | 2 | 55 | 1 | 0.964 | 0.981 |
| Optical coherence tomography | 1 | 0 | 37 | 38 | 1 | 0.026 | 0.051 |
| Slit Lamp | 8 | 0 | 7 | 15 | 1 | 0.533 | 0.696 |
| Avastin/Bevacizumab | 15 | 0 | 0 | 15 | 1 | 1 | 1 |
| Brimonidine/Alphagan | 26 | 0 | 4 | 30 | 1 | 0.867 | 0.929 |
| Latanoprost/Xalatan | 22 | 0 | 3 | 25 | 1 | 0.880 | 0.936 |
| Timolol/Timoptic | 20 | 0 | 1 | 21 | 1 | 0.952 | 0.976 |
| Keratoplasty | 12 | 0 | 4 | 16 | 1 | 0.750 | 0.857 |
| Pars plana vitrectomy | 15 | 0 | 1 | 16 | 1 | 0.938 | 0.968 |
| Cataract surgery/Cataract extraction | 39 | 0 | 6 | 45 | 1 | 0.867 | 0.929 |
| TOTAL | 734 | 9 | 195 | 938 | 0.988 | 0.790 | 0.878 |
Table 4. CLAMP Validation Results for Free-Text Clinical Note Sections.
TP = True Positive, FP = False Positive, FN = False Negative
| Category | TP | FP | FN | Total | Precision | Recall | F1 |
|---|---|---|---|---|---|---|---|
| *Allergy | 50 | 0 | 0 | 50 | 1 | 1 | 1 |
| Assessment and Plan | 122 | 5 | 14 | 141 | 0.961 | 0.897 | 0.928 |
| *Attestation | 11 | 0 | 0 | 11 | 1 | 1 | 1 |
| Chief Complaint/ History of Present Illness | 122 | 0 | 1 | 123 | 1 | 0.992 | 0.999 |
| *Consent | 18 | 0 | 0 | 18 | 1 | 1 | 1 |
| Exam | 194 | 28 | 3 | 225 | 0.874 | 0.985 | 0.9269 |
| *Family and Social History | 153 | 49 | 0 | 202 | 0.757 | 1 | 0.862 |
| History | 143 | 25 | 6 | 174 | 0.851 | 0.960 | 0.902 |
| *Instruction | 19 | 0 | 0 | 19 | 1 | 1 | 1 |
| Interpretation | 17 | 1 | 0 | 18 | 0.944 | 1 | 0.971 |
| Medication | 117 | 10 | 1 | 128 | 0.921 | 0.992 | 0.955 |
| Review of Systems | 84 | 0 | 0 | 84 | 1 | 1 | 1 |
| TOTAL | 1050 | 118 | 25 | 1193 | 0.899 | 0.977 | 0.936 |
Sections that were deleted during pre-processing of CLAMP outputs
3.2. Model Results
Figure 3 shows the receiver operating (ROC) and precision recall curves (PRC) on the held-out test set for the structured, text, and combination models. Supplementary Figure B shows the binormal smoothened ROC and PRC curves. The word embedding Model B had the best AUROC (0.819) followed by the combination Model E (0.817). For AUPRC, we can see that Model E had the best value (0.776) with Model B as a close second (0.776). Table 5 reports all performance metrics for each model, with 95% confidence intervals.
Figure 3. Receiver Operating Curve (ROC) and Precision Recall Curves (PRC) for Predictive Models.
