The human coronavirus HCoV-229E S-protein structure and receptor binding
- PMID: 31650956
- PMCID: PMC6970540
- DOI: 10.7554/eLife.51230
The human coronavirus HCoV-229E S-protein structure and receptor binding
Abstract
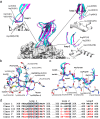
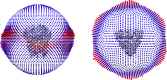
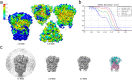
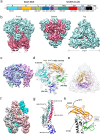
The coronavirus S-protein mediates receptor binding and fusion of the viral and host cell membranes. In HCoV-229E, its receptor binding domain (RBD) shows extensive sequence variation but how S-protein function is maintained is not understood. Reported are the X-ray crystal structures of Class III-V RBDs in complex with human aminopeptidase N (hAPN), as well as the electron cryomicroscopy structure of the 229E S-protein. The structures show that common core interactions define the specificity for hAPN and that the peripheral RBD sequence variation is accommodated by loop plasticity. The results provide insight into immune evasion and the cross-species transmission of 229E and related coronaviruses. We also find that the 229E S-protein can expose a portion of its helical core to solvent. This is undoubtedly facilitated by hydrophilic subunit interfaces that we show are conserved among coronaviruses. These interfaces likely play a role in the S-protein conformational changes associated with membrane fusion.
Keywords: HCoV-229E; biochemistry; chemical biology; coronavirus; electron cryomicroscopy; spike protein; virus; x-ray crystallography.
© 2019, Li et al.
Conflict of interest statement
ZL, AT, AW, DZ, MD, PT, SB, JR, JR No competing interests declared
Figures





Similar articles
-
Molecular determinants of species specificity in the coronavirus receptor aminopeptidase N (CD13): influence of N-linked glycosylation.J Virol. 2001 Oct;75(20):9741-52. doi: 10.1128/JVI.75.20.9741-9752.2001. J Virol. 2001. PMID: 11559807 Free PMC article.
-
Receptor-binding loops in alphacoronavirus adaptation and evolution.Nat Commun. 2017 Nov 23;8(1):1735. doi: 10.1038/s41467-017-01706-x. Nat Commun. 2017. PMID: 29170370 Free PMC article.
-
Crystal structure of the post-fusion core of the Human coronavirus 229E spike protein at 1.86 Å resolution.Acta Crystallogr D Struct Biol. 2018 Sep 1;74(Pt 9):841-851. doi: 10.1107/S2059798318008318. Epub 2018 Sep 3. Acta Crystallogr D Struct Biol. 2018. PMID: 30198895 Free PMC article.
-
Towards a coronavirus-based HIV multigene vaccine.Clin Dev Immunol. 2006 Jun-Dec;13(2-4):353-60. doi: 10.1080/17402520600579168. Clin Dev Immunol. 2006. PMID: 17162377 Free PMC article. Review.
-
A structural view of coronavirus-receptor interactions.Virus Res. 2014 Dec 19;194:3-15. doi: 10.1016/j.virusres.2014.10.005. Epub 2014 Oct 14. Virus Res. 2014. PMID: 25451063 Free PMC article. Review.
Cited by
-
Complete combinatorial mutational enumeration of a protein functional site enables sequence-landscape mapping and identifies highly-mutated variants that retain activity.Protein Sci. 2024 Aug;33(8):e5109. doi: 10.1002/pro.5109. Protein Sci. 2024. PMID: 38989563
-
Spike deep mutational scanning helps predict success of SARS-CoV-2 clades.Nature. 2024 Jul;631(8021):617-626. doi: 10.1038/s41586-024-07636-1. Epub 2024 Jul 3. Nature. 2024. PMID: 38961298 Free PMC article.
-
Genetic characterization of the first Deltacoronavirus from wild birds around Qinghai Lake.Front Microbiol. 2024 Jun 12;15:1423367. doi: 10.3389/fmicb.2024.1423367. eCollection 2024. Front Microbiol. 2024. PMID: 38933020 Free PMC article.
-
Enterocin DD14 can inhibit the infection of eukaryotic cells with enveloped viruses.Arch Microbiol. 2024 May 20;206(6):269. doi: 10.1007/s00203-024-04002-7. Arch Microbiol. 2024. PMID: 38767708
-
The Dual-Targeted Fusion Inhibitor Clofazimine Binds to the S2 Segment of the SARS-CoV-2 Spike Protein.Viruses. 2024 Apr 20;16(4):640. doi: 10.3390/v16040640. Viruses. 2024. PMID: 38675980 Free PMC article.
References
-
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica Section D Biological Crystallography. 2012;68:352–367. doi: 10.1107/S0907444912001308. - DOI - PMC - PubMed
-
- Corman VM, Baldwin HJ, Tateno AF, Zerbinati RM, Annan A, Owusu M, Nkrumah EE, Maganga GD, Oppong S, Adu-Sarkodie Y, Vallo P, da Silva Filho LVRF, Leroy EM, Thiel V, van der Hoek L, Poon LLM, Tschapka M, Drosten C, Drexler JF. Evidence for an Ancestral Association of Human Coronavirus 229E with Bats. Journal of Virology. 2015;89:11858–11870. doi: 10.1128/JVI.01755-15. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases


