A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion
- PMID: 9531558
- PMCID: PMC2132722
- DOI: 10.1083/jcb.141.1.187
A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion
Abstract
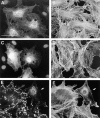
Members of the Rho GTPase family regulate the organization of the actin cytoskeleton in response to extracellular growth factors. We have identified three proteins that form a distinct branch of the Rho family: Rnd1, expressed mostly in brain and liver; Rnd2, highly expressed in testis; and Rnd3/RhoE, showing a ubiquitous low expression. At the subcellular level, Rnd1 is concentrated at adherens junctions both in confluent fibroblasts and in epithelial cells. Rnd1 has a low affinity for GDP and spontaneously exchanges nucleotide rapidly in a physiological buffer. Furthermore, Rnd1 lacks intrinsic GTPase activity suggesting that in vivo, it might be constitutively in a GTP-bound form. Expression of Rnd1 or Rnd3/RhoE in fibroblasts inhibits the formation of actin stress fibers, membrane ruffles, and integrin-based focal adhesions and induces loss of cell-substrate adhesion leading to cell rounding (hence Rnd for "round"). We suggest that these proteins control rearrangements of the actin cytoskeleton and changes in cell adhesion.
Figures

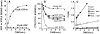

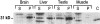




Similar articles
-
Socius is a novel Rnd GTPase-interacting protein involved in disassembly of actin stress fibers.Mol Cell Biol. 2002 May;22(9):2952-64. doi: 10.1128/MCB.22.9.2952-2964.2002. Mol Cell Biol. 2002. PMID: 11940653 Free PMC article.
-
Small GTPase RhoD suppresses cell migration and cytokinesis.Oncogene. 1999 Apr 15;18(15):2431-40. doi: 10.1038/sj.onc.1202604. Oncogene. 1999. PMID: 10229194
-
Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization.J Cell Biol. 1997 Aug 25;138(4):913-26. doi: 10.1083/jcb.138.4.913. J Cell Biol. 1997. PMID: 9265656 Free PMC article.
-
Rho family proteins and regulation of the actin cytoskeleton.Prog Mol Subcell Biol. 1999;22:1-22. doi: 10.1007/978-3-642-58591-3_1. Prog Mol Subcell Biol. 1999. PMID: 10081062 Review. No abstract available.
-
Ras-related GTPases and the cytoskeleton.Mol Biol Cell. 1992 May;3(5):475-9. doi: 10.1091/mbc.3.5.475. Mol Biol Cell. 1992. PMID: 1611153 Free PMC article. Review.
Cited by
-
FOXA2 Activates RND1 to Regulate Arachidonic Acid Metabolism Pathway and Suppress Cisplatin Resistance in Lung Squamous Cell Carcinoma.Clin Respir J. 2024 Aug;18(8):e13814. doi: 10.1111/crj.13814. Clin Respir J. 2024. PMID: 39129202 Free PMC article.
-
Molecular basis and current insights of atypical Rho small GTPase in cancer.Mol Biol Rep. 2024 Jan 18;51(1):141. doi: 10.1007/s11033-023-09140-7. Mol Biol Rep. 2024. PMID: 38236467 Review.
-
Transcriptomic patterns in early-secretory and mid-secretory endometrium in a natural menstrual cycle immediately before in vitro fertilization and embryo transfer.Obstet Gynecol Sci. 2023 Sep;66(5):417-429. doi: 10.5468/ogs.22315. Epub 2023 Jul 17. Obstet Gynecol Sci. 2023. PMID: 37460099 Free PMC article.
-
Transcriptome profiling of colorectal tumors from patients with sepsis reveals an ethnic basis for viral infection risk and sepsis progression.Sci Rep. 2022 Nov 30;12(1):20646. doi: 10.1038/s41598-022-24489-8. Sci Rep. 2022. PMID: 36450776 Free PMC article.
-
Essential role of Rnd1 in innate immunity during viral and bacterial infections.Cell Death Dis. 2022 Jun 2;13(6):520. doi: 10.1038/s41419-022-04954-y. Cell Death Dis. 2022. PMID: 35654795 Free PMC article.
References
-
- Aberle H, Bierkamp C, Torchard D, Serova O, Wagner T, Natt E, Wirshing J, Heidkamper C, Montagna M, Lynch HT, et al. The human plakoglobin gene localizes on chromosome 17 q21 and is subjected to loss of heterozygosity in breast and ovarian cancers. Proc Natl Acad Sci USA. 1995;92:6384–6388. - PMC - PubMed
-
- Aktories K, Just I. In vitro ADP-ribosylation of Rho by bacterial ADP-ribosyl transferases. Methods Enzymol. 1995;256:184–195. - PubMed
-
- Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–20249. - PubMed
-
- Bar-Sagi D, Feramisco JR. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by Ras proteins. Science. 1986;233:1061–1068. - PubMed
-
- Cerione RA, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;8:216–222. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases


