Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies
- PMID: 32106567
- PMCID: PMC7150947
- DOI: 10.3390/v12030254
Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies
Abstract
The beginning of 2020 has seen the emergence of COVID-19 outbreak caused by a novel coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). There is an imminent need to better understand this new virus and to develop ways to control its spread. In this study, we sought to gain insights for vaccine design against SARS-CoV-2 by considering the high genetic similarity between SARS-CoV-2 and SARS-CoV, which caused the outbreak in 2003, and leveraging existing immunological studies of SARS-CoV. By screening the experimentally-determined SARS-CoV-derived B cell and T cell epitopes in the immunogenic structural proteins of SARS-CoV, we identified a set of B cell and T cell epitopes derived from the spike (S) and nucleocapsid (N) proteins that map identically to SARS-CoV-2 proteins. As no mutation has been observed in these identified epitopes among the 120 available SARS-CoV-2 sequences (as of 21 February 2020), immune targeting of these epitopes may potentially offer protection against this novel virus. For the T cell epitopes, we performed a population coverage analysis of the associated MHC alleles and proposed a set of epitopes that is estimated to provide broad coverage globally, as well as in China. Our findings provide a screened set of epitopes that can help guide experimental efforts towards the development of vaccines against SARS-CoV-2.
Keywords: 2019 novel coronavirus; 2019-nCoV; B cell epitopes; COVID-19; Coronavirus; MERS-CoV; SARS-CoV; SARS-CoV-2; T cell epitopes; vaccine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

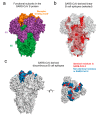
Similar articles
-
Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): Immunoinformatics approach.J Med Virol. 2020 Jun;92(6):618-631. doi: 10.1002/jmv.25736. Epub 2020 Mar 5. J Med Virol. 2020. PMID: 32108359 Free PMC article.
-
Immune and bioinformatics identification of T cell and B cell epitopes in the protein structure of SARS-CoV-2: A systematic review.Int Immunopharmacol. 2020 Sep;86:106738. doi: 10.1016/j.intimp.2020.106738. Epub 2020 Jun 28. Int Immunopharmacol. 2020. PMID: 32683296 Free PMC article.
-
Epitope-based peptide vaccines predicted against novel coronavirus disease caused by SARS-CoV-2.Virus Res. 2020 Oct 15;288:198082. doi: 10.1016/j.virusres.2020.198082. Epub 2020 Jul 1. Virus Res. 2020. PMID: 32621841 Free PMC article.
-
Protective Immunity against SARS Subunit Vaccine Candidates Based on Spike Protein: Lessons for Coronavirus Vaccine Development.J Immunol Res. 2020 Jul 18;2020:7201752. doi: 10.1155/2020/7201752. eCollection 2020. J Immunol Res. 2020. PMID: 32695833 Free PMC article. Review.
-
The genetic sequence, origin, and diagnosis of SARS-CoV-2.Eur J Clin Microbiol Infect Dis. 2020 Sep;39(9):1629-1635. doi: 10.1007/s10096-020-03899-4. Epub 2020 Apr 24. Eur J Clin Microbiol Infect Dis. 2020. PMID: 32333222 Free PMC article. Review.
Cited by
-
COVID-19 vaccines: current and future challenges.Front Pharmacol. 2024 Nov 6;15:1434181. doi: 10.3389/fphar.2024.1434181. eCollection 2024. Front Pharmacol. 2024. PMID: 39568586 Free PMC article. Review.
-
The characteristics of TCR CDR3 repertoire in COVID-19 patients and SARS-CoV-2 vaccine recipients.Virulence. 2024 Dec;15(1):2421987. doi: 10.1080/21505594.2024.2421987. Epub 2024 Nov 4. Virulence. 2024. PMID: 39468707 Free PMC article. Review.
-
Immunogenicity and Protective Efficacy of Baculovirus-Expressed SARS-CoV-2 Envelope Protein in Mice as a Universal Vaccine Candidate.Vaccines (Basel). 2024 Aug 28;12(9):977. doi: 10.3390/vaccines12090977. Vaccines (Basel). 2024. PMID: 39340009 Free PMC article.
-
HLA-C Peptide Repertoires as Predictors of Clinical Response during Early SARS-CoV-2 Infection.Life (Basel). 2024 Sep 19;14(9):1181. doi: 10.3390/life14091181. Life (Basel). 2024. PMID: 39337964 Free PMC article.
-
Insights into structural vaccinology harnessed for universal coronavirus vaccine development.Clin Exp Vaccine Res. 2024 Jul;13(3):202-217. doi: 10.7774/cevr.2024.13.3.202. Epub 2024 Jul 31. Clin Exp Vaccine Res. 2024. PMID: 39144127 Free PMC article. Review.
References
-
- Centers-of-Disease-Control-and-Prevention Confirmed 2019-nCoV cases globally. [(accessed on 31 January 2020)]; Available online: https://www.cdc.gov/coronavirus/2019-ncov/locations-confirmed-cases.html.
-
- World-Health-Organization Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV) [(accessed on 31 January 2020)]; Available online: https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-...
-
- World-Health-Organization Coronavirus disease (COVID-19) outbreak. [(accessed on 31 January 2020)]; Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
-
- World-Health-Organization Statement on the meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV) [(accessed on 31 January 2020)]; Available online: https://www.who.int/news-room/detail/23-01-2020-statement-on-the-meeting...
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous


