Abstract
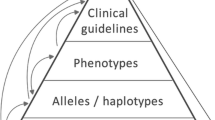
Treatment recommendations within Clinical Practice Guidelines (CPGs) are largely based on findings from clinical trials and case studies, referred to here as research studies, that are often based on highly selective clinical populations, referred to here as study cohorts. When medical practitioners apply CPG recommendations, they need to understand how well their patient population matches the characteristics of those in the study cohort, and thus are confronted with the challenges of locating the study cohort information and making an analytic comparison. To address these challenges, we develop an ontology-enabled prototype system, which exposes the population descriptions in research studies in a declarative manner, with the ultimate goal of allowing medical practitioners to better understand the applicability and generalizability of treatment recommendations. We build a Study Cohort Ontology (SCO) to encode the vocabulary of study population descriptions, that are often reported in the first table in the published work, thus they are often referred to as Table 1. We leverage the well-used Semanticscience Integrated Ontology (SIO) for defining property associations between classes. Further, we model the key components of Table 1s, i.e., collections of study subjects, subject characteristics, and statistical measures in RDF knowledge graphs. We design scenarios for medical practitioners to perform population analysis, and generate cohort similarity visualizations to determine the applicability of a study population to the clinical population of interest. Our semantic approach to make study populations visible, by standardized representations of Table 1s, allows users to quickly derive clinically relevant inferences about study populations.
Resource Website: https://tetherless-world.github.io/study-cohort-ontology/.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
- 1.
Declarative manner: in a clear, unambiguous, and computer understandable manner.
- 2.
- 3.
- 4.
- 5.
Find the list of all supported publication types at https://www.ncbi.nlm.nih.gov/books/NBK3827/table/pubmedhelp.T.publication_types/.
- 6.
We use the ontology prefixes: (1) sio: SemanticScience Integrated Ontology (2) uo: The Units of Measurement Ontology (3) chear: Children’s Health Exposure Analysis Resource Ontology (4) ncit: National Cancer Institute Thesaurus (5) provcare: ProveCaRe (6) doid: Human Disease Ontology (7) sco: Study Cohort Ontology (8) hasco: Human-Aware Science Ontology (9) prov: The PROV ontology (10) dct: Dublin Core Terms (11) vann: A vocabulary for annotating vocabulary descriptions.
- 7.
View the definition of sio:hasProperty and sio:hasAttribute relations at: https://raw.githubusercontent.com/micheldumontier/semanticscience/master/ontology/sio/release/sio-subset-labels.owl.
- 8.
More Table 1 reporting style and composition details at https://prsinfo.clinicaltrials.gov/webinars/module6/resources/BaselineCharacteristics_Handouts.pdf.
- 9.
Definition adapted from: https://en.wikipedia.org/wiki/Descriptive_statistics.
- 10.
- 11.
Dataset Information Page. https://wwwn.cdc.gov/nchs/nhanes/continuousnhanes/default.aspx?BeginYear=2015.
- 12.
- 13.
- 14.
- 15.
- 16.
- 17.
NIH Collaboratory run grand-round presentation: https://www.nihcollaboratory.org/Pages/Grand-Rounds-02-28-14.aspx.
- 18.
References
American Diabetes Association (ADA) et al.: 8. Pharmacologic approaches to glycemic treatment: Standards of medical care in diabetes - 2018. Diabetes Care 41(Suppl. 1), S73–S85 (2018)
American Diabetes Association (ADA) et al.: 9. Cardiovascular disease and risk management: standards of medical care in diabetes - 2018. Diabetes Care 41(Suppl. 1), S86–S104 (2018)
Auer, S., Kovtun, V., Prinz, M., Kasprzik, A., Stocker, M., Vidal, M.E.: Towards a knowledge graph for science. In: Proceedings of the 8th International Conference on Web Intelligence, Mining and Semantics, p. 1. ACM, Novi Sad (2018)
Bechhofer, S., et al.: OWL web ontology language reference. OWL Reference Guide. https://www.w3.org/TR/owl-ref/
Courtot, M., et al.: MIREOT: The minimum information to reference an external ontology term. Appl. Ontol. 6(1), 23–33 (2011)
