Abstract
Purpose
Computer-aided detection and diagnosis (CAD) of colonic polyps always faces the challenge of classifying imbalanced data. In this paper, three new operating point selection strategies based on receiver operating characteristic curve are proposed to address the problem.
Methods
Classification on imbalanced data performs inferiorly because of a major reason that the best differentiation threshold shifts due to the degree of data imbalance. To address this decision threshold shifting issue, three operating point selection strategies, i.e., shortest distance, harmonic mean and anti-harmonic mean, are proposed and their performances are investigated.
Results
Experiments were conducted on a class-imbalanced database, which contains 64 polyps in 786 polyp candidates. Support vector machine (SVM) and random forests (RFs) were employed as basic classifiers. Two imbalanced data correcting techniques, i.e., cost-sensitive learning and training data down sampling, were applied to SVM and RFs, and their performances were compared with the proposed strategies. Comparing to the original thresholding method, i.e., 0.488 sensitivity and 0.986 specificity for RFs and 0.526 sensitivity and 0.977 specificity for SVM, our strategies achieved more balanced results, which are around 0.89 sensitivity and 0.92 specificity for RFs and 0.88 sensitivity and 0.90 specificity for SVM. Meanwhile, their performance remained at the same level regardless of whether other correcting methods are used.
Conclusions
Based on the above experiments, the gain of our proposed strategies is noticeable: the sensitivity improved from 0.5 to around 0.88 for RFs and 0.89 for SVM while remaining a relatively high level of specificity, i.e., 0.92 for RFs and 0.90 for SVM. The performance of our proposed strategies was adaptive and robust with different levels of imbalanced data. This indicates a feasible solution to the shifting problem for favorable sensitivity and specificity in CAD of polyps from imbalanced data.
Keywords: Computer-aided detection and diagnosis (CAD), Computed tomography colonography (CTC), Random forests, Harmonic mean, Support vector machine (SVM), Receiver operating characteristic (ROC)
Introduction
According to the up-to-date statistics from American Cancer Society [1], colon and rectum cancer ranks the third most common occurrence of both cancer deaths and new cancer cases in 2012 for both men and women in the United States. It has been generally recognized that early detection and removal of colonic polyps prior to their malignant transformation can effectively decrease the incidence of colon cancer [2,3]. As a potential minimally invasive screening technique, computer-aided detection and diagnosis (CAD) for computed tomography colonography (CTC), which has shown several advantages over the traditional optical colonoscopy (OC), is gaining more attention in the field of medical imaging [4,5]. The essential task of CAD is to differentiate polyps from normal tissues and fecal residue. In implementation, CAD usually starts by looking the patient-specific colon model to mark suspicious locations, called patches or initial polyp candidates (IPCs), based on some basic geometric features, e.g., shape index and curvedness [6,7]. It is expected that the number of IPCs is large, where there are lots of false-positives (FPs) or patches of normal tissues, fecal residue, image artifacts, etc. In general, the number of true-positives (TPs) or patches of polyps is much smaller than the number of FPs in the IPCs pool, which makes the pool of IPCs or the dataset very unbalanced and leads to a challenging differentiation task for CAD.
As the name suggests, class-imbalanced data stand for dataset where the number of observations belonging to each class is different. Sometime one of the classes contributes only a very small minority of the data and makes the dataset significantly imbalanced, just as in our case. Most available classification algorithms assume or expect balanced class distributions and equal misclassification costs, so when applied to imbalanced dataset, they prefer to classify most cases to the majority class (reason will be discussed later) and often fail to predict the minority class, on which our interest usually leans. For this reason, between-class imbalance problem has been receiving increasing attention in recent years.
In this paper, we propose three strategies based on receiver operating characteristic (ROC) curve analysis to tackle the class imbalance data problem. We focus more on sensitivity (true positive (TP) rate, considering positive class only) and specificity (true negative (FN) rate, considering negative class only) and try to reveal the shifted-decision value by maximizing/minimizing a function of sensitivity and specificity in the ROC space. For evaluation purpose, we employ support vector machine (SVM) and random forests (RFs) as basic classifier. Based on the basic classifier, two common approaches to deal with imbalanced data, i.e., cost-sensitive learning and down sampling, are implemented as reference and their performance is compared with our proposed strategies.
The remainder of this paper is organized as follows. Section “Methods” introduces the quantitative measuring criteria (ROC and AUC: the area under the ROC curve), followed by a detailed description of the three proposed operating point selection strategies and the design of experiments. Section “Results” reports the experiment results. Section “Conclusion, future work and limitation discussion” draws the conclusions and remarks. Discussions on several concerns and our future work are presented in Section “Conclusion, future work and limitation discussion.”
Methods
In this section, we first give a brief introduction of ROC and AUC and then provide a detailed description of the rationale and methodology of our proposed decision threshold-chosen strategies, followed by the experiment design of our study.
Receiver operating characteristics (ROC) curve and area under the ROC curve (AUC)
ROC was first introduced in signal detection theory. It is a graphical plot of the sensitivity, or TP rate, versus 1-specificity, or FP rate in test phase, and illustrates the performance of a binary classifier system as its discrimination threshold varies. The area under the ROC curve or AUC actually depicts the probability that a randomly chosen positive example (or TP) is correctly rated (ranked) with greater suspicion than a randomly chosen negative example (or FP). In other words, the AUC measure is equivalent to the Mann–Whitney U statistic normalized by the number of possible pairings of positive and negative values, also known as the two sample Wilcoxon rank-sum statistic [8]. Detailed introduction of ROC analysis can be found in [9].
High AUC value usually reflects good differentiation capability of a classifier. So maximizing the AUC is often employed to direct sequential parameter or model searching to achieve better classification performance [10,11]. However, in these studies [10,11], only the optimized AUC is reported, the optimal operating point problem is not revealed in which we care more about. High AUC does not guarantee high accuracy of the prediction. For example, SVM using 0 threshold value achieves high AUC but has poor prediction accuracy. In such case, a strategy is needed in order to draw an optimal classification decision. The following analysis provides, a clue on the needed strategy. As mentioned above, most current classification methods aim to solve the problems in balanced data, where the overall accuracy is optimized when AUC is optimized. The relationship of overall accuracy, sensitivity and specificity is shown below:
| Predicted positive class | Predicted negative class | |
|---|---|---|
| Actual positive class | TP (true positive) | FN (false negative) |
| Actual negative class | FP (false positive) | TN (true negative) |
| (1) |
To be more specific, we represent the overall accuracy in terms of sensitivity and specificity as follows. Consider a dataset with N+ positive cases and N− negative cases. Define as the ratio of the two classes. After classification, we obtain TN, TP, FN and FP, and by definition, we have N+ = TP + FN and N− = TN + FP, then,
| (2) |
Now if we define , then we can obtain the following equation:
| (3) |
For balanced data, β is near or equal to 0.5, in which case maximizing the overall accuracy is equal to maximizing the sensitivity and specificity with the same weight. However, for imbalanced data with β approaching 0 (positive class minority), maximizing the overall accuracy will bias toward maximizing the specificity more than the specificity, and vice versa as β approaches 1, see Fig. 1. This is probably the reason why the current methods tend to classify minority cases to majority when dealing with imbalanced data.
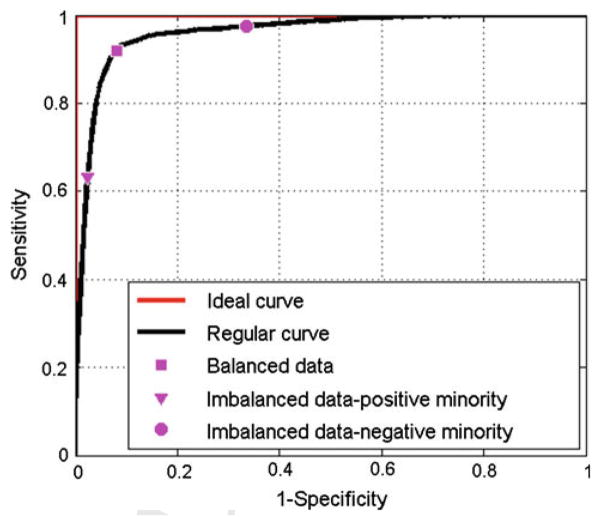
Fig. 1.
A typical representation of ROC curves. The red curve represents the ideal curve. The black curve shows an example of regular ROC curve (not ideal). The square, triangular and circle magenta markers indicate the results of maximizing the overall accuracy of data with different imbalance level
As shown in Fig. 1, the operating point used for balanced data is no longer suitable for imbalanced data, because the bias leads to low prediction accuracy in the minority class. Therefore, we need to find a strategy which can help us to determine a trade-off with balanced sensitivity and specificity instead of simply maximizing the overall accuracy. The previous method use balanced learning strategy, which maximizes the overall accuracy and generates points near top left corner when the original threshold is used. Following this idea, we try to obtain the similar results (points near top left corner) but with both high sensitivity and specificity by certain decision operating point chosen methods. In the following section, to the best of our knowledge, we propose three new strategies to choose such operating points, which are all based on the ROC space. We would want to find the best splitting threshold by minimizing or maximizing a cost function of sensitivity and specificity. More details are described below.
Three new proposed threshold-chosen strategies
Minimum distance
Intuitively, the points close to point (0, 1) on the ROC curve tend to have high sensitivity and specificity values and, therefore, are chosen to calculate the distance between point (0, 1) and the points which compose the ROC curve. The one with minimum distance is picked out, and its corresponding threshold is chosen as the final splitting threshold. This method can be illustrated by the following equation, where i is the ith point on the ROC curve, see Fig. 2a.
| (4) |
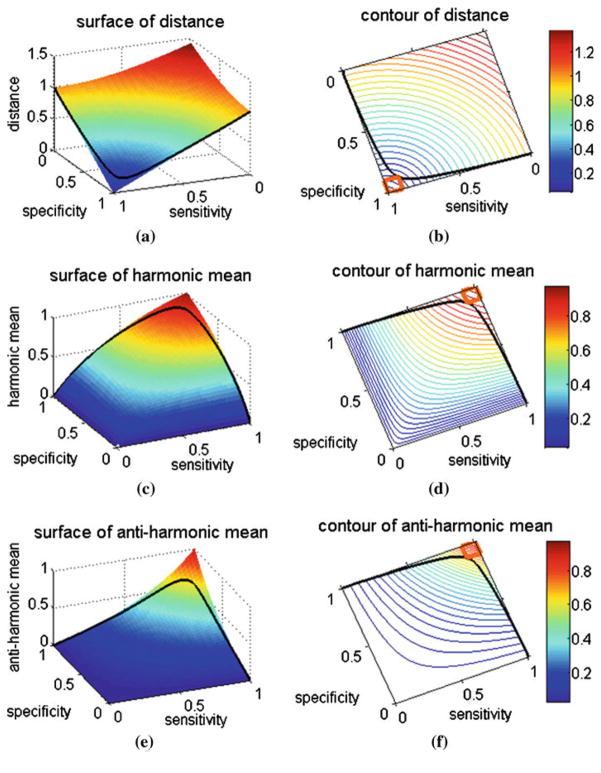
Fig. 2.
a, c and e show the 3D surface plots of the three proposed operating point selection strategies in the sensitivity–specificity domain corresponding to the shortest distance, the harmonic mean and the anti-harmonic mean measures, respectively. b, d and f show the contour plot of the corresponding strategies. The black curve in each figure represents the plot of a same regular ROC curve. For the shortest distance case (a), the surface part with blue color is preferred, while for the harmonic mean case c and the anti-harmonic mean case (e), the red areas are preferred. Region bounded with orange line in the contour plot is the “favorite region,” which means both sensitivity and specificity are larger than 0.9 in this region
Harmonic mean of sensitivity and specificity
In mathematics, the harmonic mean is one kind of averages. As it tends strongly toward the smaller element of the pair, it may mitigate the influence of the larger value and increase the influence of the small value. In other words, the larger the difference of the elements in the pair, the smaller the harmonic mean is. It pays more attention to the balance of the pair compared to the arithmetic mean. This is desired for classification of unbalanced data. From this aspect, we choose to seek the point which maximizes the harmonic mean on the ROC curve, see Fig. 2c,
| (5) |
Anti-harmonic mean of sensitivity and specificity
From the three-dimensional (3D) plot of the harmonic mean of Fig. 2c, we can see that the whole surface bends to point (0, 0, 1). This indicates a kind of “convex” surface in the 3D space. Then, a question arises that how would the result be if we find a “concave” surface which bolds to point (1, 1, 0). To answer this question, we generated another kind of mean, named as anti-harmonic mean, whose surface is “concave” just like what we want, see Fig. 2e. Following is its equation expression:
| (6) |
Figure 2b, d and f show the contour plots of these three proposed new strategies. All the contours bend to point (0, 0), which means if we constrain the sum of sensitivity and specificity to be a constant, all the proposed strategies tend to give larger values (for shortest distance, it is smaller value) for the balanced sensitivity–specificity pair. The orange square defines a “favorite region” in the ROC space (shown in the contour plots of Fig. 2), and our objective is to use the proposed strategies to locate the operating point near or in the region in the ROC plot.
Experimental design
Database
Evaluation of the three newly proposed threshold-chosen strategies was conducted on a CTC database of 49 scans from 25 patients with polyps of size from 6 to 22 mm, see Fig. 3 (where the morphology and pathology information is also included). By quickly examining each patient-specific colon wall in each scan using some geometry-based features, such as shape index and curvedness, in a CAD pipeline [6,7], we obtained 786 IPCs, among which 64 are TPs (which are confirmed by both OC and CTC and, therefore, are considered as the gold standard in this study). Twenty-one geometric features and density features [12–17] were calculated on each extracted IPC volume. To be specific, density features include statistical information, i.e., mean, variance, entropy, etc., of CT value [14,15], and geometric features include statistical information of the shape index and curvedness [12,17]. Volume-based features, e.g., number of region growing seeds, axis ratio, disk-likeness and highlighting ratio, are also included [13,16]. The class imbalance ratio (TPs/FPs) is 0.0886, which indicates our dataset or IPC pool is significantly imbalanced. All the following experiments were based on this imbalanced database.
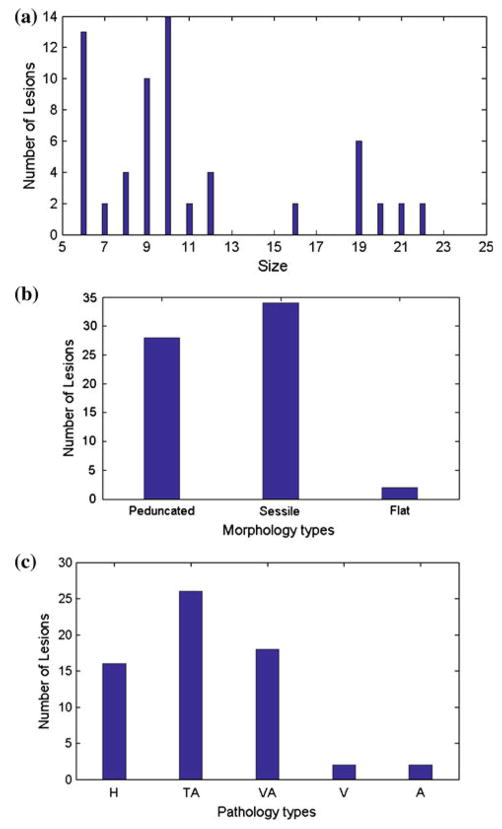
Fig. 3.
The distribution of 64 lesions in CTC database. a, b and c show the size, morphology and pathology distribution information, respectively. In (c), H, TA, VA, V and A stand for hyperplastic, tubular adenoma, tubulovillous adenoma, villous adenoma and adenocarcinoma, respectively
It is noted that the above database was downloaded from the online public domain (http://imaging.nci.nih.gov) where the CTC database was made by the Walter Reed Army Medical Center (WRAMC) after the clinical trial study [4]. All examinations were performed in adherence following standards [18] with a full cathartic bowel preparation, fecal tagging, without IV CM, and with MDCT scanners. The image data were acquired in helical mode with collimations of 1.25–5.0 mm, pitch of 1 to 2, reconstruction intervals of 1.25–5.0mm, and modulated tube current–time products ranging from 50 to 200mAs and tube voltages from 80 to 120 kVp. The indication for CTC was screening for colorectal cancer in all individuals. The human studies had been approved by appropriate ethical committee and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. All subjects gave their informed consent prior to their inclusion in the study. The subjects’ identities have been fully anonymized.
Basic classifiers
RFs [19,20] and SVM [21–23] were chosen here as the basic classifiers in this feasibility study. Both classifiers are widely used and sometimes considered as the gold standard tools in the field because of their good performances, as reported in the previous studies [7,24–30]. In implementation, the widely used SVM package LIBSVM [23] with RBF kernel was employed in this study. Following the guideline of LIBSVM, the two parameters (cost and gamma) in the RBF kernel were determined by a grid search process (fivefold cross-validation) in the training step. For RFs implementation, the widely used tuneRF function in the well-known package random forest [31] was employed with the number of tree-set to be 5,000, where the tuneRF will search for the best fitting number of variables used for each tree in the training phase (i.e., the out of bag estimation).
Two imbalanced data correcting technologies
For comparison purpose, two common approaches to tackle the imbalanced data classification problem, i.e., the cost-sensitive learning (weighted RFs [32] and weighted SVM [22]) and down-sampling technique [33,34], were implemented as references or baseline and compared with our proposed strategies. In the cost-sensitive learning, for both weighted RFs and weighted SVM, the best fitting weight was automatically searched from 0 to 1 by stepwise 0.02 in the training phase. For the down-sampling technique, we down sampled the majority (negative) class to make the dataset more balanced with different levels, i.e., 75, 50 and 25% of the negatives were sampled in the training step. In summary, for each classifier, taking RFs for example, we evaluated it with 5 different settings, i.e., un-weighted RFs with imbalanced data, weighted RFs with imbalanced data, weighted RFs with 75% down sampling, weighted RFs with 50% down sampling and weighted RFs with 25% down sampling.
Three-way cross-validation
For each experiment of those described above, the original dataset was randomly partitioned into three subsets with the same TPs/FPs ratio as the original dataset. One subset (subset 1) was taken for testing purpose, and the other two subsets (subset 2 and subset 3) were treated as the training set. For the original threshold method, we treated both subset 2 and subset 3 as training set to train the classifiers. Regular threshold (0.5 for RFs and 0 for SVM) was applied on subset 1 to draw results. For our proposed operating point chosen strategies, we used subset 2 as classifier training set and subset 3 as cut-off optimization training set and applied the selected threshold on the testing set. Then we rotated the role of the three subsets (6 permutations) and outputted the average results, which are like a three-way cross-validation. To minimize the bias of one time running, the random partition process was repeated 100 times and the average results were outputted.
Results
Figures 4 and 5 show the averaged ROC curves of RFs and SVM with different settings, and Tables 1 and 2 show the averaged AUC information of RFs and SVM. We can see from the figures and tables that RFs and SVM with different settings all achieved high AUC values (0.96 for RFs and 0.95 for SVM) and the standard deviations were relatively small (around 0.02 for both classifiers), indicating that these two classifiers are capable to classify the imbalanced data. However, the classification accuracy with the original threshold is not favorable – very low prediction accuracy in the minority TP class, as shown as the red circle marker in Figs. 4 and 5. Taking the weighted original imbalanced data for example, when using the original threshold, RFs only detected half polyps (0.52 sensitivity), i.e., 10.92 of 21 polyps, in testing set while SVM detected around 60% of polyps (0.62 sensitivity), i.e., 13.02 of 21 polyps.
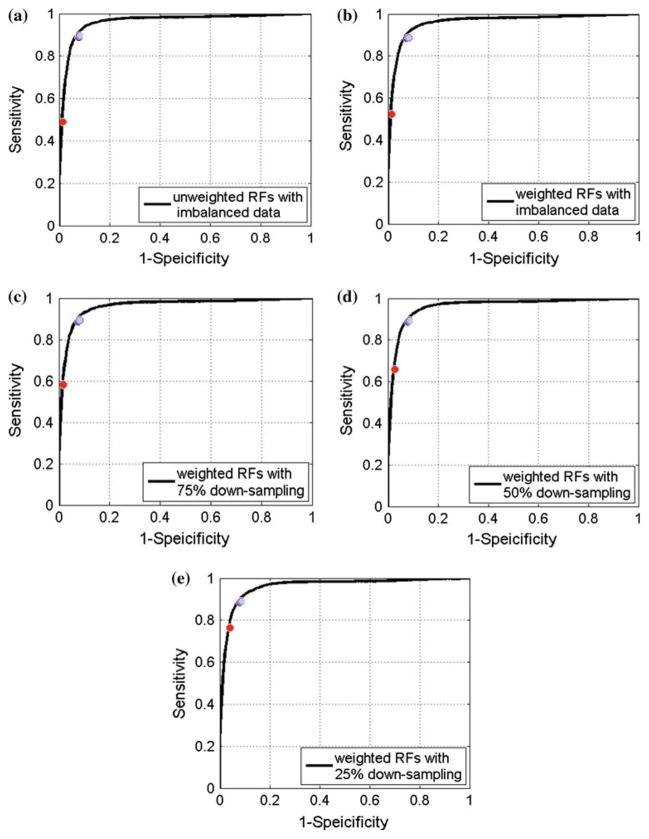
Fig. 4.
Averaged ROC curve of RFs results. a–e show averaged ROC curves of un-weighted RFs with imbalanced data, weighted RFs with imbalanced data, weighted RFs with 75% down-sampling data, weighted RFs with 50% down-sampling data and weighted RFs with 25% down-sampling data. The red, blue, magenta and cyan circle marker represent results of regular 0.5 threshold, shortest distance, harmonic mean and anti-harmonic mean, respectively. The averaged ROC curves was conducted according to the horizontal axis, where the linear interpolation was employed when needed
Fig. 5.