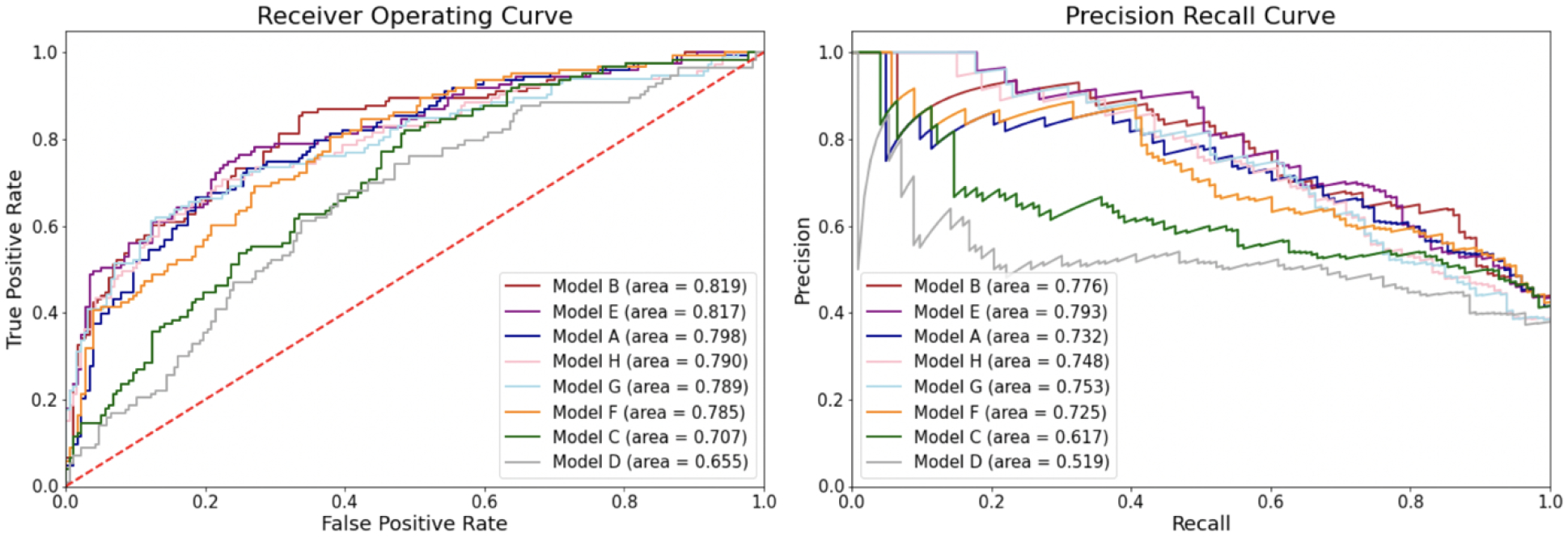
These figures reveal the ROC and PRC curves, as well as the area under the curves. The red line in the ROC curve reflects the points at which true positive rates equal true negative rates, which is only as good as a random classifier
Table 5. Performance metrics for deep learning models in predicting low vision prognosis.
Bolded values reflect the best value for that performance metric. Values in parentheses are 95% confidence intervals. Threshold values reflect the threshold that was chosen to generate the F1 scores, precision, and recall. PPV = Positive Predictive Value. NPV = Negative Predictive Value
| Value (95% Confidence Interval) | AUROC | AUPRC | F1 | Sensitivity (Recall) | Specificity | PPV (Precision) | NPV | Accuracy | Threshold |
|---|---|---|---|---|---|---|---|---|---|
| (A) Structured Model | 0.80 (0.75–0.85) | 0.73 (0.64–0.81) | 0.70 (0.61–0.74) | 0.73 (0.62–0.78) | 0.76 (0.68–0.81) | 0.68 (0.57–0.74) | 0.80 (0.72–0.84) | 0.73 (0.68–0.78) | 0.40 |
| (B) CNN Word Embedding Text Model | 0.82 (0.77–0.87) | 0.78 (0.70–0.84) | 0.67 (0.60–0.74) | 0.67 (0.60–0.76) | 0.77 (0.70–0.83) | 0.67 (0.59–0.75) | 0.77 (0.71–0.83) | 0.74 (0.68–0.78) | 0.45 |
| (C) CUI One-Hot Text Model | 0.71 (0.65–0.76) | 0.62 (0.53–0.70) | 0.64 (0.57–0.70) | 0.79 (0.71–0.86) | 0.52 (0.45–0.59) | 0.53 (0.46–0.60) | 0.78 (0.70–0.85) | 0.66 (0.57–0.68) | 0.35 |
| (D) CUI Cui2vec Text Model | 0.66 (0.59–0.72) | 0.52 (0.43–0.62) | 0.58 (0.51–0.64) | 0.80 (0.72–0.87) | 0.42 (0.35–0.49) | 0.45 (0.38–0.52) | 0.77 (0.69–0.85) | 0.65 (0.50–0.62) | 0.20 |
| (E) A+B Combined Model | 0.82 (0.76–0.87) | 0.79 (0.72–0.85) | 0.69 (0.63–0.75) | 0.79 (0.71–0.86) | 0.66 (0.58–0.72) | 0.61 (0.54–0.69) | 0.82 (0.75–0.88) | 0.74 (0.66–0.76) | 0.35 |
| (F) A+C Combined Model | 0.79 (0.73–0.83) | 0.73 (0.64–0.80) | 0.63 (0.56–0.70) | 0.64 (0.56–0.73) | 0.73 (0.67–0.80) | 0.63 (0.54–0.71) | 0.75 (0.68–0.81) | 0.73 (0.64–0.75) | 0.40 |
| (G) A+D Combined Model | 0.79 (0.73–0.84) | 0.75 (0.68–0.82) | 0.67 (0.59–0.74) | 0.66 (0.57–0.75) | 0.80 (0.74–0.86) | 0.67 (0.58–0.76) | 0.80 (0.74–0.85) | 0.77 (0.70–0.80) | 0.45 |
| (H)A+D + FC Word Embedding Model | 0.79 (0.73–0.84) | 0.75 (0.67–0.82) | 0.66 (0.59–0.73) | 0.66 (0.57–0.75) | 0.79 (0.73–0.85) | 0.66 (0.57–0.74) | 0.80 (0.74–0.85) | 0.76 (0.69–0.79) | 0.45 |
4. Discussion