Cyganiak, R., Field, S., Gregory, A., Halb, W., Tennison, J.: Semantic statistics: bringing together SDMX and SCOVO. In: Proceedings of the Linked Data on the Web Workshop (LDOW 2010), Raleigh, North Carolina, USA, 27 April 2010 (2010). http://ceur-ws.org/Vol-628/. Accessed 26 Mar 2019
Garijo, D., Poveda-VillalÃşn, M.: A checklist for complete vocabulary metadata. List of Desirable Ontology Best-Practices. http://dgarijo.github.io/Widoco/doc/bestPractices/index-en.html
Graham, R., et al.: Trustworthy clinical practice guidelines: challenges and potential. In: Clinical Practice Guidelines We Can Trust, pp. 53–75. National Academies Press (US), Washington D.C. (2011)
Hurtado, C.A., Poulovassilis, A., Wood, P.T.: Query relaxation in RDF. J. Data Semant. X 4900, 31–61 (2008)
Ontarget Investigators: Telmisartan, ramipril, or both in patients at high risk for vascular events. N. Engl. J. Med. 358(15), 1547–1559 (2008)
Jang, M., Jahanshad, N., Espiritu, R.: The cohort ontology. Enigma Knowledge Capture and Discovery Project. https://knowledgecaptureanddiscovery.github.io/EnigmaOntology/release/cohort/1.0.0/index-en.html
Masic, I., Miokovic, M., Muhamedagic, B.: Evidence based medicine-new approaches and challenges. Acta Inform. Med. 16(4), 219 (2008)
National Institute of Health (NIH): Rigor and Reproducibility. Introduction and need for principles. https://www.nih.gov/research-training/rigor-reproducibility
New, A., Rashid, S.M., Erickson, J.S., McGuinness, D.L., Bennett, K.P.: Semantically-aware population health risk analyses. Presented as a Poster at Machine Learning for Health (ML4H) Workshop, NeurIPS, Montreal, Canada (2018). https://arxiv.org/abs/1811.11190. Accessed 20 Mar 2019
NIH Colloboratory: Table 1 project. Rethinking Clinical Trials. https://sites.duke.edu/rethinkingclinicaltrials/ehr-phenotyping/table-1-project/
Noy, N.F., et al.: BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res. 37(suppl\(_2\)), W170–W173 (2009)
Patel, C., et al.: Matching patient records to clinical trials using ontologies. In: Aberer, K., et al. (eds.) ISWC 2007. LNCS, vol. 4825, pp. 816–829. Springer, Heidelberg (2007). https://doi.org/10.1007/978-3-540-76298-0_59
Reinhardt, S.: Property reification vocabulary. A Strawman Draft. https://www.w3.org/wiki/PropertyReificationVocabulary
Shankar, R.D., Martins, S.B., O’Connor, M.J., Parrish, D.B., Das, A.K.: Epoch: an ontological framework to support clinical trials management. In: Proceedings of the International Workshop on Healthcare Information and Knowledge Management, pp. 25–32. ACM, Arlington (2006)
Sim, I., et al.: The ontology of clinical research (OCRe): an informatics foundation for the science of clinical research. J. Biomed. Inform. 52, 78–91 (2014)
Tu, S.W., et al.: A practical method for transforming free-text eligibility criteria into computable criteria. J. Biomed. Inform. 44(2), 239–250 (2011)
Valdez, J., Kim, M., Rueschman, M., Socrates, V., Redline, S., Sahoo, S.S.: Provcare semantic provenance knowledgebase: evaluating scientific reproducibility of research studies. In: AMIA Annual Symposium Proceedings, vol. 2017, p. 1705. American Medical Informatics Association, Washington D.C., USA (2017)
Xiang, Z., Courtot, M., Brinkman, R.R., Ruttenberg, A., He, Y.: OntoFox: web-based support for ontology reuse. BMC Res. Notes 3(1), 175 (2010)
Younesi, E.: A knowledge-based integrative modeling approach for in-silico identification of mechanistic targets in neurodegeneration with focus on Alzheimer’s disease. Ph.D. thesis, Department of Mathematics and Natural Sciences, Universitäts-und Landesbibliothek Bonn, Bonn, Germany (2014)
Acknowledgements
This work is partially supported by IBM Research AI through the AI Horizons Network. We thank our colleagues from IBM Research, Dan Gruen, Morgan Foreman and Ching-Hua Chen, and from RPI, John Erickson, Alexander New, and Rebecca Cowan, who greatly assisted the research.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Chari, S. et al. (2019). Making Study Populations Visible Through Knowledge Graphs. In: Ghidini, C., et al. The Semantic Web – ISWC 2019. ISWC 2019. Lecture Notes in Computer Science(), vol 11779. Springer, Cham. https://doi.org/10.1007/978-3-030-30796-7_4
Download citation
DOI: https://doi.org/10.1007/978-3-030-30796-7_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-30795-0
Online ISBN: 978-3-030-30796-7
eBook Packages: Computer ScienceComputer Science (R0)