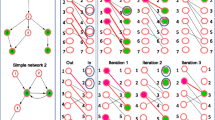
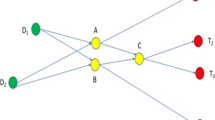
Abstract
Biological networks describe the relationships among molecular elements and help in the deep understanding of the biological mechanisms and functions. One of the common problems is to identify the set of biomolecules that could be targeted by drugs to drive the state transition of the cells from disease states to health states called desired states as the realization of the therapy of complex diseases. Most previous studies based on the output control determine the set of steering nodes without considering available biological information. In this study, we propose a strategy by using the additionally available information like the FDA-approved drug targets to restrict the range for choosing steering nodes in output control instead, where we call it the Set Preference Output Control (SPOC) problem. A graphic-theoretic algorithm is proposed to approximately tackle it by using the Maximum Weighted Complete Matching (MWCM). The computation experiment results from two biological networks illustrate that our proposed SPOC strategy outperforms the full control and output control strategies to identify drug targets. Finally, the case studies further demonstrate the role of the combination therapy in two biological networks, which reveals that our proposed SPOC strategy is potentially applicable for more complicated cases.
This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC), the National Natural Science Foundation of China [61772552], and the Program of Independent Exploration Innovation in Central South University (2019zzts959), the Fundamental Research Funds for the Central Universities, CSU (2282019SYLB004).
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Barabási, A.L., et al.: Network Science. Cambridge University Press, Cambridge (2016)
Barabási, A.L., Gulbahce, N., Loscalzo, J.: Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 12(1), 56 (2011)
Jeong, H., Mason, S.P., Barabási, A.L., Oltvai, Z.N.: Lethality and centrality in protein networks. Nature 411(6833), 41 (2001)
Winzeler, E.A., et al.: Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285(5429), 901–906 (1999)
Zeng, M., Zhang, F., Wu, F.X., Li, Y., Wang, J., Li, M.: Protein-protein interaction site prediction through combining local and global features with deep neural networks. Bioinformatics 36(4), 1114–1120 (2020)
Li, M., Gao, H., Wang, J., Wu, F.X.: Control principles for complex biological networks. Brief. Bioinform. 20(6), 2253–2266 (2019)
Guo, W.F., Zhang, S.W., Zeng, T., Akutsu, T., Chen, L.: Network control principles for identifying personalized driver genes in cancer. Brief. Bioinform. (2019)
Liu, Y.Y., Slotine, J.J., Barabási, A.L.: Controllability of complex networks. Nature 473(7346), 167 (2011)
Wu, L., Shen, Y., Li, M., Wu, F.X.: Network output controllability-based method for drug target identification. IEEE Trans. Nanobiosci. 14(2), 184–191 (2015)
Gao, J., Liu, Y.Y., D’souza, R.M., Barabási, A.L.: Target control of complex networks. Nat. Commun. 5, 5415 (2014)
Wu, L., Li, M., Wang, J., Wu, F.X.: Minimum steering node set of complex networks and its applications to biomolecular networks. IET Syst. Biol. 10(3), 116–123 (2016)
Guo, W.F., et al.: Discovering personalized driver mutation profiles of single samples in cancer by network control strategy. Bioinformatics 34(11), 1893–1903 (2018)
Hu, Y., et al.: Optimal control nodes in disease-perturbed networks as targets for combination therapy. Nat. Commun. 10(1), 2180 (2019)
Wu, L., Tang, L., Li, M., Wang, J., Wu, F.X.: Biomolecular network controllability with drug binding information. IEEE Trans. Nanobiosci. 16(5), 326–332 (2017)
Guo, W.F., et al.: Constrained target controllability of complex networks. J. Stat. Mech: Theory Exp. 2017(6), 063402 (2017)
Müller, F.J., Schuppert, A.: Few inputs can reprogram biological networks. Nature 478(7369), E4 (2011)
Kailath, T.: Linear Systems, vol. 156. Prentice-Hall, Englewood Cliffs (1980)
Lin, C.T.: Structural controllability. IEEE Trans. Autom. Control 19(3), 201–208 (1974)
Jungnickel, D.: Graphs, Networks and Algorithms. Springer, Heidelberg (2005). https://doi.org/10.1007/b138283
Crouse, D.F.: On implementing 2D rectangular assignment algorithms. IEEE Trans. Aerosp. Electron. Syst. 52(4), 1679–1696 (2016)
Grieco, L., Calzone, L., Bernard-Pierrot, I., Radvanyi, F., Kahn-Perles, B., Thieffry, D.: Integrative modelling of the influence of mapk network on cancer cell fate decision. PLoS Comput. Biol. 9(10), e1003286 (2013)
Wishart, D.S., et al.: Drugbank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 36(suppl–1), D901–D906 (2007)
Frenzel, A., Grespi, F., Chmelewskij, W., Villunger, A.: Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis 14(4), 584–596 (2009). https://doi.org/10.1007/s10495-008-0300-z
Lafitte, M., et al.: FGFR3 has tumor suppressor properties in cells with epithelial phenotype. Mol. Cancer 12(1), 83 (2013)
Sigismund, S., Avanzato, D., Lanzetti, L.: Emerging functions of the EGFR in cancer. Mol. Oncol. 12(1), 3–20 (2018)
Vleugel, M.M., Greijer, A.E., Bos, R., van der Wall, E., van Diest, P.J.: c-jun activation is associated with proliferation and angiogenesis in invasive breast cancer. Hum. Pathol. 37(6), 668–674 (2006)
Raimondi, C., Falasca, M.: Targeting PDK1 in cancer. Curr. Med. Chem. 18(18), 2763–2769 (2011)
Gómez Tejeda Zañudo, J., Scaltriti, M., Albert, R.: A network modeling approach to elucidate drug resistance mechanisms and predict combinatorial drug treatments in breast cancer. Cancer Converg. 1(1), 1–25 (2017). https://doi.org/10.1186/s41236-017-0007-6
Eischen, C., Adams, C., Clark-Garvey, S., Porcu, P.: Targeting the Bcl-2 family in B cell lymphoma. Front. Oncol. 8, 636 (2018)
Zou, M., et al.: Knockdown of the Bcl-2 gene increases sensitivity to EGFR tyrosine kinase inhibitors in the H1975 lung cancer cell line harboring T790m mutation. Int. J. Oncol. 42(6), 2094–2102 (2013)
Royce, M.E., Osman, D.: Everolimus in the treatment of metastatic breast cancer. Breast Cancer: Basic Clin. Res. 9, BCBCR–S29268 (2015)
Kornblum, N., et al.: Randomized phase ii trial of fulvestrant plus everolimus or placebo in postmenopausal women with hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer resistant to aromatase inhibitor therapy: results of pre0102. ASCO (2018)
Piha-Paul, S.A., et al.: Phase i study of the pan-HER inhibitor neratinib given in combination with everolimus, palbociclib or trametinib in advanced cancer subjects with EGFR mutation/amplification, HER2 mutation/amplification or HER3/4 mutation (2018)
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Gao, H., Li, M., Wu, FX. (2020). SPOC: Identification of Drug Targets in Biological Networks via Set Preference Output Control. In: Cai, Z., Mandoiu, I., Narasimhan, G., Skums, P., Guo, X. (eds) Bioinformatics Research and Applications. ISBRA 2020. Lecture Notes in Computer Science(), vol 12304. Springer, Cham. https://doi.org/10.1007/978-3-030-57821-3_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-57821-3_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-57820-6
Online ISBN: 978-3-030-57821-3
eBook Packages: Computer ScienceComputer Science (R0)