In this study, we developed and evaluated deep learning predictive models for low vision prognosis, comparing several different approaches to integrating structured and unstructured free-text data from ophthalmology electronic health records. We compared three different representations of ophthalmology clinical text, including 1) using previously developed ophthalmology domain-specific word embeddings, and representing medical concepts from notes as 2) named entities represented by one-hot vectors and 3) named entities represented by embedding vectors. We also validated and customized a biomedical NER pipeline (CLAMP) for ophthalmology notes and concepts. Word embedding models (Model B and E), the only ones that outperformed the single-modality model based on structured inputs, appeared to outperform NER models (Model C) on AUROC of 0.82 and 0.71 respectively. These models may ultimately form the basis of clinical decision support systems to aid physicians in their workflow to refer patients to low-vision rehabilitation services.
Our work represents efforts unique in the ophthalmology field to incorporate multimodal data types from electronic health records, including free-text data types, into predictive algorithms. Despite the increased interest in using machine learning techniques in ophthalmology over the past few years [22], most studies have been focused on image interpretation [23]. There have been few studies that have incorporated electronic health records, and very limited studies that have predicted development of clinical eye diseases [24]. Lin et. al predicted myopia development in children using refraction data from the electronic health records [25], while Alexeeff et. al predicted visual acuity after cataract surgery [26]. Both of these studies utilized only structured data without input from the information-rich text in the electronic health records. Our study differs in incorporating multiple modalities of data from the electronic health record, including free-text as well as structured data.
Prior works in other medical domains have noted the superiority of utilizing data fusion models that combine structured EHR data with representations of free-text through word embeddings and/or CUIs extracted by biomedical NER pipelines such as CLAMP [27]. However, it was unclear whether such approaches could work in the highly specialized domain of ophthalmology. We first showed that CLAMP, a biomedical NER tool that was trained on general clinical notes, was able to extract relevant terms from ophthalmology notes and map them to the corresponding CUI’s. We found that the total F1 score for a selection of common ophthalmology-specific terms was 0.88. We already know, from prior works, that CLAMP’s ability to capture general medical features allows it to be a starting point for feature extraction from free-text. But through our validation here, we further suggest that CLAMP may also be appropriate for use on ophthalmology notes.
However, in our models predicting low vision outcomes, we observed that representing free-text using domain-specific word embeddings (Models B and E) still led to better performance than representing free-text using CLAMP-extracted named entities mapped to CUIs (Models C and D). Adding CUI’s to the structured data model (Models F and G) did not appear to significantly improve the performance. One explanation is possible overlap of information between CUI’s and structured data. Both our structured data and our NER pipeline extracted concepts including medications and procedures, resulting in possible redundancy of data. Another explanation may relate to use of the pretrained cui2vec [18] to translate CUI’s into vectorized embeddings. Because cui2vec was trained on the general medical domain and not on ophthalmology-specific text, we found that 51.5% of the identified CUI’s from our text were not represented in cui2vec, which may have decreased the performance of the cui2vec text representation model. Wang et. al used a similar approach with cui2vec embeddings to predict distant recurrence of breast cancer, achieving an AUROC of 0.84 [27], which is somewhat higher than the AUROC of 0.79 from our combined Model H (structured + cui2vec embedding + word embedding). Incorporating text representation using ophthalmology domain-specific embeddings [10] did improve performance over the structured model, suggesting that domain-specific text representations may be especially important to the performance of highly domain specific ophthalmology predictive models. Future work to improve the cui2vec approach could include training domain-specific CUI embeddings to improve CUI coverage. These approaches could be combined with CLAMP to augment ophthalmology-specific features with general medical terms. Researchers in other highly specialized medical subdomains who wish to incorporate clinical free-text into predictive models may also wish to develop more domain-specific representations of their text.
Even though this iteration of representing free-text using NER did not outperform word embeddings, there is still value in further developing this approach. Extraction of important concepts from medical notes is inherently more interpretable than mapping notes to word embeddings. We can determine which types of diagnoses, treatments, and findings the predictive models rely upon through the types of entities extracted and inputted into the models. Future work could focus more formally on explainability studies, using local interpretable model-agnostic explanations (LIME) [29] to determine which model features helped contribute to the predictions. Understanding how to perform explainability studies for complex combination models with multiple data modalities is an area of active research in the field. These studies not only would provide additional insights into the medical context surrounding low vision prognosis, but also would increase interpretability of the results, increasing clinician’s trust in these models [30–32].
Another approach to representation of free text that has grown in popularity recently is the use of context-aware word embeddings learned in transformer-based approaches [30–32]. Future work can experiment with tuning transformer-based models for the ophthalmology domain [31], and using multiple hierarchical levels of text representation [33]. Despite these newer advances in NLP, understanding how to utilize and customize NER and word embeddings for ophthalmology is still valuable, as these simpler and less computationally intensive approaches might not be outperformed by transformer-based approaches. In addition, NER and word embedding approaches do not have the additional limitations that are present with transformer-based approaches such as slower training, larger data requirements, and shorter limits on lengths of text inputs.
There are several limitations to our study. Our study cohort is from a single academic center, which limits the variability and composition of the patients and notes. This cohort included very few patients identifying as American Indian/Alaska Native or Native Hawaiian/Pacific Islander; the patients’ gender is documented only as male and female, which does not cover the non-binary spectrum. Future works could explore conducting sub-analyses on race and gender. In addition, NER is a high-level NLP task that often faces challenges with clinical data, arising from variations in word and phrase ordering, derivation, synonymy, etc. [34]. Ophthalmology written notes tend to harbor many abbreviations that often represent different terms in different fields. CLAMP was able to recognize some abbreviations, but missed others, which led us to remove all abbreviations from the CLAMP pipeline. This could have resulted in under-capture of some information, which may have contributed to the suboptimal model performance.
5. Conclusion
In conclusion, we developed and compared models predicting low vision prognosis which combined multiple modalities of data (structured and free-text) from electronic health records. In addition to exploring different representations of ophthalmology clinical text, we validated CLAMP’s named entity recognition on ophthalmology-specific terminologies. We ultimately showed that models incorporating text represented by domain-specific word embeddings outperformed the single-modality model using structured inputs and outperformed all single- and multiple-modality models representing text with biomedical concepts extracted through NER pipelines. This study is a first step towards development of models using multiple modalities of data to predict ophthalmology outcomes. Researchers in other highly specialized biomedical domains may wish to carefully consider how to incorporate free-text into predictive models and favor domain-specific representations of text for best performance.
Supplementary Material
Summary Table.
What was already known:
Structured data from ophthalmology electronic health records have been used to successfully predict biomedical outcomes.
We have previously shown that representing ophthalmology clinical notes using domain-specific word embeddings leads to predictive models for low vision outcomes that are superior to those using general domain word embeddings.
Works in other medical domains have noted the superiority of utilizing combination or “data fusion” models that combine structured EHR data with representations of free-text through word embeddings and/or CUIs extracted by biomedical named entity recognition pipelines.
What this study added:
We demonstrated multiple ways to combine structured data and free text notes in combination models to predict low vision outcomes and are the first to build these “data fusion” models in ophthalmology.
We found that domain-specific representations of clinical text through neural word embeddings resulted in better performing predictive models compared to the more general approach of representing text through extraction of biomedical concepts.
We customized and validated a biomedical named entity recognition pipeline for ophthalmology to facilitate future natural language processing research in this field.
Funding Sources and Conflict of Interest
This work was supported by: Stanford MedScholars program (HG); National Eye Institute 1K23EY03263501(SYW); Career Development Award from Research to Prevent Blindness (SYW); unrestricted departmental grant from Research to Prevent Blindness (SYW, BT, WH); departmental grant National Eye Institute P30-EY026877 (SYW, BT, WH). There is no conflict of interest to report.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
References
- [1].Vision Rehabilitation PPP - 2017, (2017). https://www.aao.org/preferred-practice-pattern/vision-rehabilitation-ppp-2017 (accessed March 21, 2021).
- [2].Scott IU, Smiddy WE, Schiffman J, Feuer WJ, Pappas CJ, Quality of life of low-vision patients and the impact of low-vision services, Am. J. Ophthalmol 128 (1999) 54–62. 10.1016/s0002-9394(99)00108-7. [DOI] [PubMed] [Google Scholar]
- [3].de Boer MR, Pluijm SMF, Lips P, Moll AC, Völker-Dieben HJ, Deeg DJH, van Rens GHMB, Different aspects of visual impairment as risk factors for falls and fractures in older men and women, J. Bone Miner. Res 19 (2004) 1539–1547. 10.1359/JBMR.040504. [DOI] [PubMed] [Google Scholar]
- [4].Toyoshima A, Martin P, Sato S, Poon LW, The relationship between vision impairment and well-being among centenarians: findings from the Georgia Centenarian Study, Int. J. Geriatr. Psychiatry 33 (2018) 414–422. 10.1002/gps.4763. [DOI] [PubMed] [Google Scholar]
- [5].Zhang T, Jiang W, Song X, Zhang D, The association between visual impairment and the risk of mortality: a meta-analysis of prospective studies, J. Epidemiol. Community Health 70 (2016) 836–842. 10.1136/jech-2016-207331. [DOI] [PubMed] [Google Scholar]
- [6].Ho VWT, Chen C, Merchant RA, Cumulative Effect of Visual Impairment, Multimorbidity, and Frailty on Intrinsic Capacity in Community-Dwelling Older Adults, J. Aging Health 32 (2020) 670–676. 10.1177/0898264319847818. [DOI] [PubMed] [Google Scholar]
- [7].Virgili G, Acosta R, Bentley SA, Giacomelli G, Allcock C, Evans JR, Reading aids for adults with low vision, Cochrane Database Syst. Rev 4 (2018) CD003303. 10.1002/14651858.CD003303.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Assale M, Dui LG, Cina A, Seveso A, Cabitza F, The Revival of the Notes Field: Leveraging the Unstructured Content in Electronic Health Records, Front. Med 6 (2019) 66. 10.3389/fmed.2019.