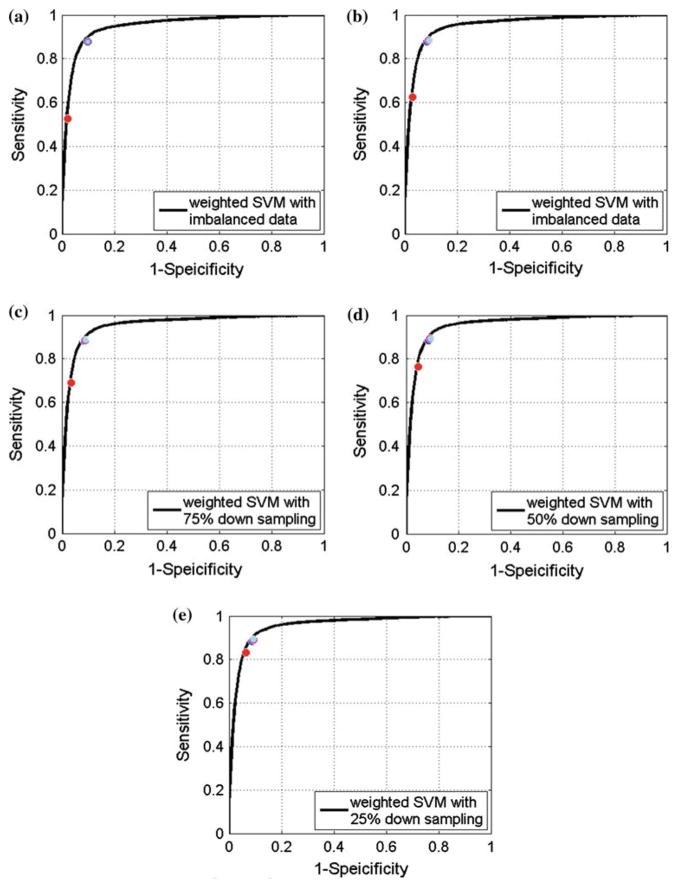
Averaged ROC curve of SVM results. a–e show averaged ROC curves of un-weighted SVM with imbalanced data, weighted SVM with imbalanced data, weighted SVM with 75% down-sampling data, weighted SVM with 50% down-sampling data and weighted SVM with 25% down-sampling data. The red, blue, magenta and cyan circle marker represent results of regular 0 threshold, shortest distance, harmonic mean and anti-harmonic mean, respectively. The averaged ROC curves was conducted according to the horizontal axis, where the linear interpolation was employed when needed
Table 1.
Averaged RFs AUC information of the 100 runs
| Area under ROC curve | ||
|---|---|---|
|
| ||
| Mean | Std. | |
| Imbalanced data, un-weighted | 0.9614 | 0.0201 |
| Imbalanced data, weighted | 0.9608 | 0.0197 |
| 75% down sampling, weighted | 0.9613 | 0.0194 |
| 50% down sampling, weighted | 0.9600 | 0.0201 |
| 25% down sampling, weighted | 0.9600 | 0.0204 |
Table 2.
Averaged SVM AUC information of the 100 runs
| Area under ROC curve | ||
|---|---|---|
|
| ||
| Mean | Std. | |
| Imbalanced data, un-weighted | 0.9493 | 0.0247 |
| Imbalanced data, weighted | 0.9523 | 0.0240 |
| 75% down sampling, weighted | 0.9547 | 0.0215 |
| 50% down sampling, weighted | 0.9561 | 0.0208 |
| 25% down sampling, weighted | 0.9543 | 0.0208 |
Tables 3 and 4 show the classification results of RFs and SVM with different thresholding methods, where the classification results, i.e., sensitivity, specificity and overall accuracy, with the original threshold and the proposed strategies are outputted and compared. The threshold (averaged over 100 runs) chosen by the new strategies is also listed in the tables. As we expected, it is observed from the last column of Tables 3 and 4 that the operating point did shift due to the imbalance of the data, and the original threshold was no longer suitable to draw the decision. From both tables, it was also observed that both the weighted classifier (i.e., the cost sensitivity learning) and the down-sampling technique did help to some degree in improving the sensitivity (compared to the original threshold). For example, for the weighted classifier, only moderate improvement can be observed, about 0.04 increasing for RFs and 0.10 for SVM, for the outcomes of 0.52 for RFs and 0.62 for SVM (see the first two rows arranged along the left column). The down-sampling technique showed improvements as the down-sampling level went up, and the classification result with 25% down sampling (see the last row arranged along the left column) is very close to our proposed strategies (similar results can be observed in [35]). However, there are concerns with the down-sampling technique: First one is the concern about the information loss because it only uses part of the training data; second one is the concern that there is not any rule to determine how much we should down sample the majority class.
Table 3.
Averaged RFs classification results with the original threshold and cutoff chosen by the proposed three strategies over the 100 runs
| Averaged results | |||||
|---|---|---|---|---|---|
|
| |||||
| Sensitivity | Specificity | Accuracy | Threshold | ||
| Imbalanced data, un-weighted | Original_thres | 0.4884±0.1177 | 0.9855±0.0081 | 0.9450±0.0104 | 0.5 |
| Shortest_dist | 0.8895±0.0835 | 0.9235±0.0412 | 0.9208±0.0357 | 0.1271±0.0603 | |
| Harmonic | 0.8957±0.0802 | 0.9186±0.0425 | 0.9167±0.0370 | 0.1198±0.0601 | |
| Anti_harmonic | 0.8969±0.0803 | 0.9172±0.0434 | 0.9155±0.0378 | 0.1185±0.0600 | |
| Imbalanced data, weighted | Original_thres | 0.5243±0.1226 | 0.9862±0.0079 | 0.9486±0.0106 | 0.5 |
| Shortest_dist | 0.8835±0.0802 | 0.9239±0.0330 | 0.9206±0.0281 | 0.1177±0.0610 | |
| Harmonic | 0.8876±0.0796 | 0.9174±0.0384 | 0.9150±0.0328 | 0.1115±0.0627 | |
| Anti_harmonic | 0.8873±0.0803 | 0.9166±0.0397 | 0.9142±0.0339 | 0.1113±0.0633 | |
| 75% down sampling, weighted | Original_thres | 0.5835±0.1151 | 0.9825±0.0090 | 0.9500±0.0110 | 0.5 |
| Shortest_dist | 0.8876±0.0779 | 0.9230±0.0338 | 0.9202±0.0290 | 0.1545±0.0728 | |
| Harmonic | 0.8927±0.0756 | 0.9187±0.0374 | 0.9165±0.0321 | 0.1484±0.0749 | |
| Anti_harmonic | 0.8941±0.0760 | 0.9177±0.0386 | 0.9158±0.0332 | 0.1473±0.0749 | |
| 50% down sampling, weighted | Original_thres | 0.6591±0.1198 | 0.9720±0.0126 | 0.9465±0.0126 | 0.5 |
| Shortest_dist | 0.8876±0.0816 | 0.9198±0.0358 | 0.9172±0.0305 | 0.2298±0.0862 | |
| Harmonic | 0.8933±0.0818 | 0.9136±0.0392 | 0.9120±0.0334 | 0.2184±0.0894 | |
| Anti_harmonic | 0.8944±0.0818 | 0.9124±0.0403 | 0.9110±0.0343 | 0.2168±0.0898 | |
| 25% down sampling, weighted | Original_thres | 0.7649±0.1026 | 0.9592±0.0164 | 0.9434±0.0147 | 0.5 |
| Shortest_dist | 0.8857±0.0805 | 0.9196±0.0341 | 0.9168±0.0296 | 0.3348±0.1001 | |
| Harmonic | 0.8907±0.0805 | 0.9139±0.0387 | 0.9120±0.0334 | 0.3245±0.1066 | |
| Anti_harmonic | 0.8907±0.0829 | 0.9119±0.0407 | 0.9102±0.0350 | 0.3220±0.1090 | |
Format: mean ± standard deviation
Table 4.
Averaged SVM classification results with the original threshold and cutoff chosen by the proposed three strategies over the 100 runs
| Averaged results | |||||
|---|---|---|---|---|---|
|
| |||||
| Sensitivity | Specificity | Accuracy | Threshold | ||
| Imbalanced data, un-weighted | Original_thres | 0.5257±0.1645 | 0.9772±0.0111 | 0.9404±0.0117 | 0 |
| Shortest_dist | 0.8724±0.0879 | 0.9115±0.0358 | 0.9083±0.0309 | −0.8004±0.2039 | |
| Harmonic | 0.8756±0.0860 | 0.9083±0.0388 | 0.9056±0.0337 | −0.8339±0.2054 | |
| Anti_harmonic | 0.8768±0.0868 | 0.9075±0.0397 | 0.9050±0.0344 | −0.8356±0.2093 | |
| Imbalanced data, weighted | Original_thres | 0.6252±0.1559 | 0.9700±0.0136 | 0.9419±0.0121 | 0 |
| Shortest_dist | 0.8772±0.0883 | 0.9133±0.0368 | 0.9103±0.0320 | −0.6353±0.2668 | |
| Harmonic | 0.8848±0.0872 | 0.9088±0.0398 | 0.9068±0.0347 | −0.6572±0.2647 | |
| Anti_harmonic | 0.8850±0.0914 | 0.9075±0.0408 | 0.9057±0.0355 | −0.6606±0.2680 | |
| 75% down sampling, weighted | Original_thres | 0.6908±0.1390 | 0.9647±0.0136 | 0.9424±0.0124 | 0 |
| Shortest_dist | 0.8817±0.0882 | 0.9147±0.0352 | 0.9120±0.0307 | −0.5188±0.2613 | |
| Harmonic | 0.8854±0.0918 | 0.9109±0.0392 | 0.9088±0.0339 | −0.5366±0.2635 | |
| Anti_harmonic | 0.8863±0.0914 | 0.9101±0.0406 | 0.9082±0.0352 | −0.5382±0.2676 | |
| 50% down sampling, weighted | Original_thres | 0.7642±0.1252 | 0.9547±0.0168 | 0.9392±0.0139 | 0 |
| Shortest_dist | 0.8852±0.0887 | 0.9137±0.0356 | 0.9114±0.0307 | −0.3767±0.2818 | |
| Harmonic | 0.8928±0.0880 | 0.9087±0.0406 | 0.9066±0.0352 | −0.4069±0.2866 | |
| Anti_harmonic | 0.8936±0.0883 | 0.9071±0.0409 | 0.9060±0.0354 | −0.4103±0.2844 | |
| 25% down sampling, weighted | Original_thres | 0.8307±0.1086 | 0.9367±0.0249 | 0.9281±0.0211 | 0 |
| Shortest_dist | 0.8832±0.0880 | 0.9131±0.0373 | 0.9106±0.0326 | −0.1650±0.2766 | |
| Harmonic | 0.8918±0.0860 | 0.9077±0.0420 | 0.9064±0.0369 | −0.1954±0.2799 | |
| Anti_harmonic | 0.8929±0.0861 | 0.9059±0.0440 | 0.9049±0.0386 | −0.2033±0.2846 | |
Format: mean ± standard deviation
The gain by our proposed strategies is noticeable as demonstrated by Tables 3 and 4. Taking the weighted original imbalanced data for example, when using our proposed operating point selection strategies, the sensitivity improved from 0.52 to nearly 0.885 (on average 18.59 of 21 polyps are detected) for RFs and for SVM it was from 0.625 to nearly 0.88 (on average 18.48 of 21 polyps are detected); meanwhile, a relatively high specificity level remained: above 0.90 (result in 0.903 more FPs/scan) for both classifiers. The corresponding points shown in Figs. 4 and 5 indicate that our proposed strategies put the operating points very close to the “favorite region.”
In all the experiments, we always used the original class ratio in all the testing sets (TPs/FPs=0.0886) with different levels of class ratio in the training sets, i.e., the original imbalanced data (ratio=0.0886), 75% down sampling (ratio=0.1182), 50% down sampling (ratio=0.1772), 25% down sampling (ratio=0.3546). The outcomes showed that our proposed strategies deliver consistent good performances, i.e., around 0.88 sensitivity with 0.90 specificity for both classifiers. This indicates that the performance of our strategies is robust to different class ratios. Meanwhile, it also indicates that our strategies have the ability to be combined with other imbalanced data correcting techniques, such as the cost-sensitive learning, based on the fact that the outcome from the un-weighted data also showed improvement.
In general, we want to find, by a sound CAD scheme, as many TPs as possible while keeping a relatively low FPs rate. This means that the sensitivity is somehow more important than the specificity for the task, especially in dealing with the imbalanced data which biases to the negative majority class. For that purpose, the results above indicate that our proposed strategies mitigated noticeably the problems caused by the data imbalance. Furthermore, the results indicate that our proposed strategies improved the prediction of the minority class, while keeping a relatively high accuracy in prediction of the majority class.
Conclusion, future work and limitation discussion
In this paper, we presented three ROC graph-based operating point chosen strategies in order to mitigate the classification problem caused by class imbalance data. The problem, i.e., low predication accuracy in minority class, is well addressed while retaining relatively high predication accuracy in majority class. In other words, our proposed strategies can be considered as a new classification accuracy evaluation measure (similar to the overall accuracy merit), especially for the cases of class imbalance data. Meanwhile, their robust performances under different circumstances indicate that our presented strategies are feasible to combine with other techniques, e.g., the two existing imbalance data correcting methods (of cost sensitivity learning and down sampling) and the AUC optimization technique [10,11].
We notice that the number of TPs in our database is somehow small. We should expand our database to include more polyps for further investigation on the three new strategies. Evaluating our proposed strategies on more variations of datasets is one of our next tasks. Constructing a ROC tree-based random forest [8] is another one of our next tasks. Furthermore, it would be more valuable if we can find a family of surfaces, which include the harmonic mean, anti-harmonic mean and other un-explored surfaces. In addition, we want to find the family, which can be described by some kinds of mathematical functions.
We also notice that there are some limitations of this feasibility study. The first one is that the three proposed strategies had very similar performance and, to our best knowledge, there is no preference which one performs the best. Because sensitivity and specificity trade off each other, we cannot simply choose the one with the largest sensitivity, since its corresponding specificity may be the smallest. The second one is that we observed the standard deviation shown in Tables 3 and 4 is kind of large. This is partly due to the small sample size. Meanwhile, we also observed that the standard deviation level of sensitivity and specificity is very similar to those from the original thresholding method, which indicates that our result is reliable despite the small sample size. The third one is that our dataset contains 49 CTC scans from 25 patients, which means there is one patient who has only on scan. We missed the OC confirmation report of the prone position for that patient. Considering our relatively small sample size, each sample case is precious to us, so we decided to keep that CTC scan of this patient in this study. The fourth one (may not be the last one) is that we only employed SVM and RFs in this study. While the fourth one was based on the fact that SVM and RFs are very commonly used classifiers with very mature implementation and if any improvement can be made on them then it will benefit largely to the field or community, we will definitely expand our research scope to employ more classification algorithms in our future work.
Acknowledgments
This work was supported in part by the NIH/NCI under Grants #CA082402 and #CA143111.
Footnotes
Conflict of interest Bowen Song, Guopeng Zhang, WeiZhu and Zhen-grong Liang declare that they have no conflict of interest.
Contributor Information
Bowen Song, Departments of Radiology, Stony Brook University, Stony Brook, NY 11790, USA. Applied Mathematics and Statistics, Stony Brook University, Stony Brook, NY 11790, USA.
Guopeng Zhang, Department of Biomedical Engineering, Fourth Military Medical University, Xi’an 710032, Shaanxi, China.
Wei Zhu, Applied Mathematics and Statistics, Stony Brook University, Stony Brook, NY 11790, USA.
Zhengrong Liang, Email: jerome@mil.sunysb.edu, Departments of Radiology, Stony Brook University, Stony Brook, NY 11790, USA.
References
- 1.American Cancer Society. Cancer facts & figures 2012. American Cancer Society; Atlanta: 2012. [Google Scholar]
- 2.Eddy D. Screening for colorectal cancer. Ann Intern Med. 1990;113:373–384. doi: 10.7326/0003-4819-113-5-373. [DOI] [PubMed] [Google Scholar]
- 3.Gluecker T, Johnson C, Harmsen W, Offord K, Harris A, Wilson L, Ahlquist D. Colorectal cancer screening with CT colonography, colonoscopy, and double-contrast barium enema examination: prospective assessment of patient perceptions and preferences. Radiology. 2003;227(2):378–384. doi: 10.1148/radiol.2272020293. [DOI] [PubMed] [Google Scholar]
- 4.Pickhardt P, Choi J, Hwang I, Butler J, Puckett M, Hildebrandt H, Wong R, Nugent P, Mysliwiec P, Schindler W. Computed tomographic virtual colonoscopy to screen for colorectal neoplasia in asymptomatic adults. N Engl J Med. 2003;349:2191–2200. doi: 10.1056/NEJMoa031618. [DOI] [PubMed] [Google Scholar]
- 5.Summers RM, Yao J, Pickhardt P, Franaszek M, Bitter I, Brickman D, Krishna V, Choi R. Computed tomographic virtual colonoscopy computer-aided polyp detection in a screening population. Gastroenterology. 2005;129:1832–1844. doi: 10.1053/j.gastro.2005.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S, Zhu H, Lu H, Liang Z. Volume-based feature analysis of mucosa for automatic initial polyp detection in virtual colonoscopy. Int J Comput Assist Radiol Surg. 2008;3(1–2):131–142. doi: 10.1007/s11548-008-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu H, Fan Y, Lu H, Liang Z. Improving initial polyp candidate extraction for CT colonography. Phys Med Biol. 2010;55:2087–2102. doi: 10.1088/0031-9155/55/7/019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hossain M, Hassan M, Kirley M, Bailey J. ROC-tree: a novel decision tree induction algorithm based on receiver operating characteristics to classify gene expression data. Proceedings of the 2008 SIAM international conference on data mining (SDM); 2008. pp. 455–465. [Google Scholar]
- 9.Fawcett T. An introduction to ROC analysis. Pattern Recognit Lett. 2006;27:861–874. [Google Scholar]
- 10.Rakotomamonjy A. Optimizing area under ROC curve with SVMs. ROC Analysis in Artificial Intelligence. 2004:71–80. [Google Scholar]
- 11.Zhao P, Hoi SCH, Jin R, Yang T. Online AUC maximization. Proceeding of international conference of machine learning.2011. [Google Scholar]
- 12.Yoshida H, Nappi J. Three-dimensional computer-aided diagnosis scheme for detection of colonic polyps. IEEE Trans Med Imag. 2001;20(12):1261–1274. doi: 10.1109/42.974921. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Liang Z, Li L, Li X, Li B, Anderson J, Harrington D. Reduction of false positives by internal features for polyp detection in CT-based virtual colonoscopy. Med Phys. 2005;32(12):3602–3616. doi: 10.1118/1.2122447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Yao J, Summers R. Scale-based scatter correction for computer-aided polyp detection in CT colonography. Med Phys. 2008;35(12):5664–5671. doi: 10.1118/1.3013552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu H, Duan C, Pickhardt P, Wang S, Liang Z. CAD of colonic polyps with level set-based adaptive convolution in volumetric mucosa to advance CT colonography toward a screening modality. J Cancer Manag Res DOVE Med Press. 2009;1:1–13. doi: 10.2147/cmar.s4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marelo F, Musé P, Aguirre S, Sapiro G. Automatic colon polyp flagging via geometric and texture features. Engineering in Medicine and Biology Society (EMBC), 2010 Annual International Conference of the IEEE; 2010. pp. 3170–3173. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H, Fan Y, Lu H, Liang Z. Improved curvature estimation for computer-aided detection of colonic polyps in CT colonography. Acad Radiol. 2011;18(8):1024–1034. doi: 10.1016/j.acra.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.American College of Radiology. ACR practice guideline for the performance of computed tomography (CT) colonography in adults. ACR Pract Guidel. 2005;29:295–298. [Google Scholar]
- 19.Breiman L. Bagging predictors. Mach Learn. 1996;24:123–140. [Google Scholar]
- 20.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 21.Vapnik V. Statistical learning theory. Wiley; New York: 1998. [Google Scholar]
- 22.Morik K, Brokhausen P, Joachims T. Combining statistical learning with a knowledge-based approach—a case study in intensive care monitoring. Proceedings 16th international conference on machine learning.1999. [Google Scholar]
- 23.Chang C, Lin C. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol. 2011;2(27):1–27. 27. Software available at http://www.csie.ntu.edu.tw/~jlin/libsvm. [Google Scholar]
- 24.Osuna E, Freund R, Girosi F. Training support vector machines: an application to face detection. Proceedings computer vision and pattern recognition; 1997. pp. 130–136. [Google Scholar]
- 25.Pontil M, Verri A. Object recognition with support vector machines. IEEE Trans Pattern Anal Mach Intell. 1998;20:637–646. [Google Scholar]
- 26.Diaz-Uriarte R, Alvarez de Andres S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006 doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexandre LA, et al. Polyp detection in endoscopic video using SVMs. Lect Notes Comput Sci. 2007;4702:358–365. [Google Scholar]
- 28.Zhu H, Liang Z, Barish M, Pickhardt P, You J, Wang S, Fan Y, Lu H, Richards R, Posniak E, Cohen H. Increasing computer-aided detection specificity by projection features for CT colonography. Med Phys. 2010;37(4):1468–1481. doi: 10.1118/1.3302833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M, et al. Robust large scale prone-supine polyp matching using local features: a metric learning approach. The 14th international conference on medical image computing and computer assisted intervention (MICCAI); 2011. [DOI] [PubMed] [Google Scholar]
- 30.Liu M, et al. Sparse classification for computer aided diagnosis using learned dictionaries. The 14th international conference on medical image computing and computer assisted intervention (MICCAI); 2011. [DOI] [PubMed] [Google Scholar]
- 31.http://cran.r-project.org/web/packages/randomForest/index.html
- 32.Chen C, Liaw A, Breiman L. Using random forest to learn Imbalanced data. 2004. Technical Report of Dept. of Stat., UC, Berkeley. [Google Scholar]
- 33.He H, Garcia E. Learning from imbalanced data. IEEE Trans Knowl Data Eng. 2009;21(9):1263–1284. [Google Scholar]
- 34.Blagus R, Lusa L. Class prediction for high-dimensional class-imbalanced data. BMC Bioinformatics. 2010;11:523–539. doi: 10.1186/1471-2105-11-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maloof M. Learning when data sets are imbalanced and when cost are unequal and unknown. Proceedings ICML workshop learn imbalanced data sets; 2003. pp. 73–80. [Google Scholar]