The mechanistic target of rapamycin pathway downregulates bone morphogenetic protein signaling to promote oligodendrocyte differentiation
- PMID: 31904150
- PMCID: PMC7368967
- DOI: 10.1002/glia.23776
The mechanistic target of rapamycin pathway downregulates bone morphogenetic protein signaling to promote oligodendrocyte differentiation
Abstract
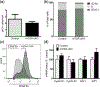
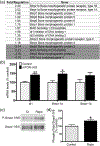
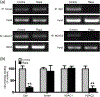
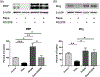
Oligodendrocyte precursor cells (OPCs) differentiate and mature into oligodendrocytes, which produce myelin in the central nervous system. Prior studies have shown that the mechanistic target of rapamycin (mTOR) is necessary for proper myelination of the mouse spinal cord and that bone morphogenetic protein (BMP) signaling inhibits oligodendrocyte differentiation, in part by promoting expression of inhibitor of DNA binding 2 (Id2). Here we provide evidence that mTOR functions specifically in the transition from early stage OPC to immature oligodendrocyte by downregulating BMP signaling during postnatal spinal cord development. When mTOR is deleted from the oligodendrocyte lineage, expression of the FK506 binding protein 1A (FKBP12), a suppressor of BMP receptor activity, is reduced, downstream Smad activity is increased and Id2 expression is elevated. Additionally, mTOR inhibition with rapamycin in differentiating OPCs alters the transcriptional complex present at the Id2 promoter. Deletion of mTOR in oligodendroglia in vivo resulted in fewer late stage OPCs and fewer newly formed oligodendrocytes in the spinal cord with no effect on OPC proliferation or cell cycle exit. Finally, we demonstrate that inhibiting BMP signaling rescues the rapamycin-induced deficit in myelin protein expression. We conclude that mTOR promotes early oligodendrocyte differentiation by suppressing BMP signaling in OPCs.
Keywords: BMP signaling; FKBP12; Id2; OPC; mTOR.
© 2020 Wiley Periodicals, Inc.
Figures




Similar articles
-
mTORC2 Loss in Oligodendrocyte Progenitor Cells Results in Regional Hypomyelination in the Central Nervous System.J Neurosci. 2023 Jan 25;43(4):540-558. doi: 10.1523/JNEUROSCI.0010-22.2022. Epub 2022 Dec 2. J Neurosci. 2023. PMID: 36460463 Free PMC article.
-
Astrocytes from the contused spinal cord inhibit oligodendrocyte differentiation of adult oligodendrocyte precursor cells by increasing the expression of bone morphogenetic proteins.J Neurosci. 2011 Apr 20;31(16):6053-8. doi: 10.1523/JNEUROSCI.5524-09.2011. J Neurosci. 2011. PMID: 21508230 Free PMC article.
-
Mammalian target of rapamycin promotes oligodendrocyte differentiation, initiation and extent of CNS myelination.J Neurosci. 2014 Mar 26;34(13):4453-65. doi: 10.1523/JNEUROSCI.4311-13.2014. J Neurosci. 2014. PMID: 24671992 Free PMC article.
-
Bone Morphogenetic Proteins: Inhibitors of Myelination in Development and Disease.Vitam Horm. 2015;99:195-222. doi: 10.1016/bs.vh.2015.05.005. Vitam Horm. 2015. PMID: 26279377 Review.
-
ID2: A negative transcription factor regulating oligodendroglia differentiation.J Neurosci Res. 2012 May;90(5):925-32. doi: 10.1002/jnr.22826. Epub 2012 Jan 18. J Neurosci Res. 2012. PMID: 22253220 Review.
Cited by
-
Therapeutic potential and impact of nanoengineered patient-derived mesenchymal stem cells in a murine resection and recurrence model of human glioblastoma.Bioeng Transl Med. 2024 May 7;9(6):e10675. doi: 10.1002/btm2.10675. eCollection 2024 Nov. Bioeng Transl Med. 2024. PMID: 39545093 Free PMC article.
-
The role and possible mechanism of the ferroptosis-related SLC7A11/GSH/GPX4 pathway in myocardial ischemia-reperfusion injury.BMC Cardiovasc Disord. 2024 Oct 1;24(1):531. doi: 10.1186/s12872-024-04220-3. BMC Cardiovasc Disord. 2024. PMID: 39354361 Free PMC article.
-
mTORC1 is required for differentiation of germline stem cells in the Drosophila melanogaster testis.PLoS One. 2024 Mar 21;19(3):e0300337. doi: 10.1371/journal.pone.0300337. eCollection 2024. PLoS One. 2024. PMID: 38512882 Free PMC article.
-
The AMPK activator metformin improves recovery from demyelination by shifting oligodendrocyte bioenergetics and accelerating OPC differentiation.Front Cell Neurosci. 2023 Oct 12;17:1254303. doi: 10.3389/fncel.2023.1254303. eCollection 2023. Front Cell Neurosci. 2023. PMID: 37904733 Free PMC article.
-
Signaling mechanisms involved in the regulation of remyelination in multiple sclerosis: a mini review.J Mol Med (Berl). 2023 Jun;101(6):637-644. doi: 10.1007/s00109-023-02312-9. Epub 2023 Apr 21. J Mol Med (Berl). 2023. PMID: 37084092 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous