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Peissig PL, Rasmussen LV, Berg RL, Linneman JG, McCarty CA, Waudby C, Chen L, Denny JC, Wilke RA, Pathak J, Carrell D, Kho AN, Starren JB, Importance of multi-modal approaches to effectively identify cataract cases from electronic health records, J. Am. Med. Inform. Assoc 19 (2012) 225–234. 10.1136/amiajnl-2011-000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang SY, Tseng B, Hernandez-Boussard T, Development and Evaluation of Novel Ophthalmology Domain-Specific Neural Word Embeddings to Predict Visual Prognosis, International Journal of Medical Informatics, under Review (n.d.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].López-Úbeda P, Díaz-Galiano MC, Martín-Noguerol T, Luna A, Ureña-López LA, Martín-Valdivia MT, COVID-19 detection in radiological text reports integrating entity recognition, Comput. Biol. Med 127 (2020) 104066. 10.1016/j.compbiomed.2020.104066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hassanpour S, Langlotz CP, Information extraction from multi-institutional radiology reports, Artif. Intell. Med 66 (2016) 29–39. 10.1016/j.artmed.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Senior M, Burghart M, Yu R, Kormilitzin A, Liu Q, Vaci N, Nevado-Holgado A, Pandit S, Zlodre J, Fazel S, Identifying Predictors of Suicide in Severe Mental Illness: A Feasibility Study of a Clinical Prediction Rule (Oxford Mental Illness and Suicide Tool or OxMIS), Front. Psychiatry 11 (2020) 268. 10.3389/fpsyt.2020.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lowe HJ, Ferris TA, Hernandez PM, Weber SC, STRIDE--An integrated standards-based translational research informatics platform, AMIA Annu. Symp. Proc. 2009 (2009) 391–395. https://www.ncbi.nlm.nih.gov/pubmed/20351886. [PMC free article] [PubMed] [Google Scholar]
- [15].Wang SY, Pershing S, Tran E, Hernandez-Boussard T, Automated extraction of ophthalmic surgery outcomes from the electronic health record, Int. J. Med. Inform 133 (2020) 104007. 10.1016/j.ijmedinf.2019.104007. [DOI] [PubMed] [Google Scholar]
- [16].Elliott DB, The good (logMAR), the bad (Snellen) and the ugly (BCVA, number of letters read) of visual acuity measurement, Ophthalmic Physiol. Opt 36 (2016) 355–358. 10.1111/opo.12310. [DOI] [PubMed] [Google Scholar]
- [17].Soysal E, Wang J, Jiang M, Wu Y, Pakhomov S, Liu H, Xu H, CLAMP - a toolkit for efficiently building customized clinical natural language processing pipelines, J. Am. Med. Inform. Assoc 25 (2018) 331–336. 10.1093/jamia/ocx132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Beam AL, Kompa B, Schmaltz A, Fried I, Weber G, Palmer N, Shi X, Cai T, Kohane IS, Clinical Concept Embeddings Learned from Massive Sources of Multimodal Medical Data, Pac. Symp. Biocomput 25 (2020) 295–306. 10.1142/9789811215636_0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang SY, eyelovedata/lowva-ner-textcnn: v1.0.0, 2021. 10.5281/zenodo.5655872. [DOI] [Google Scholar]
- [20].Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M, pROC: an open-source package for R and S+ to analyze and compare ROC curves, BMC Bioinformatics. 12 (2011) 77. 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lasko TA, Bhagwat JG, Zou KH, Ohno-Machado L, The use of receiver operating characteristic curves in biomedical informatics, J. Biomed. Inform 38 (2005) 404–415. 10.1016/j.jbi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- [22].Lee A, Taylor P, Kalpathy-Cramer J, Tufail A, Machine Learning Has Arrived!, Ophthalmology. 124 (2017) 1726–1728. 10.1016/j.ophtha.2017.08.046. [DOI] [PubMed] [Google Scholar]
- [23].Ting DSW, Pasquale LR, Peng L, Campbell JP, Lee AY, Raman R, Tan GSW, Schmetterer L, Keane PA, Wong TY, Artificial intelligence and deep learning in ophthalmology, Br. J. Ophthalmol 103 (2019) 167–175. 10.1136/bjophthalmol-2018-313173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ting DSW, Peng L, Varadarajan AV, Keane PA, Burlina PM, Chiang MF, Schmetterer L, Pasquale LR, Bressler NM, Webster DR, Abramoff M, Wong TY, Deep learning in ophthalmology: The technical and clinical considerations, Prog. Retin. Eye Res 72 (2019) 100759. 10.1016/j.preteyeres.2019.04.003. [DOI] [PubMed] [Google Scholar]
- [25].Lin H, Long E, Ding X, Diao H, Chen Z, Liu R, Huang J, Cai J, Xu S, Zhang X, Wang D, Chen K, Yu T, Wu D, Zhao X, Liu Z, Wu X, Jiang Y, Yang X, Cui D, Liu W, Zheng Y, Luo L, Wang H, Chan C-C, Morgan IG, He M, Liu Y, Prediction of myopia development among Chinese school-aged children using refraction data from electronic medical records: A retrospective, multicentre machine learning study, PLoS Med. 15 (2018) e1002674. 10.1371/journal.pmed.1002674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Alexeeff SE, Uong S, Liu L, Shorstein NH, Carolan J, Amsden LB, Herrinton LJ, Development and Validation of Machine Learning Models: Electronic Health Record Data To Predict Visual Acuity After Cataract Surgery, Perm. J 25 (2020) 1. 10.7812/TPP/20.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang H, Li Y, Khan SA, Luo Y, Prediction of breast cancer distant recurrence using natural language processing and knowledge-guided convolutional neural network, Artif. Intell. Med 110 (2020) 101977. 10.1016/j.artmed.2020.101977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Palatnik de Sousa I, Maria M Vellasco Bernardes Rebuzzi, da Silva E. Costa, Local Interpretable Model-Agnostic Explanations for Classification of Lymph Node Metastases, Sensors . 19 (2019). 10.3390/s19132969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Stiglic G, Kocbek P, Fijacko N, Zitnik M, Verbert K, Cilar L, Interpretability of machine learning based prediction models in healthcare, Wiley Interdiscip. Rev. Data Min. Knowl. Discov 10 (2020) e1379. 10.1002/widm.1379. [DOI] [Google Scholar]
- [30].Liu Y, Ott M, Goyal N, Du J, Joshi M, Chen D, Levy O, Lewis M, Zettlemoyer L, Stoyanov V, RoBERTa: A Robustly Optimized BERT Pretraining Approach, ArXiv [Cs.CL]. (2019). http://arxiv.org/abs/1907.11692. [Google Scholar]
- [31].Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, Kang J, BioBERT: a pre-trained biomedical language representation model for biomedical text mining, Bioinformatics. 36 (2020) 1234–1240. 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sanh V, Debut L, Chaumond J, Wolf T, DistilBERT, a distilled version of BERT: smaller, faster, cheaper and lighter, ArXiv [Cs.CL]. (2019). http://arxiv.org/abs/1910.01108. [Google Scholar]
- [33].Yang Z, Yang D, Dyer C, He X, Smola A, Hovy E, Hierarchical attention networks for document classification, in: Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, aclweb.org, 2016: pp. 1480–1489. https://www.aclweb.org/anthology/N16-1174.pdf. [Google Scholar]
- [34].Nadkarni PM, Ohno-Machado L, Chapman WW, Natural language processing: an introduction, J. Am. Med. Inform. Assoc 18 (2011) 544–551. 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Soysal E, Warner JL, Wang J, Jiang M, Harvey K, Jain SK, Dong X, Song H-Y, Siddhanamatha H, Wang L, Dai Q, Chen Q, Du X, Tao C, Yang P, Denny JC, Liu H, Xu H, Developing Customizable Cancer Information Extraction Modules for Pathology Reports Using CLAMP, Stud. Health Technol. Inform 264 (2019) 1041–1045. 10.3233/SHTI190383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Harris DR, Henderson DW, Corbeau A, Improving the Utility of Tobacco-Related Problem List Entries Using Natural Language Processing, AMIA Annu. Symp. Proc 2020 (2020) 534–543. https://www.ncbi.nlm.nih.gov/pubmed/33936427. [PMC free article] [PubMed] [Google Scholar]
- [37].Kim Y, Convolutional Neural Networks for Sentence Classification, in: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), 2014: pp. 1746–1751. 10.3115/v1/D14-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



