Sialic Acid-Like Sugars in Archaea: Legionaminic Acid Biosynthesis in the Halophile Halorubrum sp. PV6
- PMID: 30245679
- PMCID: PMC6137143
- DOI: 10.3389/fmicb.2018.02133
Sialic Acid-Like Sugars in Archaea: Legionaminic Acid Biosynthesis in the Halophile Halorubrum sp. PV6
Abstract
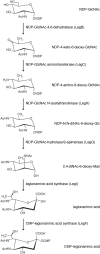
N-glycosylation is a post-translational modification that occurs in all three domains. In Archaea, however, N-linked glycans present a degree of compositional diversity not observed in either Eukarya or Bacteria. As such, it is surprising that nonulosonic acids (NulOs), nine-carbon sugars that include sialic acids, pseudaminic acids, and legionaminic acids, are routinely detected as components of protein-linked glycans in Eukarya and Bacteria but not in Archaea. In the following, we report that the N-linked glycan attached to the S-layer glycoprotein of the haloarchaea Halorubrum sp. PV6 includes an N-formylated legionaminic acid. Analysis of the Halorubrum sp. PV6 genome led to the identification of sequences predicted to comprise the legionaminic acid biosynthesis pathway. The transcription of pathway genes was confirmed, as was the co-transcription of several of these genes. In addition, the activities of LegI, which catalyzes the condensation of 2,4-di-N-acetyl-6-deoxymannose and phosphoenolpyruvate to generate legionaminic acid, and LegF, which catalyzes the addition of cytidine monophosphate (CMP) to legionaminic acid, both heterologously expressed in Haloferax volcanii, were demonstrated. Further genome analysis predicts that the genes encoding enzymes of the legionaminic acid biosynthetic pathway are clustered together with sequences seemingly encoding components of the N-glycosylation pathway in this organism. In defining the first example of a legionaminic acid biosynthesis pathway in Archaea, the findings reported here expand our insight into archaeal N-glycosylation, an almost universal post-translational modification in this domain of life.
Keywords: Halorubrum; N-formylation; N-glycosylation; S-layer glycoprotein; archaea; halophile; legionaminic acid.
Figures



Similar articles
-
Analysis of putative nonulosonic acid biosynthesis pathways in Archaea reveals a complex evolutionary history.FEMS Microbiol Lett. 2013 Aug;345(2):110-20. doi: 10.1111/1574-6968.12193. Epub 2013 Jun 27. FEMS Microbiol Lett. 2013. PMID: 23746269
-
Diversity in prokaryotic glycosylation: an archaeal-derived N-linked glycan contains legionaminic acid.Mol Microbiol. 2012 May;84(3):578-93. doi: 10.1111/j.1365-2958.2012.08045.x. Epub 2012 Apr 11. Mol Microbiol. 2012. PMID: 22435790 Free PMC article.
-
A pseudaminic acid or a legionaminic acid derivative transferase is strain-specifically implicated in the general protein O-glycosylation system of the periodontal pathogen Tannerella forsythia.Glycobiology. 2017 Jun 1;27(6):555-567. doi: 10.1093/glycob/cwx019. Glycobiology. 2017. PMID: 28334934 Free PMC article.
-
Protein glycosylation in Archaea: sweet and extreme.Glycobiology. 2010 Sep;20(9):1065-76. doi: 10.1093/glycob/cwq055. Epub 2010 Apr 5. Glycobiology. 2010. PMID: 20371512 Review.
-
Hot and sweet: protein glycosylation in Crenarchaeota.Biochem Soc Trans. 2013 Feb 1;41(1):384-92. doi: 10.1042/BST20120296. Biochem Soc Trans. 2013. PMID: 23356316 Review.
Cited by
-
N-glycosylation in Archaea - Expanding the process, components and roles of a universal post-translational modification.BBA Adv. 2024 Aug 29;6:100120. doi: 10.1016/j.bbadva.2024.100120. eCollection 2024. BBA Adv. 2024. PMID: 39296579 Free PMC article.
-
N-linked protein glycosylation in Nanobdellati (formerly DPANN) archaea and their hosts.J Bacteriol. 2024 Sep 19;206(9):e0020524. doi: 10.1128/jb.00205-24. Epub 2024 Aug 28. J Bacteriol. 2024. PMID: 39194224
-
Osmoregulation in freshwater anaerobic methane-oxidizing archaea under salt stress.ISME J. 2024 Jan 8;18(1):wrae137. doi: 10.1093/ismejo/wrae137. ISME J. 2024. PMID: 39030685 Free PMC article.
-
Sialylation and Sulfation of Anionic Glycoconjugates Are Common in the Extracellular Polymeric Substances of Both Aerobic and Anaerobic Granular Sludges.Environ Sci Technol. 2023 Sep 5;57(35):13217-13225. doi: 10.1021/acs.est.2c09586. Epub 2023 Aug 21. Environ Sci Technol. 2023. PMID: 37604486 Free PMC article.
-
Influence of N-Glycosylation on Virus-Host Interactions in Halorubrum lacusprofundi.Viruses. 2023 Jun 28;15(7):1469. doi: 10.3390/v15071469. Viruses. 2023. PMID: 37515157 Free PMC article.
References
-
- Abu-Qarn M., Yurist-Doutsch S., Giordano A., Trauner A., Morris H. R., Hitchen P., et al. (2007). Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J. Mol. Biol. 374 1224–1236. 10.1016/j.jmb.2007.10.042 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials


