A restriction map of the bacteriophage T4 genome
- PMID: 6258018
- PMCID: PMC2868831
- DOI: 10.1007/BF00425473
A restriction map of the bacteriophage T4 genome
Abstract
We report a detailed restriction map of the bacteriophage T4 genome and the alignment of this map with the genetic map. The sites cut by the enzymes Bg/II, XhoI, KpnI, SalI, PstI, EcoRI and HindIII have been localized. Several novel approaches including two-dimensional (double restriction) electrophoretic separations were used.
Figures


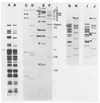
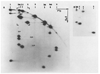
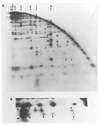
Similar articles
-
[Restriction mapping of T4 bacteriophage late gene region which contains the origins of DNA replication].Mol Biol (Mosk). 1981 Jul-Aug;15(4):883-93. Mol Biol (Mosk). 1981. PMID: 6268966 Russian.
-
A physical map of bacteriophage T4 including the positions of strong promoters and terminators recognized in vitro.Mol Gen Genet. 1984;194(1-2):232-40. doi: 10.1007/BF00383522. Mol Gen Genet. 1984. PMID: 6328215
-
The complete maps of BglII, SalI and XhoI restriction sites on T4 dC-DNA.Mol Gen Genet. 1979 Nov;176(3):417-25. doi: 10.1007/BF00333106. Mol Gen Genet. 1979. PMID: 293460
-
Bacteriophage T3 and bacteriophage T7 virus-host cell interactions.Microbiol Rev. 1981 Mar;45(1):9-51. doi: 10.1128/mr.45.1.9-51.1981. Microbiol Rev. 1981. PMID: 6261110 Free PMC article. Review. No abstract available.
-
Linkage map of Escherichia coli K-12, edition 10: the physical map.Microbiol Mol Biol Rev. 1998 Sep;62(3):985-1019. doi: 10.1128/MMBR.62.3.985-1019.1998. Microbiol Mol Biol Rev. 1998. PMID: 9729612 Free PMC article. Review.
Cited by
-
Evolutionary Dynamics between Phages and Bacteria as a Possible Approach for Designing Effective Phage Therapies against Antibiotic-Resistant Bacteria.Antibiotics (Basel). 2022 Jul 7;11(7):915. doi: 10.3390/antibiotics11070915. Antibiotics (Basel). 2022. PMID: 35884169 Free PMC article. Review.
-
Acid tolerance and morphological characteristics of five Weissella cibaria bacteriophages isolated from kimchi.Food Sci Biotechnol. 2019 Dec 23;29(6):873-878. doi: 10.1007/s10068-019-00723-4. eCollection 2020 Jun. Food Sci Biotechnol. 2019. PMID: 32523797 Free PMC article.
-
From Host to Phage Metabolism: Hot Tales of Phage T4's Takeover of E. coli.Viruses. 2018 Jul 21;10(7):387. doi: 10.3390/v10070387. Viruses. 2018. PMID: 30037085 Free PMC article. Review.
-
Isolation of the TRP2 and the TRP3 genes of Saccharomyces cerevisiae by functional complementation in yeast.Curr Genet. 1982 May;5(1):39-46. doi: 10.1007/BF00445739. Curr Genet. 1982. PMID: 24186086
-
Plasmid-like DNAs in the commercially important mushroom genus Agaricus.Curr Genet. 1984 Oct;8(8):615-9. doi: 10.1007/BF00395707. Curr Genet. 1984. PMID: 24178001
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


