Evidence for host-microbiome co-evolution in apple
- PMID: 34823272
- PMCID: PMC9299473
- DOI: 10.1111/nph.17820
Evidence for host-microbiome co-evolution in apple
Abstract
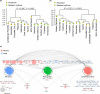
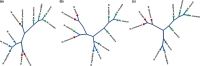
Plants evolved in association with a diverse community of microorganisms. The effect of plant phylogeny and domestication on host-microbiome co-evolutionary dynamics are poorly understood. Here we examined the effect of domestication and plant lineage on the composition of the endophytic microbiome of 11 Malus species, representing three major groups: domesticated apple (M. domestica), wild apple progenitors, and wild Malus species. The endophytic community of M. domestica and its wild progenitors showed higher microbial diversity and abundance than wild Malus species. Heirloom and modern cultivars harbored a distinct community composition, though the difference was not significant. A community-wide Bayesian model revealed that the endophytic microbiome of domesticated apple is an admixture of its wild progenitors, with clear evidence for microbiome introgression, especially for the bacterial community. We observed a significant correlation between the evolutionary distance of Malus species and their microbiome. This study supports co-evolution between Malus species and their microbiome during domestication. This finding has major implications for future breeding programs and our understanding of the evolution of plants and their microbiomes.
Keywords: bacterial community; endophytes; fungal community; microbial introgression; microbiota; phylosymbiosis.
© 2021 The Authors. New Phytologist © 2021 New Phytologist Foundation.
Figures



Similar articles
-
Phenotypic divergence between the cultivated apple (Malus domestica) and its primary wild progenitor (Malus sieversii).PLoS One. 2022 Mar 23;17(3):e0250751. doi: 10.1371/journal.pone.0250751. eCollection 2022. PLoS One. 2022. PMID: 35320270 Free PMC article.
-
Seed-Derived Microbial Colonization of Wild Emmer and Domesticated Bread Wheat (Triticum dicoccoides and T. aestivum) Seedlings Shows Pronounced Differences in Overall Diversity and Composition.mBio. 2020 Nov 17;11(6):e02637-20. doi: 10.1128/mBio.02637-20. mBio. 2020. PMID: 33203759 Free PMC article.
-
Chloroplast diversity in the genus Malus: new insights into the relationship between the European wild apple (Malus sylvestris (L.) Mill.) and the domesticated apple (Malus domestica Borkh.).Mol Ecol. 2006 Jul;15(8):2171-82. doi: 10.1111/j.1365-294X.2006.02924.x. Mol Ecol. 2006. PMID: 16780433
-
The domestication and evolutionary ecology of apples.Trends Genet. 2014 Feb;30(2):57-65. doi: 10.1016/j.tig.2013.10.002. Epub 2013 Nov 27. Trends Genet. 2014. PMID: 24290193 Review.
-
Effects of Domestication on Plant-Microbiome Interactions.Plant Cell Physiol. 2022 Nov 22;63(11):1654-1666. doi: 10.1093/pcp/pcac108. Plant Cell Physiol. 2022. PMID: 35876043 Review.
Cited by
-
Unravelling the microbiome of wild flowering plants: a comparative study of leaves and flowers in alpine ecosystems.BMC Microbiol. 2024 Oct 19;24(1):417. doi: 10.1186/s12866-024-03574-0. BMC Microbiol. 2024. PMID: 39425049 Free PMC article.
-
Implications of Domestication in Theobroma cacao L. Seed-Borne Microbial Endophytes Diversity.Microb Ecol. 2024 Aug 28;87(1):108. doi: 10.1007/s00248-024-02409-9. Microb Ecol. 2024. PMID: 39196422 Free PMC article.
-
The functional identification and evaluation of endophytic bacteria sourced from the roots of tolerant Achyranthes bidentata to overcome monoculture problems of Rehmannia glutinosa.Front Microbiol. 2024 Jul 16;15:1399406. doi: 10.3389/fmicb.2024.1399406. eCollection 2024. Front Microbiol. 2024. PMID: 39081886 Free PMC article.
-
Seed or soil: Tracing back the plant mycobiota primary sources.Environ Microbiol Rep. 2024 Jun;16(3):e13301. doi: 10.1111/1758-2229.13301. Environ Microbiol Rep. 2024. PMID: 38924368 Free PMC article.
-
Phylosymbiosis shapes skin bacterial communities and pathogen-protective function in Appalachian salamanders.ISME J. 2024 Jan 8;18(1):wrae104. doi: 10.1093/ismejo/wrae104. ISME J. 2024. PMID: 38861457 Free PMC article.
References
-
- Abarenkov K, Henrik Nilsson R, Larsson K‐H, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T et al. 2010. The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytologist 186: 281–285. - PubMed
-
- Abdelfattah A, Wisniewski M, Schena L, Tack AJM. 2021b. Experimental evidence of microbial inheritance in plants and transmission routes from seed to phyllosphere and root. Environmental Microbiology 23: 2199–2214. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources


