CRISPRdisco: An Automated Pipeline for the Discovery and Analysis of CRISPR-Cas Systems
- PMID: 31021201
- PMCID: PMC6636876
- DOI: 10.1089/crispr.2017.0022
CRISPRdisco: An Automated Pipeline for the Discovery and Analysis of CRISPR-Cas Systems
Abstract
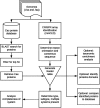
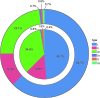
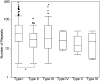
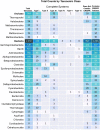
CRISPR-Cas adaptive immune systems of bacteria and archaea have catapulted into the scientific spotlight as genome editing tools. To aid researchers in the field, we have developed an automated pipeline, named CRISPRdisco (CRISPR discovery), to identify CRISPR repeats and cas genes in genome assemblies, determine type and subtype, and describe system completeness. All six major types and 23 currently recognized subtypes and novel putative V-U types are detected. Here, we use the pipeline to identify and classify putative CRISPR-Cas systems in 2,777 complete genomes from the NCBI RefSeq database. This allows comparison to previous publications and investigation of the occurrence and size of CRISPR-Cas systems. Software available at http://github.com/crisprlab/CRISPRdisco provides reproducible, standardized, accessible, transparent, and high-throughput analysis methods available to all researchers in and beyond the CRISPR-Cas research community. This tool opens new avenues to enable classification within a complex nomenclature and provides analytical methods in a field that has evolved rapidly.
Conflict of interest statement
A.B.C. and R.B. are inventors on several patents related to CRISPR technology and derived applications. R.B. is a shareholder of Caribou Biosciences, a co-founder and SAB member of Intellia Therapeutics, and co-founder and chairman of the SAB for Locus Biosciences. R.B. is the Editor-in-Chief of
Figures


Similar articles
-
Diversity and evolution of class 2 CRISPR-Cas systems.Nat Rev Microbiol. 2017 Mar;15(3):169-182. doi: 10.1038/nrmicro.2016.184. Epub 2017 Jan 23. Nat Rev Microbiol. 2017. PMID: 28111461 Free PMC article.
-
CRISPR-Cas: biology, mechanisms and relevance.Philos Trans R Soc Lond B Biol Sci. 2016 Nov 5;371(1707):20150496. doi: 10.1098/rstb.2015.0496. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27672148 Free PMC article. Review.
-
The roles of CRISPR-Cas systems in adaptive immunity and beyond.Curr Opin Immunol. 2015 Feb;32:36-41. doi: 10.1016/j.coi.2014.12.008. Epub 2015 Jan 6. Curr Opin Immunol. 2015. PMID: 25574773 Review.
-
In Silico Processing of the Complete CRISPR-Cas Spacer Space for Identification of PAM Sequences.Biotechnol J. 2018 Sep;13(9):e1700595. doi: 10.1002/biot.201700595. Epub 2018 Aug 23. Biotechnol J. 2018. PMID: 30076736
-
Investigation of direct repeats, spacers and proteins associated with clustered regularly interspaced short palindromic repeat (CRISPR) system of Vibrio parahaemolyticus.Mol Genet Genomics. 2019 Feb;294(1):253-262. doi: 10.1007/s00438-018-1504-8. Epub 2018 Oct 24. Mol Genet Genomics. 2019. PMID: 30357478
Cited by
-
Structural determinants of DNA cleavage by a CRISPR HNH-Cascade system.Mol Cell. 2024 Aug 22;84(16):3154-3162.e5. doi: 10.1016/j.molcel.2024.07.026. Epub 2024 Aug 6. Mol Cell. 2024. PMID: 39111310 Free PMC article.
-
Reprogramming the endogenous type I CRISPR-Cas system for simultaneous gene regulation and editing in Haloarcula hispanica.mLife. 2022 Mar 17;1(1):40-50. doi: 10.1002/mlf2.12010. eCollection 2022 Mar. mLife. 2022. PMID: 38818324 Free PMC article.
-
A highly efficient method for genomic deletion across diverse lengths in thermophilic Parageobacillus thermoglucosidasius.Synth Syst Biotechnol. 2024 May 17;9(4):658-666. doi: 10.1016/j.synbio.2024.05.009. eCollection 2024 Dec. Synth Syst Biotechnol. 2024. PMID: 38817825 Free PMC article.
-
Uncovering the functional diversity of rare CRISPR-Cas systems with deep terascale clustering.Science. 2023 Nov 24;382(6673):eadi1910. doi: 10.1126/science.adi1910. Epub 2023 Nov 23. Science. 2023. PMID: 37995242 Free PMC article.
-
Comparative Analysis and Phylogenetic Insights of Cas14-Homology Proteins in Bacteria and Archaea.Genes (Basel). 2023 Oct 6;14(10):1911. doi: 10.3390/genes14101911. Genes (Basel). 2023. PMID: 37895260 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources


