The RecD2 helicase balances RecA activities
- PMID: 35234892
- PMCID: PMC8989531
- DOI: 10.1093/nar/gkac131
The RecD2 helicase balances RecA activities
Abstract
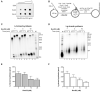
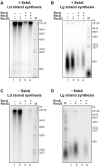
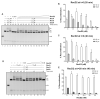
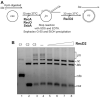
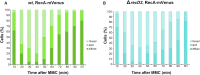
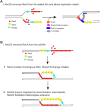
DNA helicases of the RecD2 family are ubiquitous. Bacillus subtilis RecD2 in association with the single-stranded binding protein SsbA may contribute to replication fork progression, but its detailed action remains unknown. In this work, we explore the role of RecD2 during DNA replication and its interaction with the RecA recombinase. RecD2 inhibits replication restart, but this effect is not observed in the absence of SsbA. RecD2 slightly affects replication elongation. RecA inhibits leading and lagging strand synthesis, and RecD2, which physically interacts with RecA, counteracts this negative effect. In vivo results show that recD2 inactivation promotes RecA-ssDNA accumulation at low mitomycin C levels, and that RecA threads persist for a longer time after induction of DNA damage. In vitro, RecD2 modulates RecA-mediated DNA strand-exchange and catalyzes branch migration. These findings contribute to our understanding of how RecD2 may contribute to overcome a replicative stress, removing RecA from the ssDNA and, thus, it may act as a negative modulator of RecA filament growth.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Processing of stalled replication forks in Bacillus subtilis.FEMS Microbiol Rev. 2024 Jan 12;48(1):fuad065. doi: 10.1093/femsre/fuad065. FEMS Microbiol Rev. 2024. PMID: 38052445 Free PMC article. Review.
-
Genetic recombination in Bacillus subtilis: a division of labor between two single-strand DNA-binding proteins.Nucleic Acids Res. 2012 Jul;40(12):5546-59. doi: 10.1093/nar/gks173. Epub 2012 Feb 28. Nucleic Acids Res. 2012. PMID: 22373918 Free PMC article.
-
Bacillus subtilis DprA recruits RecA onto single-stranded DNA and mediates annealing of complementary strands coated by SsbB and SsbA.J Biol Chem. 2013 Aug 2;288(31):22437-50. doi: 10.1074/jbc.M113.478347. Epub 2013 Jun 18. J Biol Chem. 2013. PMID: 23779106 Free PMC article.
-
Roles of Bacillus subtilis DprA and SsbA in RecA-mediated genetic recombination.J Biol Chem. 2014 Oct 3;289(40):27640-52. doi: 10.1074/jbc.M114.577924. Epub 2014 Aug 19. J Biol Chem. 2014. PMID: 25138221 Free PMC article.
-
The cell pole: the site of cross talk between the DNA uptake and genetic recombination machinery.Crit Rev Biochem Mol Biol. 2012 Nov-Dec;47(6):531-55. doi: 10.3109/10409238.2012.729562. Epub 2012 Oct 9. Crit Rev Biochem Mol Biol. 2012. PMID: 23046409 Free PMC article. Review.
Cited by
-
Processing of stalled replication forks in Bacillus subtilis.FEMS Microbiol Rev. 2024 Jan 12;48(1):fuad065. doi: 10.1093/femsre/fuad065. FEMS Microbiol Rev. 2024. PMID: 38052445 Free PMC article. Review.
-
Crystal Structure of DNA Replication Protein SsbA Complexed with the Anticancer Drug 5-Fluorouracil.Int J Mol Sci. 2023 Oct 4;24(19):14899. doi: 10.3390/ijms241914899. Int J Mol Sci. 2023. PMID: 37834349 Free PMC article.
-
Bacillus subtilis RadA/Sms-Mediated Nascent Lagging-Strand Unwinding at Stalled or Reversed Forks Is a Two-Step Process: RadA/Sms Assists RecA Nucleation, and RecA Loads RadA/Sms.Int J Mol Sci. 2023 Feb 25;24(5):4536. doi: 10.3390/ijms24054536. Int J Mol Sci. 2023. PMID: 36901969 Free PMC article.
-
ATPase Activity of Bacillus subtilis RecA Affects the Dynamic Formation of RecA Filaments at DNA Double Strand Breaks.mSphere. 2022 Dec 21;7(6):e0041222. doi: 10.1128/msphere.00412-22. Epub 2022 Nov 2. mSphere. 2022. PMID: 36321831 Free PMC article.
References
-
- Lo Piano A., Martinez-Jimenez M.I., Zecchi L., Ayora S.. Recombination-dependent concatemeric viral DNA replication. Virus Res. 2011; 160:1–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources


