Genomic insights into the formation of human populations in East Asia
- PMID: 33618348
- PMCID: PMC7993749
- DOI: 10.1038/s41586-021-03336-2
Genomic insights into the formation of human populations in East Asia
Abstract
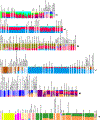
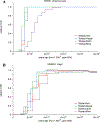
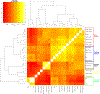
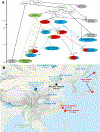
The deep population history of East Asia remains poorly understood owing to a lack of ancient DNA data and sparse sampling of present-day people1,2. Here we report genome-wide data from 166 East Asian individuals dating to between 6000 BC and AD 1000 and 46 present-day groups. Hunter-gatherers from Japan, the Amur River Basin, and people of Neolithic and Iron Age Taiwan and the Tibetan Plateau are linked by a deeply splitting lineage that probably reflects a coastal migration during the Late Pleistocene epoch. We also follow expansions during the subsequent Holocene epoch from four regions. First, hunter-gatherers from Mongolia and the Amur River Basin have ancestry shared by individuals who speak Mongolic and Tungusic languages, but do not carry ancestry characteristic of farmers from the West Liao River region (around 3000 BC), which contradicts theories that the expansion of these farmers spread the Mongolic and Tungusic proto-languages. Second, farmers from the Yellow River Basin (around 3000 BC) probably spread Sino-Tibetan languages, as their ancestry dispersed both to Tibet-where it forms approximately 84% of the gene pool in some groups-and to the Central Plain, where it has contributed around 59-84% to modern Han Chinese groups. Third, people from Taiwan from around 1300 BC to AD 800 derived approximately 75% of their ancestry from a lineage that is widespread in modern individuals who speak Austronesian, Tai-Kadai and Austroasiatic languages, and that we hypothesize derives from farmers of the Yangtze River Valley. Ancient people from Taiwan also derived about 25% of their ancestry from a northern lineage that is related to, but different from, farmers of the Yellow River Basin, which suggests an additional north-to-south expansion. Fourth, ancestry from Yamnaya Steppe pastoralists arrived in western Mongolia after around 3000 BC but was displaced by previously established lineages even while it persisted in western China, as would be expected if this ancestry was associated with the spread of proto-Tocharian Indo-European languages. Two later gene flows affected western Mongolia: migrants after around 2000 BC with Yamnaya and European farmer ancestry, and episodic influences of later groups with ancestry from Turan.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures

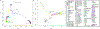
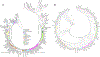

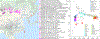
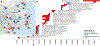
Similar articles
-
Multiple Human Population Movements and Cultural Dispersal Events Shaped the Landscape of Chinese Paternal Heritage.Mol Biol Evol. 2024 Jul 3;41(7):msae122. doi: 10.1093/molbev/msae122. Mol Biol Evol. 2024. PMID: 38885310 Free PMC article.
-
Ancient Genomes Reveal Yamnaya-Related Ancestry and a Potential Source of Indo-European Speakers in Iron Age Tianshan.Curr Biol. 2019 Aug 5;29(15):2526-2532.e4. doi: 10.1016/j.cub.2019.06.044. Epub 2019 Jul 25. Curr Biol. 2019. PMID: 31353181
-
Peopling History of the Tibetan Plateau and Multiple Waves of Admixture of Tibetans Inferred From Both Ancient and Modern Genome-Wide Data.Front Genet. 2021 Sep 3;12:725243. doi: 10.3389/fgene.2021.725243. eCollection 2021. Front Genet. 2021. PMID: 34650596 Free PMC article.
-
Human evolutionary history in Eastern Eurasia using insights from ancient DNA.Curr Opin Genet Dev. 2020 Jun;62:78-84. doi: 10.1016/j.gde.2020.06.009. Epub 2020 Jul 17. Curr Opin Genet Dev. 2020. PMID: 32688244 Review.
-
The evolutionary history of human populations in Europe.Curr Opin Genet Dev. 2018 Dec;53:21-27. doi: 10.1016/j.gde.2018.06.007. Epub 2018 Jun 28. Curr Opin Genet Dev. 2018. PMID: 29960127 Review.
Cited by
-
Genetic legacy of ancient hunter-gatherer Jomon in Japanese populations.Nat Commun. 2024 Nov 12;15(1):9780. doi: 10.1038/s41467-024-54052-0. Nat Commun. 2024. PMID: 39532881 Free PMC article.
-
A Study on the Prevalence of Osteoporosis in People with Different Altitudes in Sichuan, China.Clin Interv Aging. 2024 Nov 5;19:1819-1828. doi: 10.2147/CIA.S478020. eCollection 2024. Clin Interv Aging. 2024. PMID: 39525876 Free PMC article.
-
Bioclimatic and masticatory influences on human cranial diversity verified by analysis of 3D morphometric homologous models.Sci Rep. 2024 Nov 4;14(1):26663. doi: 10.1038/s41598-024-76715-0. Sci Rep. 2024. PMID: 39496664 Free PMC article.
-
Archaeogenetic analysis revealed East Eurasian paternal origin to the Aba royal family of Hungary.iScience. 2024 Sep 14;27(10):110892. doi: 10.1016/j.isci.2024.110892. eCollection 2024 Oct 18. iScience. 2024. PMID: 39474080 Free PMC article.
-
An Anthropometric Study of the Morphologic Facial Index of Tibetan Youth in Tibet.J Craniofac Surg. 2024 Mar-Apr 01;35(2):490-494. doi: 10.1097/SCS.0000000000009766. Epub 2023 Oct 17. J Craniofac Surg. 2024. PMID: 39445908 Free PMC article.
References
-
- HUGO Pan-Asian SNP Consortium. Mapping human genetic diversity in Asia. Science 326, 1541–1545 (2009). - PubMed
-
- Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature 522,167–172 (2015). - PubMed
-
- de Barros Damgaard P, et al.. 137 ancient human genomes from across the Eurasian steppes. Nature 557, 369–374 (2018). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials


