Glycan Profiles of gp120 Protein Vaccines from Four Major HIV-1 Subtypes Produced from Different Host Cell Lines under Non-GMP or GMP Conditions
- PMID: 31941770
- PMCID: PMC7081908
- DOI: 10.1128/JVI.01968-19
Glycan Profiles of gp120 Protein Vaccines from Four Major HIV-1 Subtypes Produced from Different Host Cell Lines under Non-GMP or GMP Conditions
Abstract
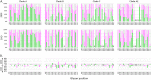
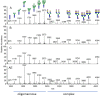
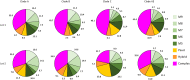
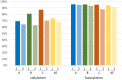
Envelope (Env) glycoprotein of human immunodeficiency virus type 1 (HIV-1) is an important target for the development of an HIV vaccine. Extensive glycosylation of Env is an important feature that both protects the virus from antibody responses and serves as a target for some highly potent broadly neutralizing antibodies. Therefore, analysis of glycans on recombinant Env proteins is highly significant. Here, we present glycosylation profiles of recombinant gp120 proteins from four major clades of HIV-1 (A, B, C, and AE), produced either as research-grade material in 293 and CHO cells or as two independent lots of clinical material under good manufacturing practice (GMP) conditions. Almost all potential N-linked glycosylation sites were at least partially occupied in all proteins. The occupancy rates were largely consistent among proteins produced under different conditions, although a few sites showed substantial variability even between the two GMP lots. Our data confirmed previous studies in the field, showing an abundance of oligomannose on Env protein, with 40 to 50% of glycans being Man5 to Man9 on all four proteins under all production conditions. Overall, the differences in occupancy and glycan forms among different Env subtypes produced under different conditions were less dramatic than anticipated, and antigenicity analysis with a panel of six monoclonal antibodies, including antibodies that recognize glycan forms, showed that all four gp120s maintained their antibody-binding profiles. Such findings have major implications for the final production of a clinical HIV vaccine with Env glycoprotein components.IMPORTANCE HIV-1 Env protein is a major target for the development of an HIV-1 vaccine. Env is covered with a large number of sugar-based glycan forms; about 50% of the Env molecular weight is composed of glycans. Glycan analysis of recombinant Env is important for understanding its roles in viral pathogenesis and immune responses. The current report presents the first extensive comparison of glycosylation patterns of recombinant gp120 proteins from four major clades of HIV-1 produced in two different cell lines, grown either under laboratory conditions or at 50-liter GMP scale in different lots. Information learned in this study is valuable for the further design and production of HIV-1 Env proteins as the critical components of HIV-1 vaccine formulations.
Keywords: Env protein; HIV-1; glycosylation; gp120; vaccine.
Copyright © 2020 Wang et al.
Figures


Similar articles
-
Glycosylation Benchmark Profile for HIV-1 Envelope Glycoprotein Production Based on Eleven Env Trimers.J Virol. 2017 Apr 13;91(9):e02428-16. doi: 10.1128/JVI.02428-16. Print 2017 May 1. J Virol. 2017. PMID: 28202756 Free PMC article.
-
HIV-1 Glycan Density Drives the Persistence of the Mannose Patch within an Infected Individual.J Virol. 2016 Nov 28;90(24):11132-11144. doi: 10.1128/JVI.01542-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27707925 Free PMC article.
-
Changes in Structure and Antigenicity of HIV-1 Env Trimers Resulting from Removal of a Conserved CD4 Binding Site-Proximal Glycan.J Virol. 2016 Sep 29;90(20):9224-36. doi: 10.1128/JVI.01116-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27489265 Free PMC article.
-
Antibody responses to the HIV-1 envelope high mannose patch.Adv Immunol. 2019;143:11-73. doi: 10.1016/bs.ai.2019.08.002. Epub 2019 Sep 11. Adv Immunol. 2019. PMID: 31607367 Free PMC article. Review.
-
The HIV glycan shield as a target for broadly neutralizing antibodies.FEBS J. 2015 Dec;282(24):4679-91. doi: 10.1111/febs.13530. Epub 2015 Oct 23. FEBS J. 2015. PMID: 26411545 Free PMC article. Review.
Cited by
-
Safety and immunogenicity of a polyvalent DNA-protein HIV vaccine with matched Env immunogens delivered as a prime-boost regimen or coadministered in HIV-uninfected adults in the USA (HVTN 124): a phase 1, placebo-controlled, double-blind randomised controlled trial.Lancet HIV. 2024 May;11(5):e285-e299. doi: 10.1016/S2352-3018(24)00036-5. Lancet HIV. 2024. PMID: 38692824 Clinical Trial.
-
ppmFixer: a mass error adjustment for pGlyco3.0 to correct near-isobaric mismatches.Glycobiology. 2024 Apr 10;34(4):cwae006. doi: 10.1093/glycob/cwae006. Glycobiology. 2024. PMID: 38263491
-
DeGlyPHER: Highly sensitive site-specific analysis of N-linked glycans on proteins.Methods Enzymol. 2023;682:137-185. doi: 10.1016/bs.mie.2022.09.004. Epub 2022 Dec 26. Methods Enzymol. 2023. PMID: 36948700 Free PMC article.
-
Strategies for Glycoengineering Therapeutic Proteins.Front Chem. 2022 Apr 13;10:863118. doi: 10.3389/fchem.2022.863118. eCollection 2022. Front Chem. 2022. PMID: 35494652 Free PMC article. Review.
-
The Characteristics of the HIV-1 Env Glycoprotein Are Linked With Viral Pathogenesis.Front Microbiol. 2022 Mar 24;13:763039. doi: 10.3389/fmicb.2022.763039. eCollection 2022. Front Microbiol. 2022. PMID: 35401460 Free PMC article.
References
-
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PNP, Spencer DIR, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109:E3268–E3277. doi:10.1073/pnas.1217207109. - DOI - PMC - PubMed
-
- Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Protocol G Principal Investigators, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi:10.1126/science.1178746. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources


